Comprehensive Genomic Characterization of the NAC Transcription Factors and Their Response to Drought Stress in Dendrobium catenatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of DcNAC Genes in D. catenatum
2.2. Phylogenetic Analysis of DcNAC Proteins
2.3. Analyses of Conserved Motif Distribution, Gene Structure, and Promoter Cis-Elements
2.4. Analysis of Spatial Expression Profiles Using RNA-Seq Data
2.5. Plant Materials and Drought Treatment
2.6. Analysis of Gene Expression under Drought Treatment
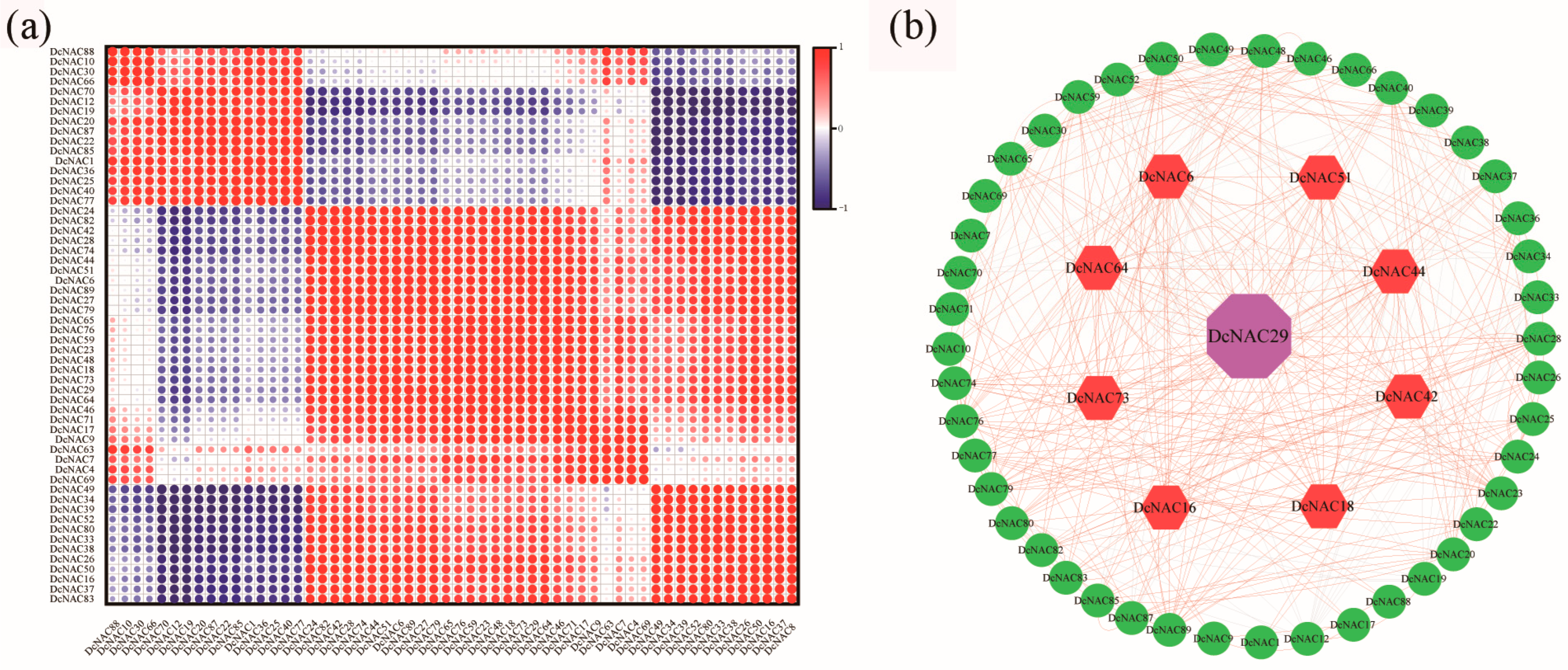
2.7. Co-Expression Network and Protein–Protein Interaction Network Analysis of DcNAC Proteins
3. Results
3.1. Identification and Characterization of NAC Genes in D. catenatum
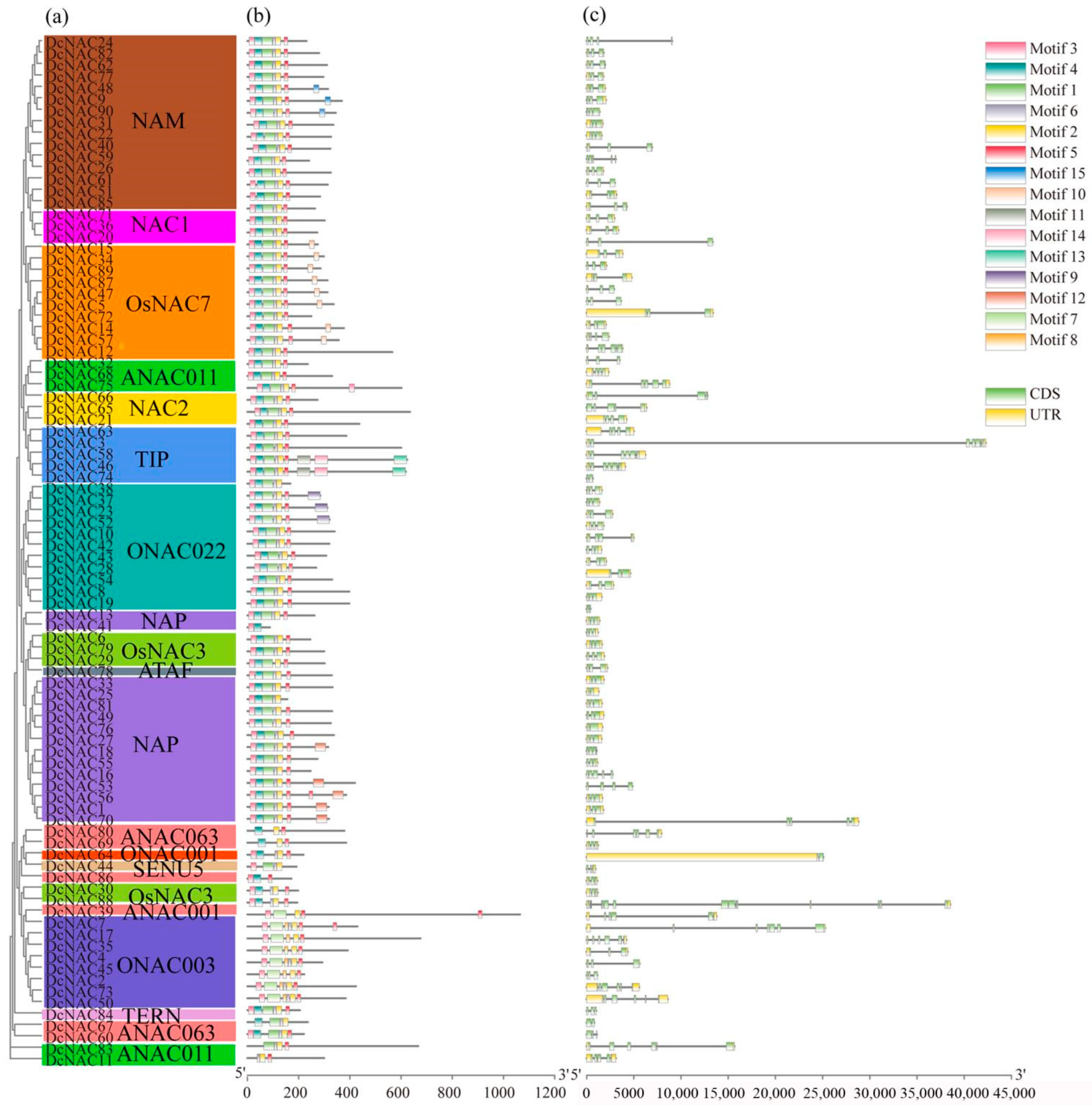
3.2. Phylogenetic Analysis and Classification of DcNAC Proteins
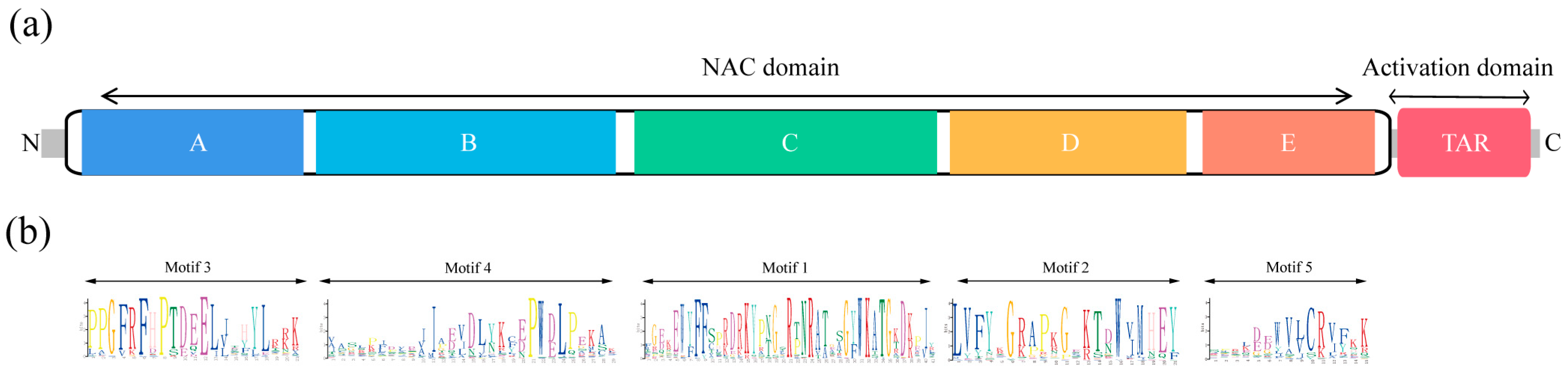
3.3. Conserved Motif and Gene Structures of DcNAC
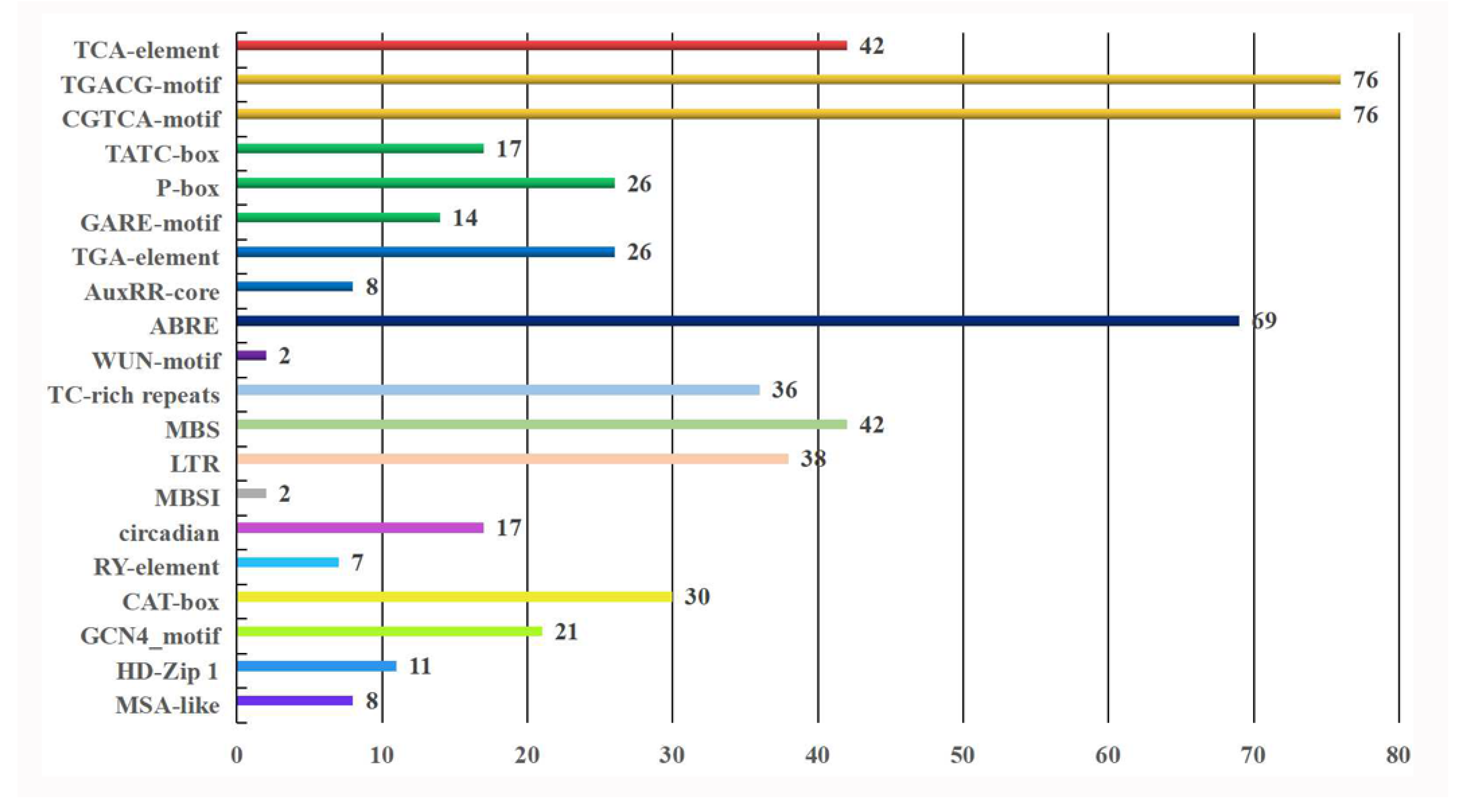
3.4. Cis-Element Analysis of the Promoter Regions of DcNAC Genes
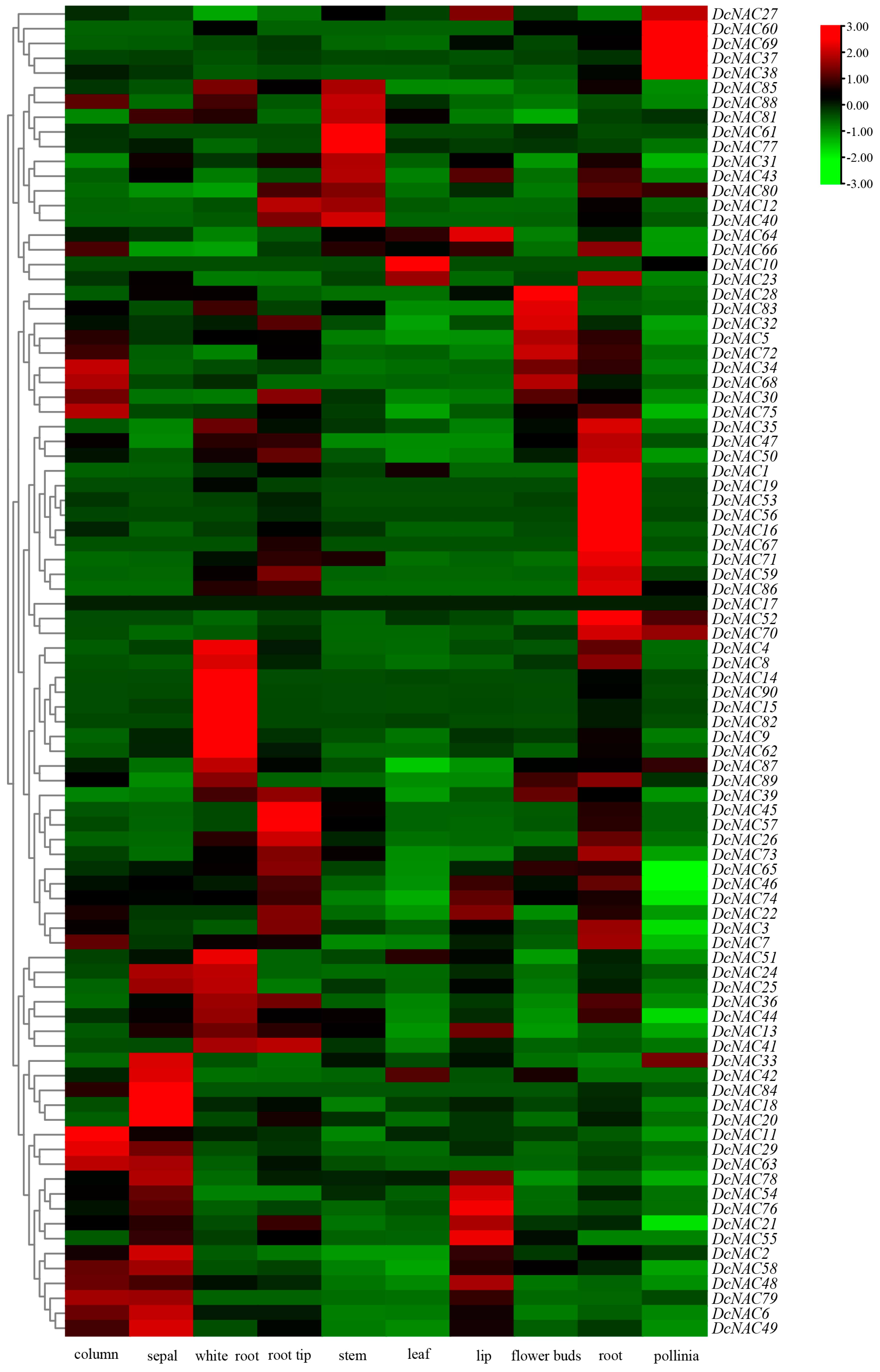
3.5. Tissue-Specific Expression Profiling of DcNAC Genes Using RNA-Seq
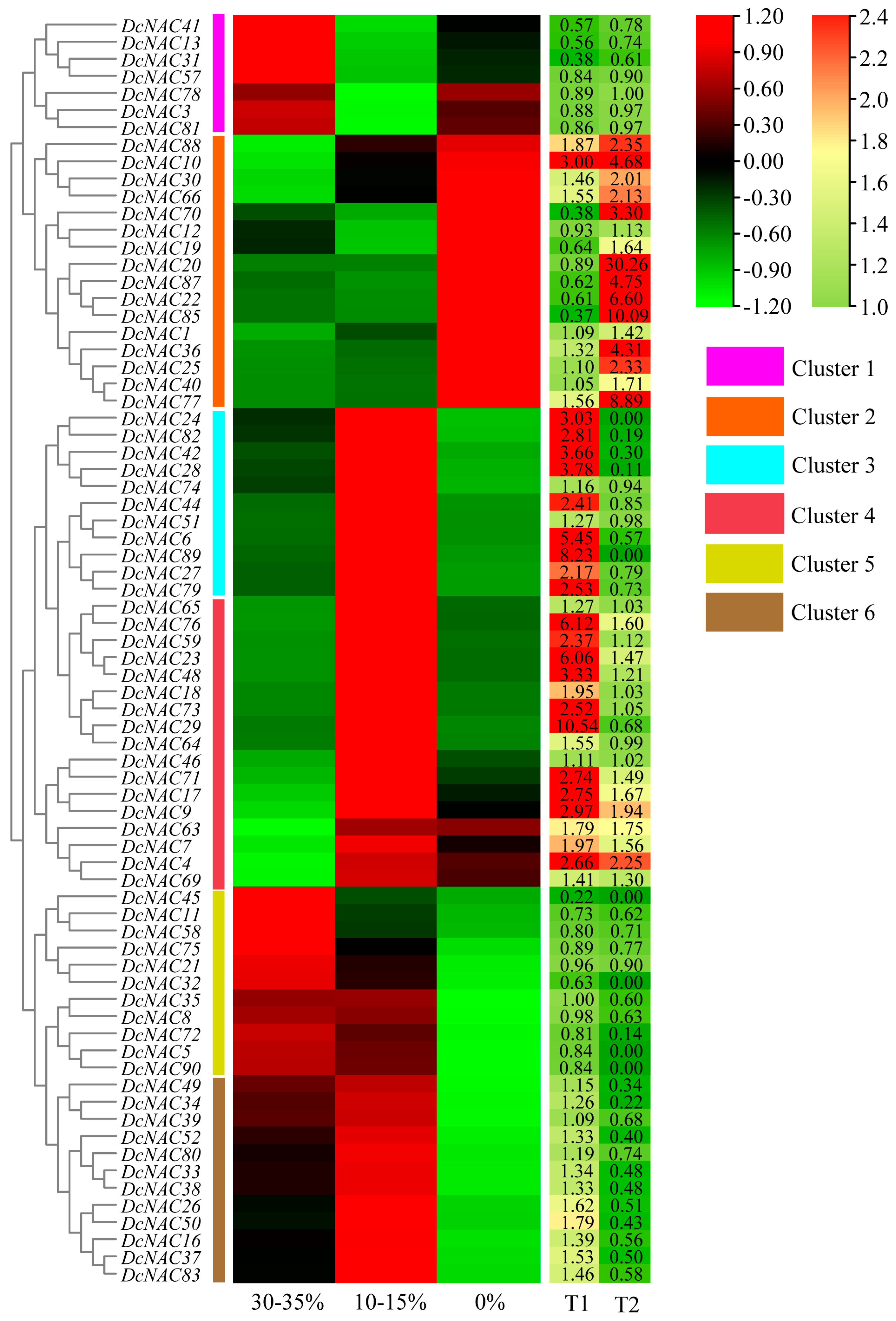
3.6. Expression of DcNACs under Drought Stress Analyzed by RNA-Seq
3.7. Comprehensive Analysis of Drought-Related DcNAC Genes
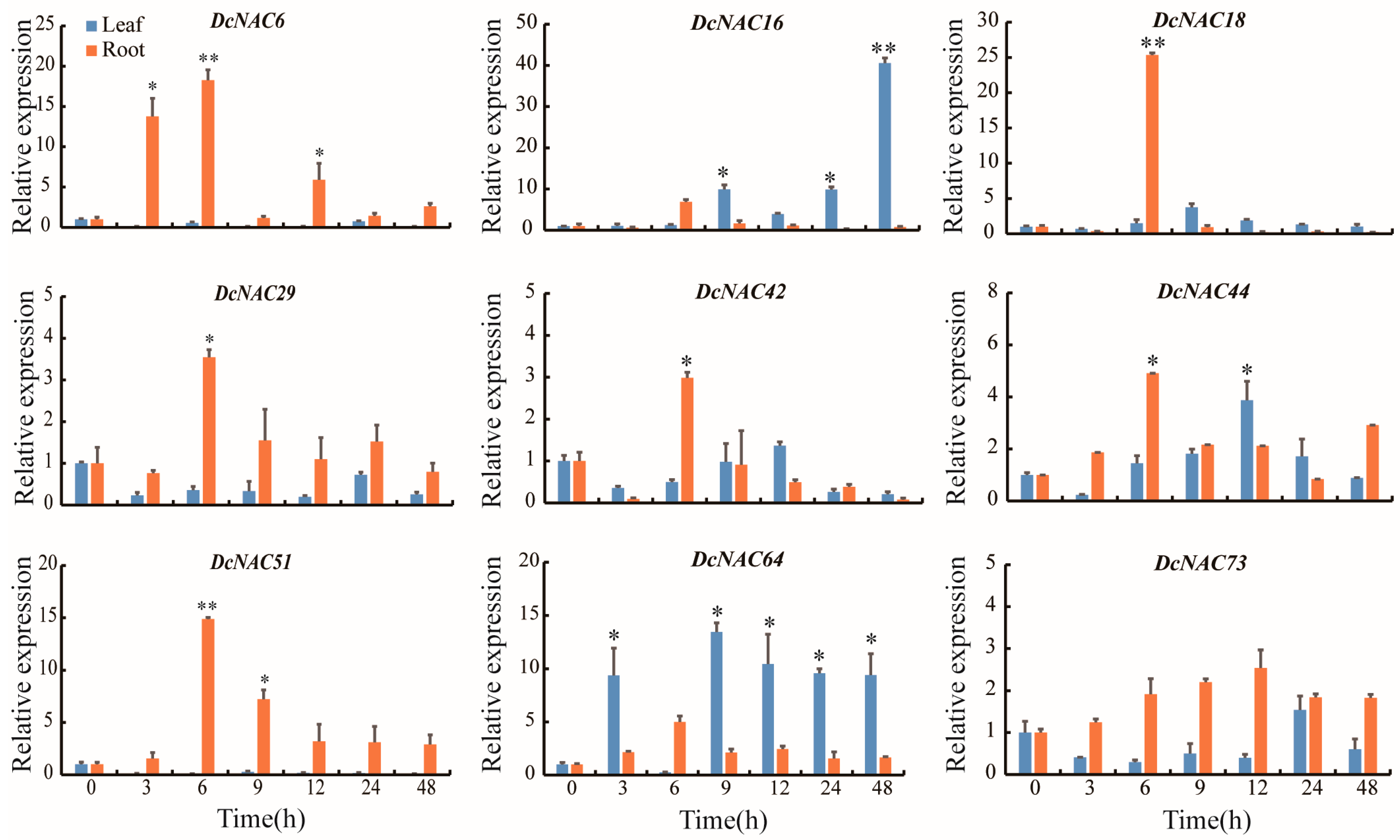
3.8. Expression Profiles of the Nine Hub DcNAC Genes under Drought Stress Using RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bechtold, U.; Field, B. Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 2018, 69, 2753–2758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Chen, W.J.; Zhu, T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004, 9, 591–597. [Google Scholar] [CrossRef]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Jin, P.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, 1182–1189. [Google Scholar] [CrossRef] [Green Version]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [Green Version]
- Mun, B.G.; Lee, S.U.; Park, E.J.; Kim, H.H.; Hussain, A.; Imran, Q.M.; Lee, I.J.; Yun, B.W. Analysis of transcription factors among differentially expressed genes induced by drought stress in Populus davidiana. 3 Biotech 2017, 3, 209. [Google Scholar] [CrossRef]
- Souer, E.; Houwelingen, V.A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–229. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Leggio, L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Q.; Xiong, L.; Lou, Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2011, 2, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 31, 239–247. [Google Scholar] [CrossRef]
- Jensen, M.K.; Rung, J.H.; Gregersen, P.L.; Gjetting, T.; Fuglsang, A.T.; Hansen, M.; Joehnk, N.; Lyngkjaer, M.F.; Collinge, D.B. The HvNAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007, 65, 137–187. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A.; Tabassum, B.; Khan, A.L.; Allah, E.F.; AI-Harrasi, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.B.L.; Reis, P.A.B.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P.B. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.P.