RNAseq Reveals Differential Gene Expression Contributing to Phytophthora nicotianae Adaptation to Partial Resistance in Tobacco
Abstract
1. Introduction
2. Materials and Methods
2.1. RNAseq Preparation
2.1.1. Collection of Pathogen Isolates
2.1.2. Pathogen Culture and Tobacco Infection
2.1.3. RNA Isolation and Transcriptome Sequencing
2.2. Analyses of RNAseq Data from P. nicotianae
2.2.1. Detection of Differentially Expressed Genes (DEGs) in P. nicotianae
2.2.2. Gene Ontology Analysis, KEGG Pathway Enrichment Analysis and PHIB-Blast
2.2.3. Detection of Differential Transcript Usage
2.2.4. Identification of Single-Nucleotide Polymorphisms (SNPs)
2.3. Analyses of RNAseq Data from N. tabacum
2.3.1. Detection of Differentially Expressed Genes (DEGs) in N. tabacum
2.3.2. Gene Ontology and KEGG Enrichment Analyses
2.4. Quantitative Real-Time PCR (qRT-PCR) Validations
3. Results
3.1. RNAseq Overview
3.1.1. Infected Tobacco Root Samples for RNAseq
3.1.2. RNA Sequencing and Sequence Mapping to the Reference Genome
3.2. Overview of DEGs in P. nicotianae
3.2.1. DEGs Identified in P. nicotianae
3.2.2. Over-Represented Gene Ontology Analysis
3.2.3. KEGG Analysis of DEGs
3.2.4. PHIB-Database Blast
3.2.5. Genes with Differential Transcript Usage in P. nicotianae
3.2.6. SNPs Identified in the Wz-Wz and Wz-H Isolates of P. nicotianae
3.3. Overview of DEGs in N. tabacum
3.3.1. DEGs Identified in N. tabacum
3.3.2. GO Analysis of DEGs in Tobacco
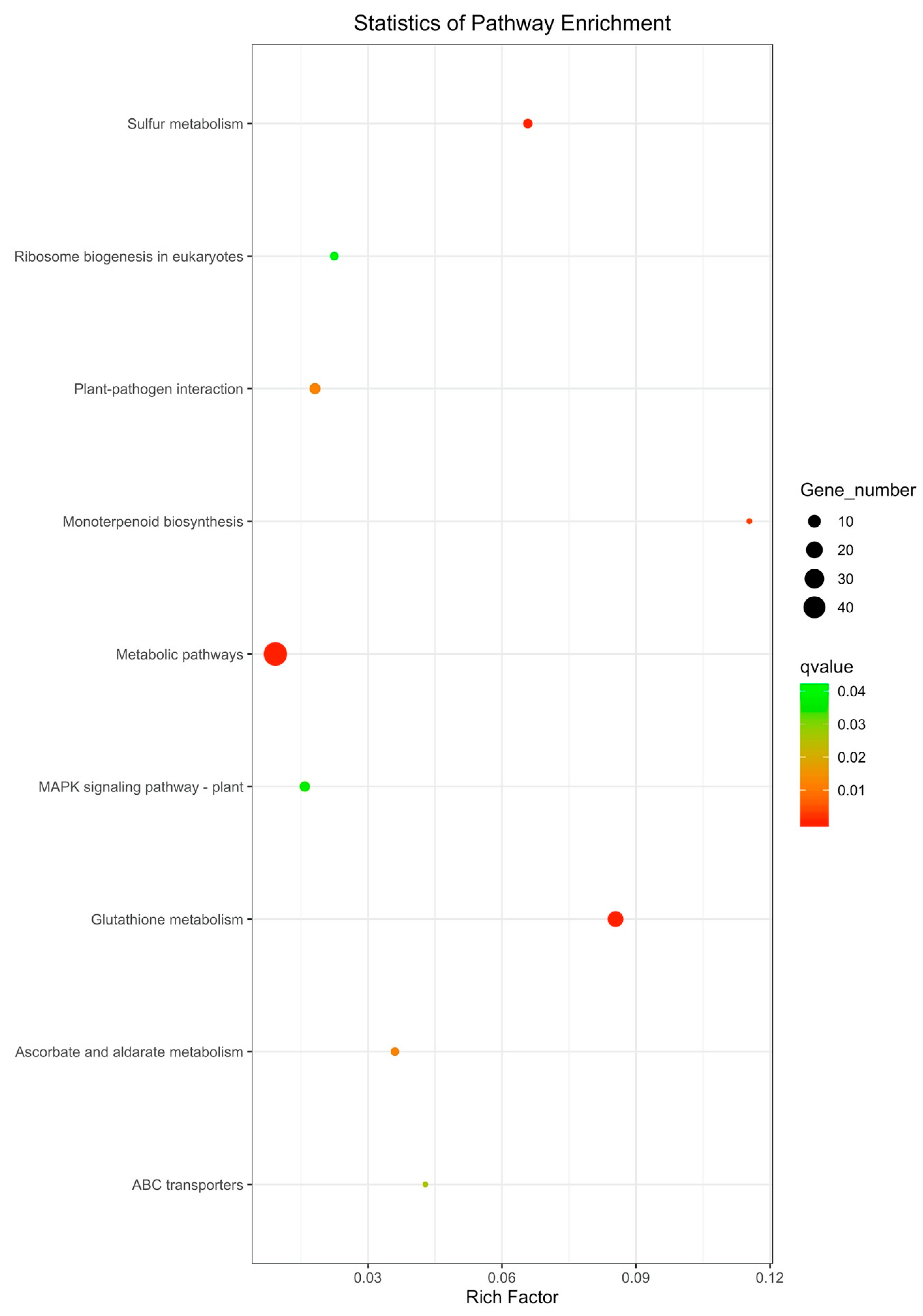
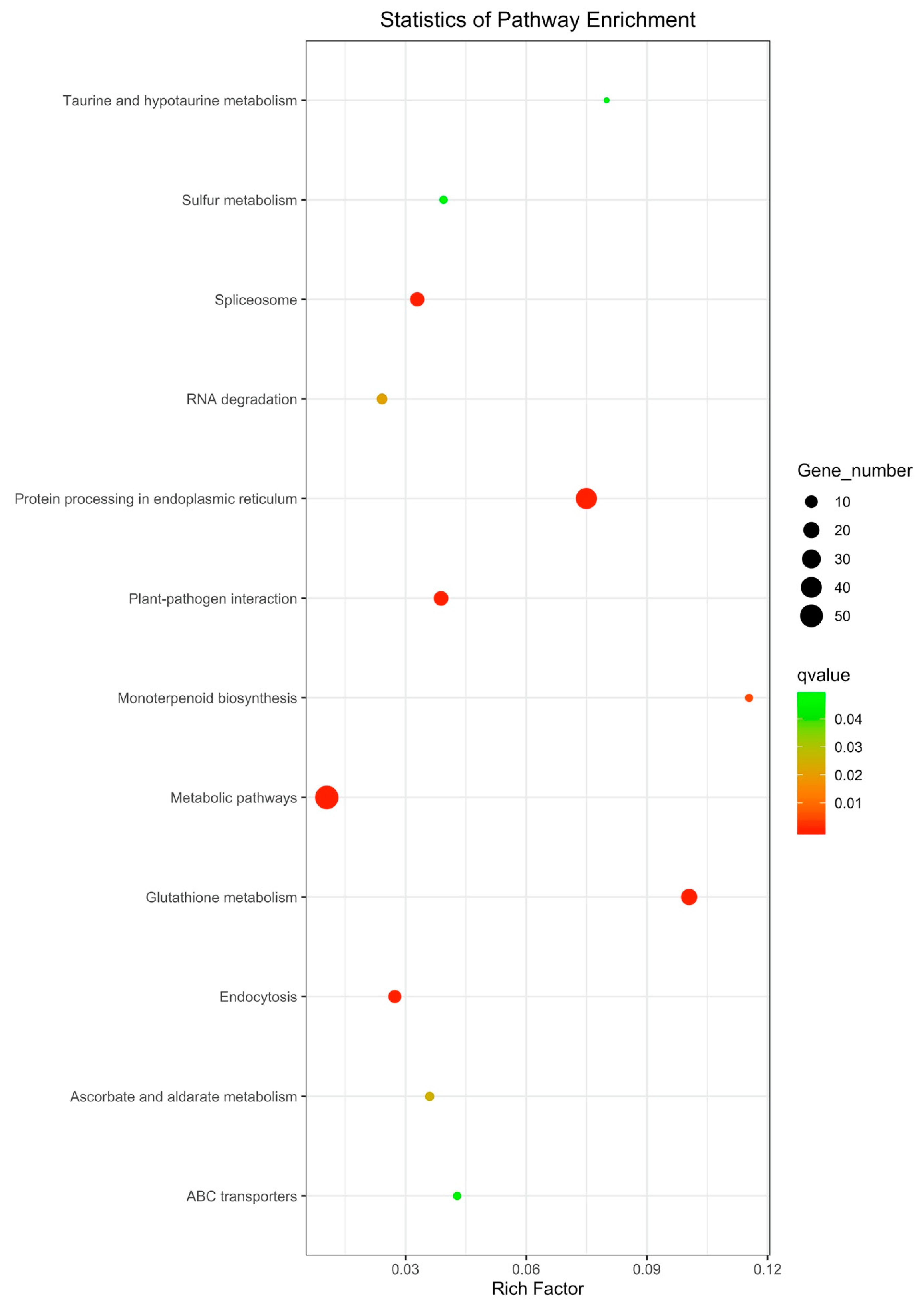
3.3.3. KEGG Pathway Enrichment Analysis of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef]
- McDonald, B.; Linde, C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 2002, 124, 163–180. [Google Scholar] [CrossRef]
- Cowger, C.; Mundt, C.C. Aggressiveness of Mycosphaerella graminicola isolates from susceptible and partially resistant wheat cultivars. Phytopathology 2002, 92, 624–630. [Google Scholar] [CrossRef]
