CLAVATA3/EMBRYO SURROUNDING REGION Genes Involved in Symbiotic Nodulation in Pisum sativum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Bacterial Strains, and Growth Conditions
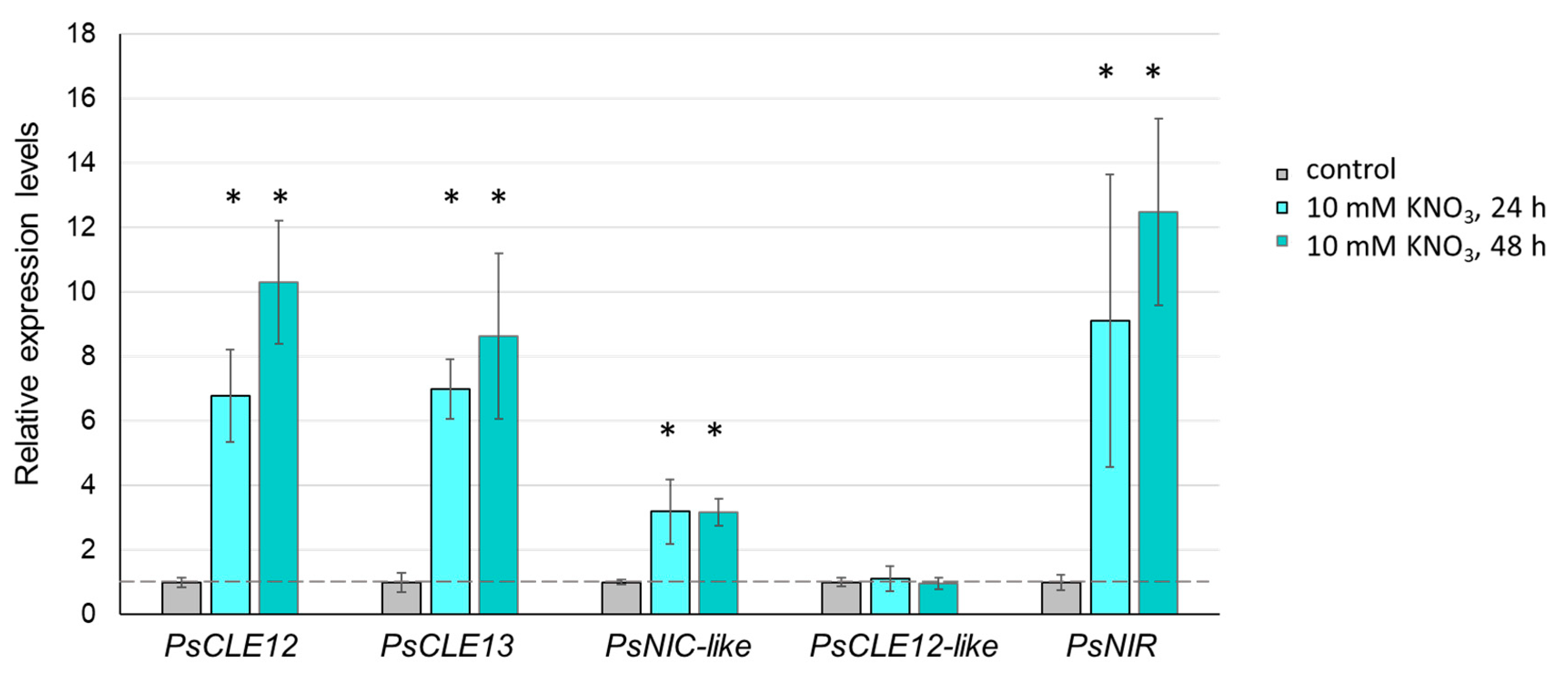
2.2. Nitrate Treatment
2.3. Molecular Cloning
2.4. Agrobacterium Rhizogene-Mediated Pea Transformation
2.5. RNA Extraction and cDNA Synthesis
2.6. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
2.7. Statistical Methods and Computer Software
3. Results
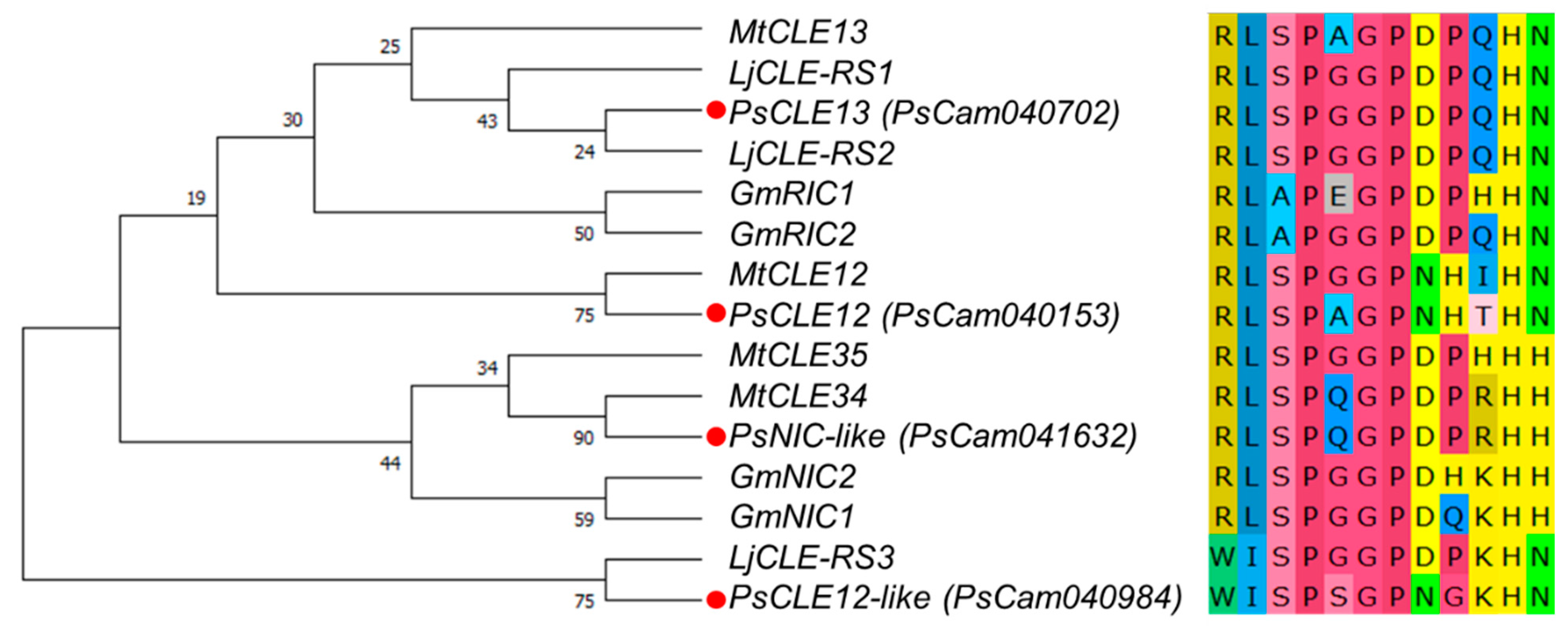
3.1. Nodulation-Related CLE Genes in Pea and their Expression Levels in Response to Rhizobial Inoculation and Nitrate Treatment
3.2. Overexpression of the PsCLE12 and PsCLE13 Genes Suppresses Nodulation
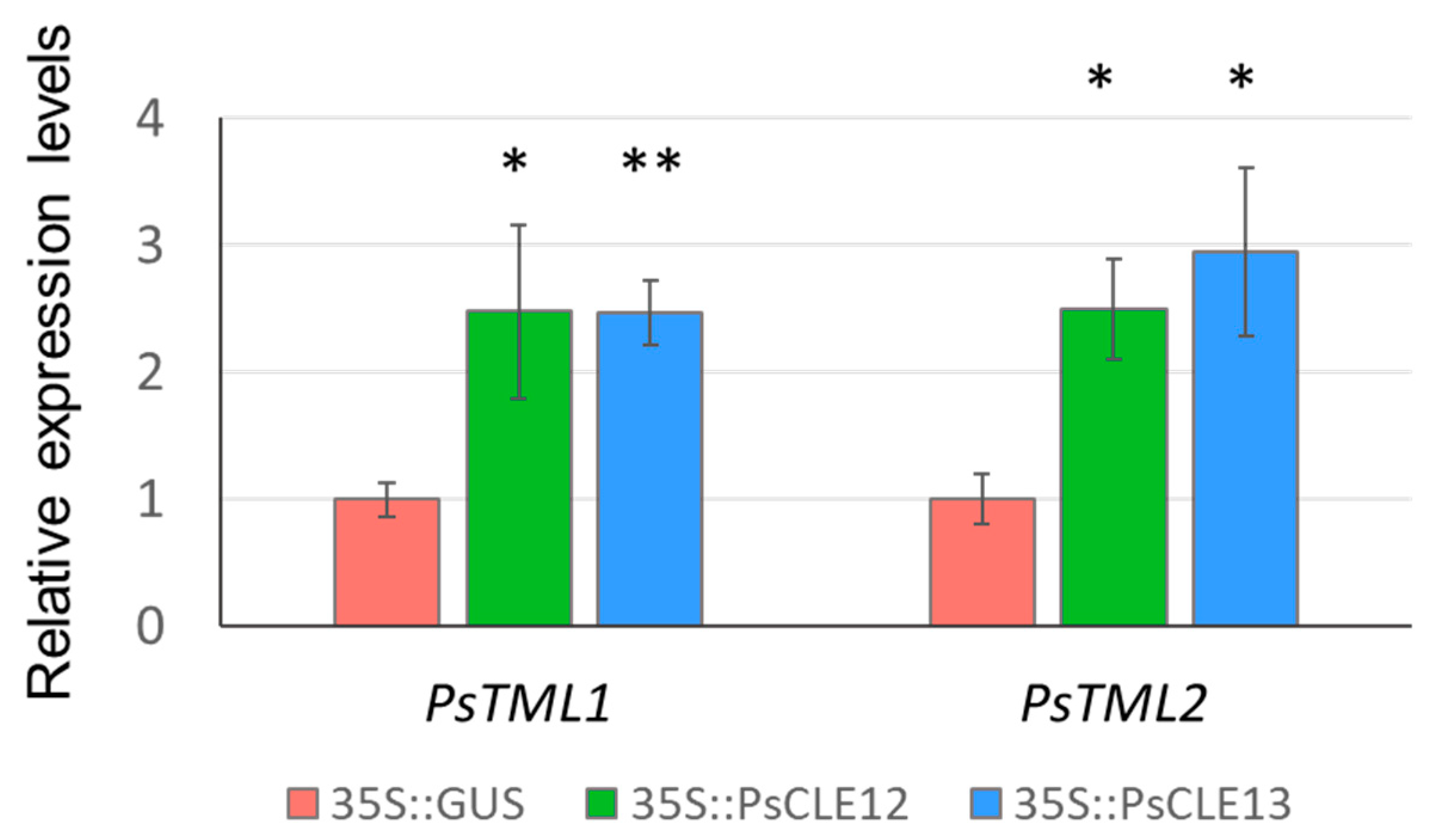
3.3. 35S::PsCLE12 and 35S::PsCLE13 Transgenic Roots Demonstrate the Upregulation of Pea Homologues of TOO MUCH LOVE Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gautrat, P.; Laffont, C.; Frugier, F.; Ruffel, S. Nitrogen systemic signaling: From symbiotic nodulation to root acquisition. Trends Plant Sci. 2021, 26, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Hirakawa, H.; Sato, S.; Hayashi, M.; Kawaguchi, M. Nodule Inception Creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. USA 2014, 111, 14607–14612. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Ivanovici, A.; Gautrat, P.; Brault, M.; Djordjevic, M.A.; Frugier, F. The NIN transcription factor coordinates CEP and CLE signaling peptides that regulate nodulation antagonistically. Nat. Commun. 2020, 11, 3167. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.E.; Ferguson, B.J.; Hayashi, S.; Lin, Y.-H.; Gresshoff, P.M. Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 2011, 108, 789–795. [Google Scholar] [CrossRef]