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M.H.S.A. Genome-wide analyses of the NAC transcription factor gene family in pepper (Capsicum annuum L.): Chromosome location, phylogeny, structure, expression patterns, cis-elements in the promoter, and interaction network. Int. J. Mol. Sci. 2018, 19, 1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigarish, M.; Chen, Y.; Chen, X.; Muhammad, A.N.; Junaid, I.; Hafiz, M.R.; Shen, X.; Lin, L.I.; Xu, X.H.; Lai, Z.X. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol. Biochem. 2020, 157, 169–184. [Google Scholar] [CrossRef]
- Li, W.; Zeng, Y.; Yin, F.; Wei, R.; Mao, X. Genome-wide identification and comprehensive analysis of the NAC transcription factor family in sunflower during salt and drought stress. Sci. Rep. 2021, 11, 19865. [Google Scholar] [CrossRef] [PubMed]
- Satheesh, V.; Jagannadham, P.T.K.; Chidambaranathan, P.; Jain, P.K.; Srinivasan, R. NAC transcription factor genes: Genome-wide identification, phylogenetic, motif and cis-regulatory element analysis in pigeonpea (Cajanus cajan (L.) Millsp.). Mol. Biol. Rep. 2014, 41, 7763–7773. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Basnayake, B.M.V.S.; Zhang, H.; Li, G.; Li, W.; Virk, N.; Mengiste, T.; Song, F.M. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe Interact. 2009, 22, 1227–1238. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohmetakagi, M.; Tran, P.L.S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Tran, L.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Kim, Y.; Han, S.; Lee, B.; Paek, N. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in rice via ABA-mediated pathways. Rice 2019, 12, 76. [Google Scholar] [CrossRef]
- Hu, H.H.; Dai, M.Q.; Yao, J.L.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L.Z. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef]
- Zhang, T.T.; Cui, Z.; Li, Y.X.; Kang, Y.Q.; Song, X.Q.; Wang, J.; Zhou, Y. Genome-Wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 2021, 26, 714696. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, Z.T.; Song, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Liu, J.X. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 2013, 76, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Wing Sze, S.C.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.H.; Li, X.F.; Wang, M.N.; Zha, X.Q.; Yang, X.F.; Liu, Z.J.; Luo, Y.B.; Luo, J.P. Comparison of hypoglycemic and antioxidative effects of polysaccharides from four different Dendrobium species. Int. J. Biol. Macromol. 2014, 64, 420–427. [Google Scholar] [CrossRef]
- Tang, H.; Zhao, T.; Sheng, Y.; Zheng, T.; Fu, L.; Zhang, Y. Dendrobium officinale Kimura et Migo: A review on its ethnopharmacology, phytochemistry, pharmacology, and industrialization. Evid.-Based Complement Altern. Med. 2017, 12, 7436259. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Guo, Y.; Fu, X.; Wang, Y.; Liu, Y.; Huo, B.; Sheng, J.; Hu, X. Dendrobium candidum inhibits MCF-7 cells proliferation by inducing cell cycle arrest at G2/M phase and regulating key biomarkers. OncoTargets Ther. 2015, 9, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [Green Version]