- Montarry, J.; Glais, I.; Corbiere, R.; Andrivon, D. Adaptation to the most abundant host genotype in an agricultural plant–pathogen system–potato late blight. J. Evol. Biol. 2008, 21, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, F.; Mestre, P.; Schneider, C.; Kassemeyer, H.H.; Kozma, P.; Richart-Cervera, S.; Rouxel, M.; Delière, L. Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: Evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect. Genet. Evol. 2014, 27, 500–508. [Google Scholar] [CrossRef]
- Na, R.; Gijzen, M. Escaping host immunity: New tricks for plant pathogens. PLoS Pathog. 2016, 12, e1005631. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, T.; Zhong, C.; Luo, S.; Xu, K.; Gu, B.; Meng, Y.; Tyler, B.M.; Shan, W. Small RNAs generated by bidirectional transcription mediate silencing of RXLR effector genes in the oomycete Phytophthora sojae. Phytopathol. Res. 2019, 1, 18. [Google Scholar] [CrossRef]
- Cline, E.T.; Farr, D.F.; Rossman, A.Y. A synopsis of Phytophthora with accurate scientific names, host range, and geographic distribution. Plant Health Prog. 2008, 9, 32. [Google Scholar] [CrossRef]
- Gallup, C.A.; Sullivan, M.J.; Shew, H.D. Black Shank of Tobacco. In The Plant Health Instructor; The American Phytopathological Society: St. Paul, MN, USA, 2006. [Google Scholar]
- Sullivan, M.J.; Parks, E.J.; Cubeta, M.A.; Gallup, C.A.; Melton, T.A.; Moyer, J.W.; Shew, H.D. An assessment of the genetic diversity in a field population of Phytophthora nicotianae with a changing race structure. Plant Dis. 2010, 94, 455–460. [Google Scholar] [CrossRef][Green Version]
- Dukes, P.D.; Apple, J.L. Influence of host passage on virulence of Phytophthora parasitica var. nicotianae. Plant Dis. Rep. 1961, 45, 362. [Google Scholar]
- Sullivan, M.J.; Melton, T.A.; Shew, H.D. Managing the race structure of Phytophthora parasitica var. nicotianae with cultivar rotation. Plant Dis. 2005, 89, 1285–1294. [Google Scholar] [CrossRef]