- Searle, I.R.; Men, A.E.; Laniya, T.S.; Buzas, D.M.; Iturbe-Ormaetxe, I.; Carroll, B.J.; Gresshoff, P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 2003, 299, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Madsen, L.H.; Sato, S.; Aubert, G.; Genua, A.; Szczyglowski, K.; Duc, G.; Kaneko, T.; Tabata, S.; De Bruijn, F. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 2002, 420, 422–426. [Google Scholar] [CrossRef]
- Schnabel, E.; Journet, E.-P.; de Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef]
- Nishimura, R.; Hayashi, M.; Wu, G.-J.; Kouchi, H.; Imaizumi-Anraku, H.; Murakami, Y.; Kawasaki, S.; Akao, S.; Ohmori, M.; Nagasawa, M. HAR1 mediates systemic regulation of symbiotic organ development. Nature 2002, 420, 426–429. [Google Scholar] [CrossRef]
- Reid, D.E.; Ferguson, B.J.; Gresshoff, P.M. Inoculation- and nitrate-Induced CLE peptides of soybean control NARK-dependent nodule formation. MPMI 2011, 24, 606–618. [Google Scholar] [CrossRef]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’haeseleer, K.; Holsters, M.; Goormachtig, S. CLE peptides control medicago truncatula nodulation locally and systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Mens, C.; Hastwell, A.H.; Su, H.; Gresshoff, P.M.; Mathesius, U.; Ferguson, B.J. Characterisation of medicago truncatula CLE34 and CLE35 in nitrate and rhizobia regulation of nodulation. New Phytol. 2021, 229, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.; Azarakhsh, M.; Yashenkova, Y.; Lutova, L. Nitrate-induced CLE Peptide systemically inhibits nodulation in medicago truncatula. Plants 2020, 9, 1456. [Google Scholar] [CrossRef]