- Atwood, J.T. The size of orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 1986, 9, 171–186. [Google Scholar]
- Zotz, G.; Winkler, U. Aerial roots of epiphytic Orchids: The velamen radicum and its role in water and nutrient uptake. Oecologia 2013, 171, 733–741. [Google Scholar] [CrossRef]
- Wan, X.; Zou, L.H.; Zheng, B.Q.; Tian, Y.Q.; Wang, Y. Transcriptomic profiling for prolonged drought in Dendrobium catenatum. Sci. Data 2018, 5, 180233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Nie, G.; Feng, G.; Han, J.; Huang, L.; Zhang, X. Genome-wide identification, characterization, and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata L.). BMC Genom. 2021, 22, 178. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Yang, X.; Yang, Z.; Zhong, M.; Zhu, Y.; Zhou, J.; Appiah, C.; Liao, Z.C.; Feng, G.Y.; Zhang, X.Q. Genome-wide investigation of the NAC transcript factor family in perennial ryegrass (Lolium perenne L.) and expression analysis under various abiotic stressor. Genomics 2020, 112, 4224–4231. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-Seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Xu, Y.; Ding, Y.D.; Yu, W.G.; Wang, J.; Lai, H.G.; Zhou, Y. Identification and expression analysis of WRKY gene family in response to abiotic stress in Dendrobium catenatum. Front. Genet. 2022, 3, 800019. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Seifert, E. OriginPro 9.1: Scientific data analysis and graphing software-software review. J. Chem. Inf. Model. 2014, 54, 1552. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonivic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Luscombe, N.M.; Austin, S.E.; Berman, H.M.; Thornton, J.M. An overview of the structures of protein-DNA complexes. Genome Biol. 2000, 1, 1–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 5499, 2105–2115. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.B. Endless forms: The evolution of gene regulation and morphological diversity. Cell 2000, 101, 577–657. [Google Scholar] [CrossRef] [Green Version]
- Girardi, C.L.; Rombaldi, C.V.; Dal Cero, J.; Nobile, P.M.; Laurens, F.; Bouzayen, M. Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci. Hortic. 2013, 151, 112–121. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–542. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–613. [Google Scholar] [CrossRef]
- Garapati, P.; Xue, G.P.; Munné-Bosch, S.; Balazadeh, S. Transcription Factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015, 168, 1122–1161. [Google Scholar] [CrossRef] [Green Version]
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.; Machado, J.P.; Brustolini, O.J.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.; et al. The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence. Plant Cell Physiol. 2016, 57, 1098–1212. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.Q.; Xi, D.D.; Chen, X.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Jiang, Y.; Li, H.; Guo, J.; Dong, M.; Zhang, J.; Liu, J.Q. Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genom. 2020, 21, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.P.; Mueller-Roeber, B. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant. 2011, 4, 346–360. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, S.V.; Martin, A.P.; Viola, I.L.; Gonzalez, D.H.; Palatnik, J.F. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 2011, 156, 1894–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhong, R.; Ye, Z.H. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 2014, 9, e105726. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kim, S.Y.; Park, C.M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 2007, 226, 647–654. [Google Scholar] [CrossRef]
- Li, P.; Zhou, H.; Shi, X.; Yu, B.; Zhou, Y.; Chen, S.; Wang, Y.F.; Peng, Y.; Meyer, R.C.; Smeekens, S.C.; et al. The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet. 2014, 10, e1004213. [Google Scholar] [CrossRef] [Green Version]
- Donze, T.; Qu, F.; Twigg, P.; Morris, T.J. Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology 2014, 449, 207–214. [Google Scholar] [CrossRef]
- Jeong, R.D.; Chandra-Shekara, A.C.; Kachroo, A.; Klessig, D.F.; Kachroo, P. HRT-mediated hypersensitive response and resistance to Turnip crinkle virus in Arabidopsis does not require the function of TIP, the presumed guardee protein. Mol. Plant Microbe Interact. 2018, 21, 1316–1324. [Google Scholar] [CrossRef] [Green Version]
- Asahina, M.; Azuma, K.; Pitaksaringkarn, W.; Yamazaki, T.; Mitsuda, N.; OhmeTakagi, M.; Yamaguchi, S.; Kamiya, Y.; Okada, K.; Nishimura, T.; et al. Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16128–16132. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.Y.; Kim, S.Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.H.; Kim, S.K.; Hong, J.C.; Hwang, D.; et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Lee, K.; Du, Q.; Yang, S.; Yuan, B.; Qi, L.; Wang, H.Z. A membraneassociated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol. 2020, 226, 59–74. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kimura, S.; Maki, H.; Britt, A.B.; Umeda, M. The role of SOG1, a plant-specific transcriptional regulator, in the DNA damage response. Plant Signal. Behav. 2014, 9, e28889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Mu, R.; Cao, W.; Zhang, Z.; Zhang, J.; Chen, S. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.L.; Chen, N.Z.; An, R.; Su, Z.; Qi, B.S.; Ren, F.; Chen, J.; Wang, X.C. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 2007, 63, 289–305. [Google Scholar] [CrossRef]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–610. [Google Scholar] [CrossRef]
- Hu, W.; Wei, Y.; Xia, Z.; Yan, Y.; Hou, X.; Zou, M.; Lu, C.; Wang, W.Q. Genome-Wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS ONE 2015, 10, e0136993. [Google Scholar] [CrossRef]