- McCorkle, K.L.; Drake-Stowe, K.; Lewis, R.S.; Shew, H.D. Characterization of Phytophthora nicotianae resistance conferred by the introgressed Nicotiana rustica region, Wz, in flue-cured tobacco. Plant Dis. 2018, 102, 309–317. [Google Scholar] [CrossRef]
- McCorkle, K.L. Characterization of the Tobacco Pathogen Phytophthora nicotianae and Its Ability to Adapt Tohost Resistance Genes. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2016. [Google Scholar]
- Jin, J.; Shew, H.D. Components of aggressiveness in Phytophthora nicotianae during adaptation to multiple sources of partial resistance in tobacco. Plant Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jin, J. Characterization of Phytophthora nicotianae Following Adaptation to Partial Resistance in Tobacco. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2020. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Inst.: Cambridge, UK, 2010. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Rutherford, K.; Irvine, A.; Pedro, H.; Pant, R.; Sadanadan, V.; Khamari, L.; Billal, S.; Mohanty, S.; et al. PHI-base: A new interface and further additions for the multi-species pathogen–host interactions database. Nucleic Acids Res. 2017, 45, D604–D610. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A. IsoformSwitchAnalyzeR: Analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics 2019, 35, 4469–4471. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.Z.; Liou, R.F. Selection of internal control genes for real-time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genet. Biol. 2006, 43, 430–438. [Google Scholar] [CrossRef]
- Iida, Y.; van‘t Hof, P.; Beenen, H.; Mesarich, C.; Kubota, M.; Stergiopoulos, I.; Mehrabi, R.; Notsu, A.; Fujiwara, K.; Bahkali, A.; et al. Novel mutations detected in avirulence genes overcoming tomato Cf resistance genes in isolates of a Japanese population of Cladosporium fulvum. PLoS ONE 2015, 10, e0123271. [Google Scholar] [CrossRef]
- Morris, P.F.; Phuntumart, V. Inventory and comparative evolution of the ABC superfamily in the genomes of Phytophthora ramorum and Phytophthora sojae. J. Mol. Evol. 2009, 68, 563–575. [Google Scholar] [CrossRef]