- Moreau, C.; Gautrat, P.; Frugier, F. Nitrate-induced CLE35 signaling peptides inhibit nodulation through the SUNN receptor and MiR2111 repression. Plant Physiol. 2021, 185, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile MicroRNA. Science 2018, 362, 233–236. [Google Scholar] [CrossRef]
- Takahara, M.; Magori, S.; Soyano, T.; Okamoto, S.; Yoshida, C.; Yano, K.; Sato, S.; Tabata, S.; Yamaguchi, K.; Shigenobu, S. Too much love, a novel kelch repeat-containing F-Box protein, functions in the long-distance regulation of the legume–rhizobium symbiosis. Plant Cell Physiol. 2013, 54, 433–447. [Google Scholar] [CrossRef]
- Gautrat, P.; Mortier, V.; Laffont, C.; De Keyser, A.; Fromentin, J.; Frugier, F.; Goormachtig, S. Unraveling new molecular players involved in the autoregulation of nodulation in medicago truncatula. J. Exp. Bot. 2019, 70, 1407–1417. [Google Scholar] [CrossRef]
- Nishida, H.; Handa, Y.; Tanaka, S.; Suzaki, T.; Kawaguchi, M. Expression of the CLE-RS3 gene suppresses root nodulation in lotus japonicus. J. Plant Res. 2016, 129, 909–919. [Google Scholar] [CrossRef]
- Samorodova, A.P.; Tvorogova, V.E.; Tkachenko, A.A.; Potsenkovskaya, E.A.; Lebedeva, M.A.; Tikhonovich, I.A.; Lutova, L.A. Agrobacterial tumors interfere with nodulation and demonstrate the expression of nodulation-induced CLE genes in pea. J. Plant Physiol. 2018, 221, 94–100. [Google Scholar] [CrossRef]
- Lullien, V.; Barker, D.G.; de Lajudie, P.; Huguet, T. Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol. Biol. 1987, 9, 469–478. [Google Scholar] [CrossRef]