- Wei, S.; Gao, L.; Zhang, Y.; Zhang, F.; Yang, X.; Huang, D. Genome wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Rep. 2016, 35, 1827–1839. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Wolf, Y.I.; Sorokin, A.V.; Mirkin, B.G.; Koonin, E.V. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003, 13, 1512–1519. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.; Kim, S.; Kim, H.J.; Shrestha, P.; Yun, J.H.; Phee, B.K.; Lee, W.; Naw, H.G.; Chang, I. The C-Domain of the NAC transcription factor ANAC019 is necessary for pH-tuned DNA binding through a histidine switch in the N-Domain. Cell Rep. 2018, 22, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2020, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Moumeni, A.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Attia, K.; Kikuchi, S. Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday Selection (drought tolerant) and IR64. Mol. Genet. Genom. 2012, 287, 389–410. [Google Scholar] [CrossRef] [Green Version]
- Ju, Y.L.; Yue, X.F.; Min, Z.; Wang, X.H.; Fang, Y.L.; Zhang, J.X. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 98–111. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, S.; Guan, C.; Kong, X.; Wang, Y.; Cui, Y.; Liu, B.; Zhou, Y.W.; Zhang, Y.N. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Kim, M.Y.; Ha, J.; Lee, S.H. Overexpression of the soybean NAC gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Duan, L.; Pruneda-Paz, J.L.; Oh, D.H.; Pound, M.; Kay, S.; Dinneny, J.R. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol. 2018, 177, 1650–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, M.; Cheng, L.; Lin, Y.; Chen, Q.; Sun, B.; Gu, X.J.; Wang, Y.; Li, M.Y.; Luo, Y.; et al. Identification of the cytosolic glucose-6-Phosphate dehydrogenase gene from strawberry involved in cold stress response. Int. J. Mol. Sci. 2020, 21, 7322. [Google Scholar] [CrossRef]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 2018, 221, 74–80. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, S.; Ma, X.; Wang, Y.; Kong, F.; Meng, Q. A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol. Plant. 2016, 158, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Theologis, A.; Ecker, J.R.; Palm, C.J.; Federspiel, N.A.; Kaul, S.; White, O.; Alonso, J.; Altafi, H.; Araujo, H.; Bowman, C.L.; et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 2000, 408, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.M.; Bhaskara, G.B.; Wen, T.N.; Lin, W.D.; Nguyen, T.T.; Chong, G.L. Phosphoproteomics of Arabidopsis highly ABA-induced1 identifies AT-Hook-Like10 phosphorylation required for stress growth regulation. Proc. Natl. Acad. Sci. USA 2019, 116, 2354–2363. [Google Scholar] [CrossRef] [PubMed]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Han, S.H.; Lee, S.H.; Hortensteiner, S.; Paek, N.C. Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep. 2016, 35, 155–166. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Locus | Length | MW(KD) | pI | No.of Transmembrane | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| DcNAC1 | LOC110104261 | NW_021319405.1:944588..946253 | 318 | 36.00 | 5.59 | - | Nuclear |
| DcNAC2 | LOC110105825 | NW_021318852.1:8900458..8901600 | 222 | 25.03 | 8.69 | - | Nuclear, Mitochondrial |
| DcNAC3 | LOC110105421 | NW_021319664.1:601710..606065 | 387 | 43.86 | 8.38 | - | Nuclear |
| DcNAC4 | LOC110095412 | NW_021319664.1:601710..606065 | 269 | 30.90 | 7.25 | - | Nuclear |
| DcNAC5 | LOC110116424 | NW_021319483.1:4428910..4432555 | 313 | 36.69 | 6.49 | - | Nuclear |