- Urban, M.; Bhargava, T.; Hamer, J.E. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 1999, 18, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Schoonbeek, H.J.; Sugiura, H.; De Waard, M.A. Multidrug resistance in Botrytis cinerea associated with decreased accumulation of the azole fungicide oxpoconazole and increased transcription of the ABC transporter gene BcatrD. Pestic. Biochem. Physiol. 2001, 70, 168–179. [Google Scholar] [CrossRef]
- Lee, Y.J.; Yamamoto, K.; Hamamoto, H.; Nakaune, R.; Hibi, T. A novel ABC transporter gene ABC2 involved in multidrug susceptibility but not pathogenicity in rice blast fungus, Magnaporthe grisea. Pestic. Biochem. Physiol. 2005, 81, 13–23. [Google Scholar] [CrossRef]
- Liu, H.; Ma, X.; Yu, H.; Fang, D.; Li, Y.; Wang, X.; Wang, W.; Dong, Y.; Xiao, B. Genomes and virulence difference between two physiological races of Phytophthora nicotianae. Gigascience 2016, 5, 3. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Lin, F.C. Autophagy during conidiation, conidial germination and turgor generation in Magnaporthe grisea. Autophagy 2007, 3, 472–473. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zhang, Z.; Shao, W.; Yang, Y.; Zhou, M.; Chen, C. The autophagy-related gene BcATG1 is involved in fungal development and pathogenesis in Botrytis cinerea. Mol. Plant Pathol. 2017, 18, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Wang, C.; Yang, N.; Que, Y.; Talbot, N.J.; Wang, Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 2017, 7, 11062. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Wang, W.; Geng, X.; Shi, Y.; Na, R.; Dou, D.; Li, H. Network and role analysis of autophagy in Phytophthora sojae. Sci. Rep. 2017, 7, 1879. [Google Scholar] [CrossRef]