- Afonin, A.M.; Gribchenko, E.S.; Zorin, E.A.; Sulima, A.S.; Zhukov, V.A. DNA methylation patterns differ between free-living Rhizobium leguminosarum RCAM1026 and bacteroids formed in symbiosis with pea (Pisum Sativum L.). Microorganisms 2021, 9, 2458. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.G.; Pfaff, T.; Moreau, D.; Groves, E.; Ruffel, S.; Lepetit, M.; Whitehand, S.; Maillet, F.; Nair, R.M.; Journet, E.P. Growing M. Truncatula: Choice of substrates and growth conditions. In The Medicago Truncatula Handbook; Samuel Roberts Noble Foundation: Ardmore, OK, USA, 2006. [Google Scholar]
- Osipova, M.A.; Mortier, V.; Demchenko, K.N.; Tsyganov, V.E.; Tikhonovich, I.A.; Lutova, L.A.; Dolgikh, E.A.; Goormachtig, S. Wuschel-Related Homeobox5 Gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 2012, 158, 1329–1341. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The imagej ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Alves-Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.-L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-length de novo assembly of RNA-Seq Data in Pea (Pisum sativum L.) Provides a Gene expression atlas and gives insights into root nodulation in this species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- Lebedeva, M.; Dvornikova, K.; Lutova, L. Nitrate-Induced MtCLE34 gene lacks the ability to reduce symbiotic nodule number and carries nonsense mutation in a few accessions of medicago truncatula. Agronomy 2022, 12, 842. [Google Scholar] [CrossRef]
- Gu, B.; Chen, Y.; Xie, F.; Murray, J.D.; Miller, A.J. Inorganic nitrogen transport and assimilation in Pea (Pisum Sativum). Genes 2022, 13, 158. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Identification of a Nitrate-responsive Cis-element in the arabidopsis NIR1 promoter defines the presence of multiple Cis-regulatory elements for nitrogen response. Plant J. 2010, 63, 269–282. [Google Scholar] [CrossRef]