| DcNAC6 | LOC110098761 | NW_021319202.1:1232820..1234194 | 274 | 31.28 | 5.42 | - | Mitochondrial, Cytoplasmic |
| DcNAC7 | LOC110109664 | NW_021319373.1:1292252..1306026 | 430 | 48.34 | 5.36 | - | Nuclear |
| DcNAC8 | LOC110114298 | NW_021319478.1:223003..227638 | 398 | 45.26 | 6.68 | - | Nuclear |
| DcNAC9 | LOC110106797 | NW_021320118.1:443972..446023 | 369 | 41.45 | 5.96 | - | Nuclear |
| DcNAC10 | LOC110104614 | NW_021319853.1:571176..573917 | 341 | 39.44 | 6.68 | - | Nuclear |
| DcNAC11 | LOC110107694 | NW_021319484.1:2340621..2343718 | 300 | 33.78 | 5.61 | - | Nuclear |
| DcNAC12 | LOC110116691 | NW_021319255.1:789964..792321 | 357 | 39.80 | 6.43 | - | Cytoplasmic |
| DcNAC13 | LOC110107876 | NW_021319309.1:5212803..5214395 | 262 | 30.18 | 5.32 | - | Nuclear, Mitochondrial |
| DcNAC14 | LOC110108485 | NW_021318618.1:1341771..1350421 | 250 | 28.56 | 8.58 | - | Nuclear |
| DcNAC15 | LOC110105195 | NW_021319341.1:1740160..1753523 | 327 | 37.21 | 6.73 | - | Mitochondrial, Cytoplasmic |
| DcNAC16 | LOC110112974 | NW_021320019.1:6917562..6918718 | 247 | 28.25 | 7.66 | - | Mitochondrial, Cytoplasmic |
| DcNAC17 | LOC110098358 | NW_021318618.1:140601..165868 | 676 | 76.21 | 6.81 | - | Nuclear |
| DcNAC18 | LOC110104359 | NW_021319405.1:898780..900362 | 316 | 35.49 | 6.80 | - | Nuclear |
| DcNAC19 | LOC110114553 | NW_021318619.1:1786474..1789318 | 398 | 45.00 | 6.55 | - | Nuclear |
| DcNAC20 | LOC110107078 | NW_021345752.1:73081..76452 | 273 | 31.91 | 6.07 | - | Cytoplasmic |
| DcNAC21 | LOC110103331 | NW_021318728.1:755755..762108 | 635 | 70.39 | 4.68 | 1 | Nuclear, Cytoplasmic |
| DcNAC22 | LOC110111768 | NW_021318693.1:2812812..2814409 | 308 | 35.24 | 6.04 | - | Nuclear |
| DcNAC23 | LOC110110716 | NW_021319682.1:11969540..11971158 | 313 | 35.85 | 7.80 | - | Nuclear |
| DcNAC24 | LOC110115981 | NW_021319056.1:515803..524871 | 231 | 26.84 | 6.66 | - | Cytoplasmic |
| DcNAC25 | LOC110110725 | NW_021319746.1:1017210..1019037 | 333 | 37.75 | 7.65 | - | Nuclear |
| DcNAC26 | LOC110107219 | NW_021318474.1:52459..54233 | 326 | 36.90 | 8.48 | - | Nuclear |
| DcNAC27 | LOC110095790 | NW_021318852.1:8686697..8688365 | 338 | 36.98 | 8.62 | - | Nuclear |
| DcNAC28 | LOC110115485 | NW_021319945.1:1292983..1294555 | 269 | 31.45 | 5.16 | - | Cytoplasmic |
| DcNAC29 | LOC110092932 | NW_021319682.1:12055658..12057289 | 300 | 34.21 | 5.37 | - | Nuclear |
| DcNAC30 | LOC110097953 | NW_021319408.1:90079..91220 | 198 | 22.50 | 5.80 | - | Cytoplasmic |
| DcNAC31 | LOC110107849 | NW_021319309.1:5177244..5178952 | 336 | 38.09 | 6.15 | - | Nuclear, Cytoplasmic |
| DcNAC32 | LOC110114255 | NW_021318952.1:407937..411747 | 566 | 64.69 | 5.74 | - | Nuclear |
| DcNAC33 | LOC110105798 | NW_021320118.1:62855..65065 | 330 | 37.90 | 6.80 | - | Nuclear |
| DcNAC34 | LOC110104809 | NW_021318693.1:5734624..5738435 | 275 | 31.70 | 6.32 | - | Nuclear |
| DcNAC35 | LOC110102585 | NW_021319099.1:1124888..1129100 | 392 | 44.26 | 5.45 | - | Nuclear |
| DcNAC36 | LOC110094938 | NW_021318963.1:481017..483962 | 302 | 34.48 | 5.73 | - | Cytoplasmic |
| DcNAC37 | LOC114580353 | NW_021318576.1:1853942..1854694 | 285 | 32.93 | 6.18 | - | Nuclear |