- Patron, N.J.; Durnford, D.G.; Kopriva, S. Sulfate assimilation in eukaryotes: Fusions, relocations and lateral transfers. BMC Evol. Biol. 2008, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Demongeot, J.; Seligmann, H. Why is AUG the start codon? Theoretical minimal RNA rings: Maximizing coded information biases 1st codon for the universal initiation codon AUG. BioEssays 2020, 42, 1900201. [Google Scholar] [CrossRef]
- Netto, L.E.S.; de Oliveira, M.A.; Monteiro, G.; Demasi, A.P.D.; Cussiol, J.R.R.; Discola, K.F.; Demasi, M.; Silva, G.M.; Alves, S.V.; Faria, V.G.; et al. Reactive cysteine in proteins: Protein folding, antioxidant defense, redox signaling and more. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 146, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.; Silva, D.N.; Várzea, V.; Paulo, O.S.; Batista, D. Genome-wide signatures of selection in Colletotrichum kahawae reveal candidate genes potentially involved in pathogenicity and aggressiveness. Front. Microbiol. 2019, 10, 1374. [Google Scholar] [CrossRef]
- Liu, Y.; Gonzàlez-Porta, M.; Santos, S.; Brazma, A.; Marioni, J.C.; Aebersold, R.; Venkitaraman, A.R.; Wickramasinghe, V.O. Impact of alternative splicing on the human proteome. Cell Rep. 2017, 20, 1229–1241. [Google Scholar] [CrossRef]
- Savory, E.A.; Zou, C.; Adhikari, B.N.; Hamilton, J.P.; Buell, C.R.; Shiu, S.H.; Day, B. Alternative splicing of a multi-drug transporter from Pseudoperonospora cubensis generates an RXLR effector protein that elicits a rapid cell death. PLoS ONE 2012, 7, e34701. [Google Scholar] [CrossRef]
- De Fine Licht, H.H. Does pathogen plasticity facilitate host shifts? PLoS Pathog. 2018, 14, e1006961. [Google Scholar] [CrossRef]
- Stassen, J.H.; Van den Ackerveken, G. How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 2011, 14, 407–414. [Google Scholar] [CrossRef]
- Torto, T.A.; Li, S.; Styer, A.; Huitema, E.; Testa, A.; Gow, N.A.; Van West, P.; Kamoun, S. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 2003, 13, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Schornack, S.; van Damme, M.; Bozkurt, T.O.; Cano, L.M.; Smoker, M.; Thines, M.; Gaulin, E.; Kamoun, S.; Huitema, E. Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA 2010, 107, 17421–17426. [Google Scholar] [CrossRef]