- Zhukov, V.A.; Zhernakov, A.I.; Kulaeva, O.A.; Ershov, N.I.; Borisov, A.Y.; Tikhonovich, I.A. De novo assembly of the pea (Pisum Sativum L.) Nodule Transcriptome. Int. J. Genom. 2015, 2015, e695947. [Google Scholar] [CrossRef]
- Hastwell, A.H.; Corcilius, L.; Williams, J.T.; Gresshoff, P.M.; Payne, R.J.; Ferguson, B.J. Triarabinosylation is required for nodulation-suppressive CLE peptides to systemically inhibit nodulation in Pisum Sativum. Plant Cell Environ. 2019, 42, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Román, B.; Nadal, S.; González-Verdejo, C.I. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebedeva, M.A.; Sadikova, D.S.; Dobychkina, D.A.; Zhukov, V.A.; Lutova, L.A. CLAVATA3/EMBRYO SURROUNDING REGION Genes Involved in Symbiotic Nodulation in Pisum sativum. Agronomy 2022, 12, 2840. https://doi.org/10.3390/agronomy12112840
Lebedeva MA, Sadikova DS, Dobychkina DA, Zhukov VA, Lutova LA. CLAVATA3/EMBRYO SURROUNDING REGION Genes Involved in Symbiotic Nodulation in Pisum sativum. Agronomy. 2022; 12(11):2840. https://doi.org/10.3390/agronomy12112840
Chicago/Turabian StyleLebedeva, Maria A., Darina S. Sadikova, Daria A. Dobychkina, Vladimir A. Zhukov, and Lyudmila A. Lutova. 2022. "CLAVATA3/EMBRYO SURROUNDING REGION Genes Involved in Symbiotic Nodulation in Pisum sativum" Agronomy 12, no. 11: 2840. https://doi.org/10.3390/agronomy12112840
APA StyleLebedeva, M. A., Sadikova, D. S., Dobychkina, D. A., Zhukov, V. A., & Lutova, L. A. (2022). CLAVATA3/EMBRYO SURROUNDING REGION Genes Involved in Symbiotic Nodulation in Pisum sativum. Agronomy, 12(11), 2840. https://doi.org/10.3390/agronomy12112840