| DcNAC38 | LOC110101656 | NW_021350263.1:1773..2430 | 168 | 19.47 | 9.99 | - | Nuclear |
| DcNAC39 | LOC110102005 | NW_021319610.1:706265..744789 | 1065 | 119.24 | 5.35 | - | Nuclear |
| DcNAC40 | LOC110106875 | NW_021320142.1:1232496..1239440 | 324 | 36.46 | 8.76 | - | Nuclear |
| DcNAC41 | LOC114578614 | NW_021412344.1:5..417 | 88 | 10.34 | 6.56 | - | Nuclear, Mitochondrial |
| DcNAC42 | LOC110115000 | NW_021319004.1:3740985..3742795 | 320 | 36.49 | 6.68 | - | Nuclear |
| DcNAC43 | LOC110092814 | NW_021319127.1:4426569..4431555 | 308 | 35.87 | 6.49 | - | Nuclear |
| DcNAC44 | LOC110107435 | NW_021319315.1:7525933..7551014 | 192 | 21.73 | 9.53 | 1 | Mitochondrial, Cytoplasmic |
| DcNAC45 | LOC110101851 | NW_021319178.1:14978035..14983656 | 293 | 32.75 | 8.37 | - | Nuclear |
| DcNAC46 | LOC110100943 | NW_021319134.1:403956..410187 | 624 | 68.83 | 4.84 | 1 | Nuclear, Cytoplasmic |
| DcNAC47 | LOC110094901 | NW_021319875.1:435195..438112 | 313 | 36.62 | 6.21 | - | Nuclear |
| DcNAC48 | LOC110108674 | NW_021318516.1:1875800..1877760 | 315 | 35.64 | 8.60 | - | Nuclear |
| DcNAC49 | LOC110092756 | NW_021319682.1:12023901..12025518 | 331 | 37.66 | 8.92 | - | Nuclear |
| DcNAC50 | LOC110103097 | NW_021318927.1:550233..558831 | 384 | 43.03 | 7.04 | - | Mitochondrial |
| DcNAC51 | LOC110104882 | NW_021319718.1:578668..596732 | 314 | 35.40 | 8.16 | - | Nuclear, Mitochondrial |
| DcNAC52 | LOC110098556 | NW_021586606.1:401626..402992 | 321 | 36.57 | 6.06 | - | Nuclear |
| DcNAC53 | LOC110112625 | NW_021318921.1:882187..884939 | 420 | 48.34 | 8.92 | - | Cytoplasmic |
| DcNAC54 | LOC110107887 | NW_021319309.1:4726707..4728783 | 313 | 36.04 | 6.55 | - | Nuclear |
| DcNAC55 | LOC110107355 | NW_021318535.1:1389..2468 | 274 | 30.68 | 8.20 | - | Nuclear |
| DcNAC56 | LOC110112624 | NW_021318921.1:866825..871675 | 386 | 43.57 | 5.45 | - | Nuclear |
| DcNAC57 | LOC110100421 | NW_021356156.1:29165..31208 | 377 | 41.97 | 6.60 | - | Nuclear |
| DcNAC58 | LOC110116407 | NW_021319956.1:483041..525326 | 601 | 67.20 | 5.77 | 1 | Nuclear |
| DcNAC59 | LOC110102675 | NW_021318663.1:278514..281625 | 241 | 27.23 | 9.16 | - | Nuclear, Cytoplasmic |
| DcNAC60 | LOC110114310 | NW_021319478.1:59280..60370 | 221 | 25.27 | 9.07 | - | Nuclear |
| DcNAC61 | LOC110101259 | NW_021319682.1:5659051..5662051 | 331 | 37.09 | 6.27 | - | Cytoplasmic |
| DcNAC62 | LOC110108396 | NW_021319952.1:264477..266434 | 311 | 35.99 | 5.54 | - | Nuclear |
| DcNAC63 | LOC110097330 | NW_021319772.1:190444..194675 | 437 | 50.03 | 6.15 | 1 | Nuclear |
| DcNAC64 | LOC110100119 | NW_021319284.1:705412..706624 | 219 | 25.00 | 9.17 | - | Nuclear |
| DcNAC65 | LOC110099898 | NW_021319550.1:257520..270338 | 539 | 60.66 | 4.50 | - | Nuclear |
| DcNAC66 | LOC110097190 | NW_021319315.1:4053808..4062584 | 602 | 67.38 | 4.78 | 1 | Nuclear, Cytoplasmic |
| DcNAC67 | LOC110114311 | NW_021319478.1:78155..78973 | 236 | 26.54 | 6.25 | - | Endoplasmic reticulum Mitochondrial |
| DcNAC68 | LOC110114265 | NW_021318952.1:398944..402450 | 236 | 27.24 | 4.94 | - | Nuclear |
| DcNAC69 | LOC110105590 | NW_021319523.1:2753877..2761806 | 386 | 43.35 | 4.73 | - | Nuclear |