- Stam, R.; Jupe, J.; Howden, A.J.; Morris, J.A.; Boevink, P.C.; Hedley, P.E.; Huitema, E. Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS ONE 2013, 8, e59517. [Google Scholar] [CrossRef]
- Frantzeskakis, L.; Kusch, S.; Panstruga, R. The need for speed: Compartmentalized genome evolution in filamentous phytopathogens. Mol. Plant Pathol. 2019, 20, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.; Goodman, R.M.; Gut-Rella, M.; Glascock, C.; Weymann, K.; Friedrich, L.; Maddox, D.; Ahl-Goy, P.; Luntz, T.; Ward, E.S. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 1993, 90, 7327–7331. [Google Scholar] [CrossRef] [PubMed]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front. Plant Sci. 2015, 6, 584. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed]
- Hernández, I.; Chacón, O.; Rodriguez, R.; Portieles, R.; López, Y.; Pujol, M.; Borrás-Hidalgo, O. Black shank resistant tobacco by silencing of glutathione S-transferase. Biochem. Biophys. Res. Commun. 2009, 387, 300–304. [Google Scholar] [CrossRef]
- Stam, R.; Howden, A.J.M.; Delgado Cerezo, M.; Amaro, T.M.; Motion, G.B.; Pham, J.; Huitema, E. Characterization of cell death inducing Phytophthora capsici CRN effectors suggests diverse activities in the host nucleus. Front. Plant Sci. 2013, 4, 387. [Google Scholar] [CrossRef] [PubMed]
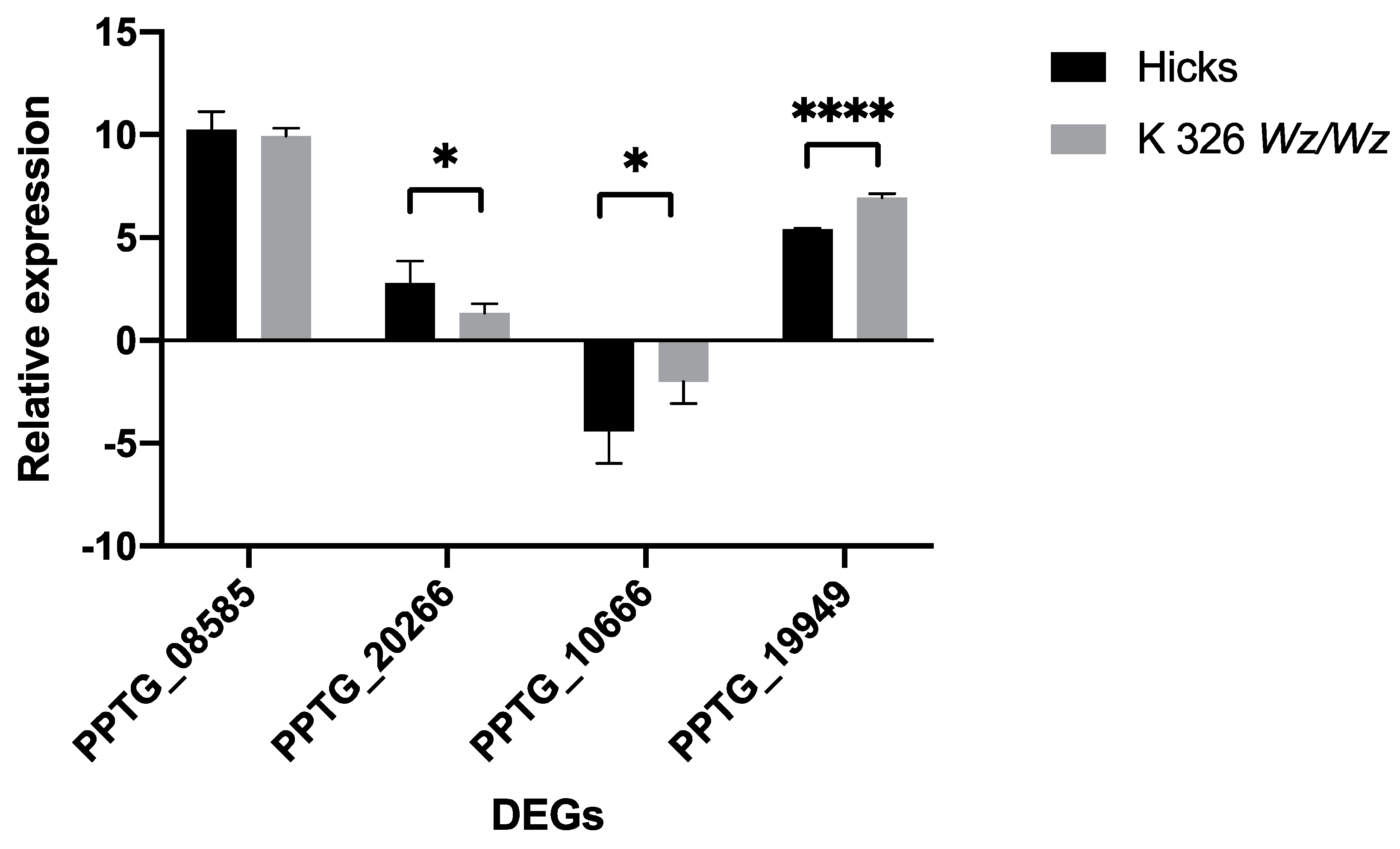

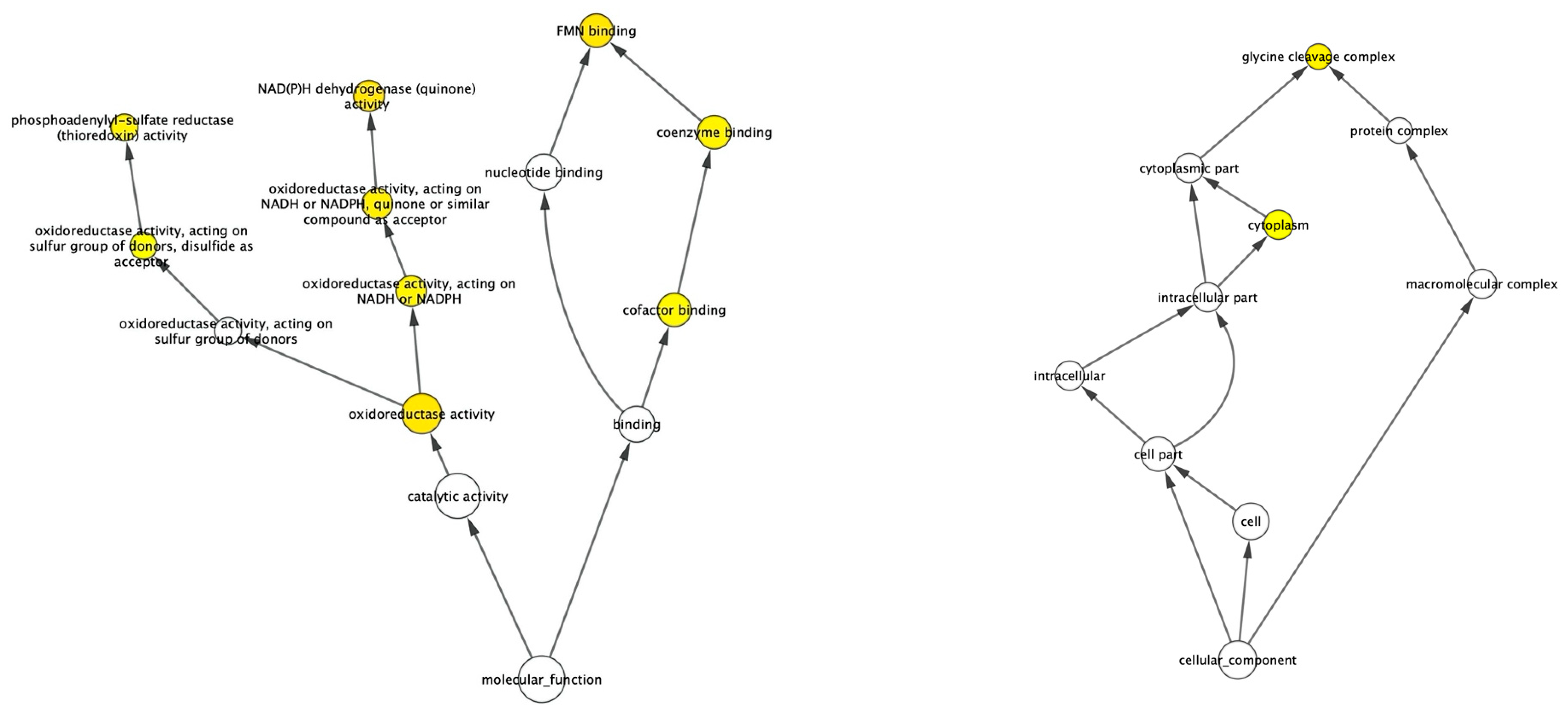
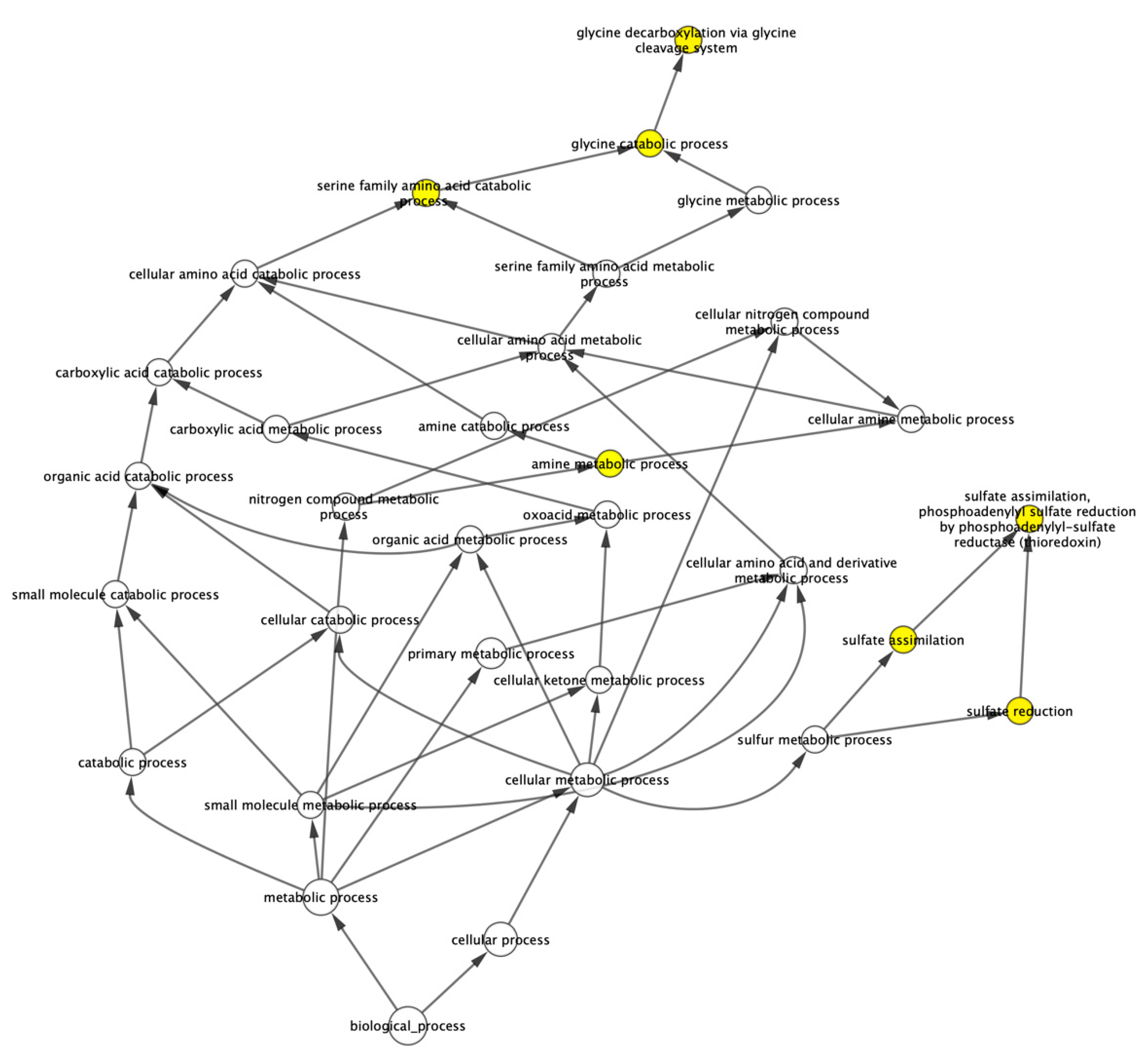
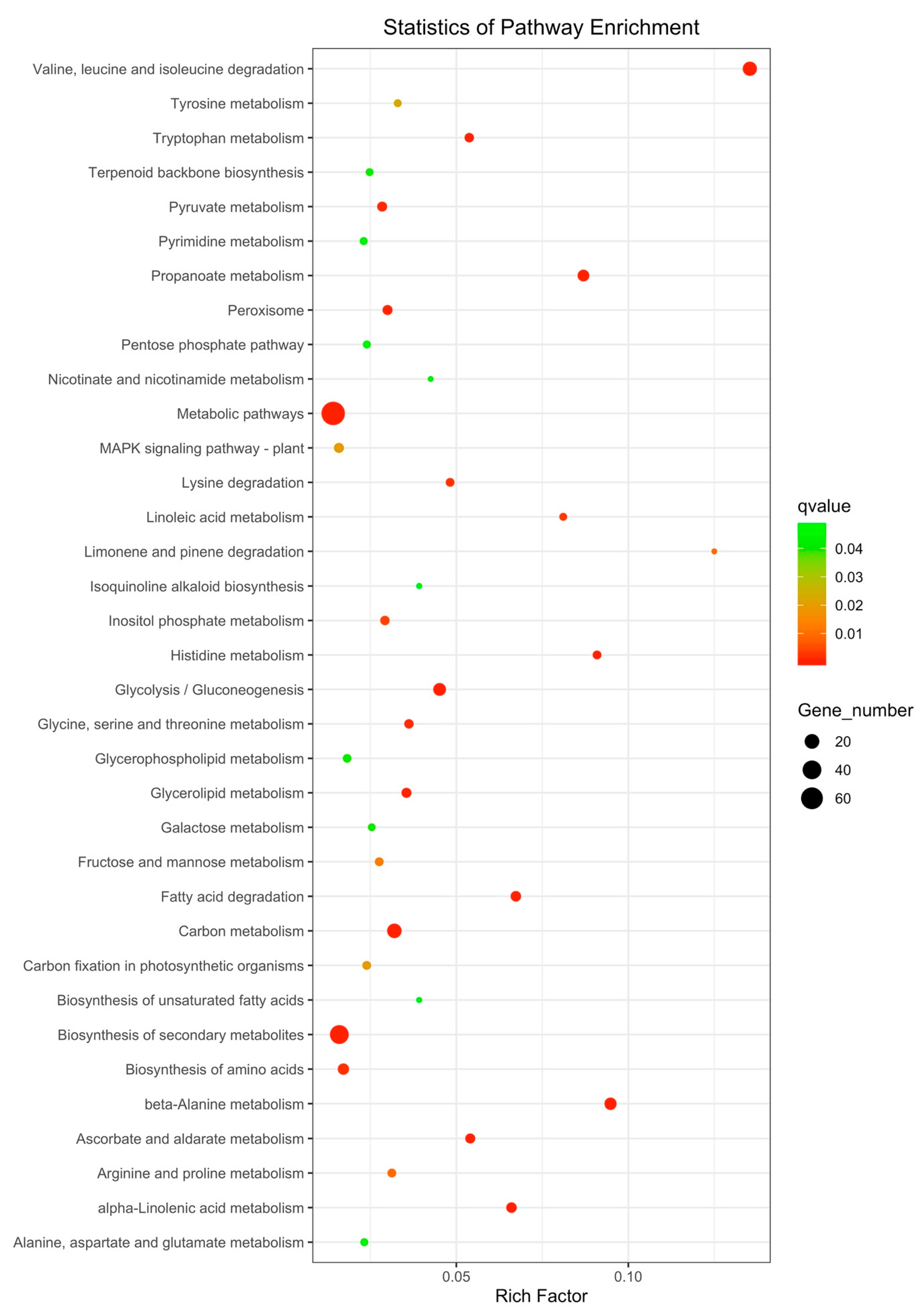

| Gene | Regulation | Forward Primer | Reverse Primer |
|---|---|---|---|
| PPTG_10666 | Up | CGTTCTCTTTTGCTCACGGA | CAGCTCCGACAAGTACACTG |
| PPTG_19949 | Up | CAACACTGTCACTGCTGGAT | GATCCAGTTGCTAGCGAGAG |
| PPTG_20266 | Down | CTCTCCGAAACAGAACCAACT | GTAGATCTCGGCAGTAACGC |
| PPTG_08585 | Down | AACACCACTACTCCAGCACT | ACAACTTCACCACATCCGTC |