| DcNAC70 | LOC110104365 | NW_021319405.1:957135..958903 | 319 | 36.16 | 5.63 | - | Cytoplasmic |
| DcNAC71 | LOC110095285 | NW_021318596.1:516144..520405 | 264 | 30.49 | 5.94 | - | Nuclear |
| DcNAC72 | LOC110104495 | NW_021318542.1:456982..470372 | 337 | 39.31 | 5.45 | - | Nuclear |
| DcNAC73 | LOC110092252 | NW_021319862.1:754733..760299 | 424 | 47.33 | 6.63 | - | Nuclear |
| DcNAC74 | LOC110100951 | NW_021319134.1:429429..433522 | 619 | 68.61 | 4.86 | 1 | Nuclear, Cytoplasmic |
| DcNAC75 | LOC110110098 | NW_021473832.1:229827..232159 | 331 | 37.78 | 5.78 | - | Nuclear |
| DcNAC76 | LOC110104101 | NW_021319154.1:640611..642422 | 326 | 37.44 | 8.80 | - | Nuclear |
| DcNAC77 | LOC110104023 | NW_021319309.1:351852..353642 | 297 | 34.27 | 9.02 | - | Nuclear |
| DcNAC78 | LOC110098637 | NW_021319551.1:1686647..1688517 | 302 | 33.68 | 8.83 | - | Nuclear |
| DcNAC79 | LOC110111459 | NW_021319876.1:468565..469773 | 246 | 28.25 | 6.01 | - | Nuclear, Mitochondrial |
| DcNAC80 | LOC110099650 | NW_021318496.1:315052..343857 | 379 | 42.02 | 4.90 | - | Nuclear |
| DcNAC81 | LOC110112020 | NW_021319961.1:27508..28792 | 156 | 18.00 | 9.76 | - | Nuclear, Mitochondrial |
| DcNAC82 | LOC110104089 | NW_021319154.1:989262..991072 | 352 | 40.80 | 5.09 | - | Nuclear, Cytoplasmic |
| DcNAC83 | LOC110110797 | NW_021441140.1:128927..144598 | 667 | 76.33 | 6.46 | - | Nuclear |
| DcNAC84 | LOC110092208 | NW_021319083.1:5535325..5536353 | 205 | 23.18 | 8.74 | - | Nuclear |
| DcNAC85 | LOC110116607 | NW_021404996.1:183594..186750 | 284 | 32.23 | 6.90 | - | Nuclear, Cytoplasmic |
| DcNAC86 | LOC114579126 | NW_021318625.1:16861..17817 | 172 | 19.31 | 5.53 | - | Nuclear |
| DcNAC87 | LOC110107637 | NW_021319168.1:1236367..1241143 | 286 | 33.75 | 7.63 | - | Nuclear |
| DcNAC88 | LOC110111207 | NW_021516239.1:66394..67565 | 195 | 22.13 | 4.97 | - | Cytoplasmic |
| DcNAC89 | LOC110107480 | NW_021319567.1:739642..741736 | 299 | 34.42 | 5.23 | - | Nuclear |
| DcNAC90 | LOC110111452 | NW_021319876.1:524351..525738 | 345 | 39.42 | 6.32 | - | Nuclear |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, T.; Xing, W.; Wang, J.; Yu, W.; Zhou, Y. Comprehensive Genomic Characterization of the NAC Transcription Factors and Their Response to Drought Stress in Dendrobium catenatum. Agronomy 2022, 12, 2753. https://doi.org/10.3390/agronomy12112753
Li Y, Zhang T, Xing W, Wang J, Yu W, Zhou Y. Comprehensive Genomic Characterization of the NAC Transcription Factors and Their Response to Drought Stress in Dendrobium catenatum. Agronomy. 2022; 12(11):2753. https://doi.org/10.3390/agronomy12112753
Chicago/Turabian StyleLi, Yuxin, Tingting Zhang, Wenting Xing, Jian Wang, Wengang Yu, and Yang Zhou. 2022. "Comprehensive Genomic Characterization of the NAC Transcription Factors and Their Response to Drought Stress in Dendrobium catenatum" Agronomy 12, no. 11: 2753. https://doi.org/10.3390/agronomy12112753
APA StyleLi, Y., Zhang, T., Xing, W., Wang, J., Yu, W., & Zhou, Y. (2022). Comprehensive Genomic Characterization of the NAC Transcription Factors and Their Response to Drought Stress in Dendrobium catenatum. Agronomy, 12(11), 2753. https://doi.org/10.3390/agronomy12112753