| Ubc (ubiquitin-conjugating enzyme) | CCACTTAGAGCACGCTAGGA | TACCGACTGTCCTTCGTTCA | |
| WS21 (40S ribosomal protein S3A) | TACGCCAAGACGGCTCAGA | TTCCATCAGACGCACCAGG |
| Host Genotype | Isolate for Inoculation | Replication | Total Reads | P. nicotianae | N. tabacum | ||
|---|---|---|---|---|---|---|---|
| No. and Rate of Reads Mapped | Overall Alignment Rate | No. and Rate of Reads Mapped | Overall Alignment Rate | ||||
| Hicks | Wz-H | 1 | 32,616,245 | 11,799,948 (36.18%) | 38.50% | 1,170,706 (3.59%) | 4.77% |
| Hicks | Wz-H | 2 | 37,590,753 | 8,954,805 (23.82%) | 25.42% | 691,568 (1.84%) | 2.55% |
| Hicks | Wz-H | 3 | 32,142,283 | 8,581,032 (26.70%) | 28.55% | 1,526,273 (4.75%) | 6.21% |
| K 326 Wz/Wz | Wz-H | 1 | 30,738,911 | 6,756,839 (21.98%) | 23.62% | 1,234,275 (4.02%) | 5.84% |
| K 326 Wz/Wz | Wz-H | 2 | 32,228,624 | 7,512,158 (23.31%) | 24.89% | 2,571,539 (7.98%) | 10.15% |
| K 326 Wz/Wz | Wz-H | 3 | 41,304,137 | 8,159,363 (19.75%) | 21.14% | 1,146,835 (2.78%) | 3.63% |
| Hicks | Wz-Wz | 1 | 32,441,520 | 8,583,661 (26.46%) | 28.35% | 1,003,943 (3.09%) | 4.19% |
| Hicks | Wz-Wz | 2 | 33,639,749 | 6,441,805 (19.15%) | 20.54% | 561,746 (1.67%) | 2.95% |
| Hicks | Wz-Wz | 3 | 30,538,468 | 10,333,429 (33.84%) | 36.10% | 557,494 (1.83%) | 2.91% |
| K 326 Wz/Wz | Wz-Wz | 1 | 32,424,684 | 9,437,437 (29.11%) | 31.12% | 5,499,898 (16.96%) | 20.63% |
| K 326 Wz/Wz | Wz-Wz | 2 | 34,550,232 | 7,129,753 (20.64%) | 22.21% | 412,999 (1.20%) | 1.98% |
| K 326 Wz/Wz | Wz-Wz | 3 | 30,323,291 | 7,477,321 (24.66%) | 26.38% | 852,994 (2.81%) | 4.20% |
| Average | 33,378,241 | 27.23% | 5.83% | ||||
| Up-Regulated Gene | logFC | FDR | Annotation in NCBI |
| PPTG_02121 | 2.72729992 | 0.04116515 | Nucleotide-Binding Domain of the sugar kinase/HSP70/actin superfamily |
| PPTG_10666 | 1.40170745 | 0.00461859 | NADB_Rossmann |
| PPTG_12300 | 1.38934498 | 0.0113136 | Elicitin protein RAL13D [Phytophthora nicotianae] |
| PPTG_06767 | 1.29785829 | 0.00975047 | Cytochrome P450 |
| PPTG_00731 | 1.14244371 | 0.02689335 | Mitochondrial succinate-semialdehyde dehydrogenase and ALDH family members 5A1 and 5F1-like |
| PPTG_08145 | 1.06166001 | 0.03655436 | 4-aminobutyrate aminotransferase or related aminotransferase |
| PPTG_01316 | 1.01159768 | 0.0113136 | Aspartate aminotransferase (AAT) superfamily (fold type I) of pyridoxal phosphate (PLP)-dependent enzymes |
| PPTG_06886 | 0.9770123 | 0.04116515 | Mitochondrial carrier protein |
| PPTG_00433 | 0.95687542 | 0.04116515 | Amino acid permease |
| PPTG_05470 | 0.94439287 | 0.00975047 | SPRY domain in Ran binding proteins, SSH4, HECT E3 and SPRYD3 |
| PPTG_19949 | 0.90621478 | 0.01386612 | Peptidase domain in the S8 and S53 families |
| PPTG_08778 | 0.89782849 | 0.0287979 | NA |
| PPTG_11182 | 0.89467137 | 0.04116515 | GAF domain |
| PPTG_10595 | 0.88099898 | 0.02633833 | Second domain of the pleiotropic drug resistance-like (PDR) subfamily G of ATP-binding cassette transporters |
| PPTG_05834 | 0.80783638 | 0.01128107 | Putative lectin [Phytophthora palmivora var. palmivora] |
| PPTG_12158 | 0.63468318 | 0.02633833 | Serine/Threonine protein kinases, catalytic domain |
| Down-Regulated Gene | logFC | FDR | Annotation in NCBI |
| PPTG_12754 | −0.6308213 | 0.04116515 | 60S ribosomal protein L38 |
| PPTG_19261 | −0.6513629 | 0.01740912 | WRKY transcription factor 19 [Phytophthora nicotianae] |
| PPTG_15145 | −0.7844201 | 0.04116515 | Scavenger mRNA decapping enzyme C-term binding |
| PPTG_00424 | −0.8320101 | 0.04116515 | Amino acid permease |
| PPTG_19041 | −0.9101204 | 0.02968312 | acetate kinase A/propionate kinase 2 |
| PPTG_00957 | −0.9268608 | 0.04550147 | NA |
| PPTG_00099 | −0.9276355 | 0.03028585 | NA |
| PPTG_11197 | −1.01735 | 0.03970974 | 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase (PAPS reductase)/FAD synthetase or related enzyme |
| PPTG_09433 | −1.2431276 | 0.02997457 | Major Facilitator Superfamily (MFS) proteins |
| PPTG_17813 | −1.2434395 | 0.04550147 | Ankyrin repeats |
| PPTG_04568 | −1.3541329 | 0.0459794 | PQ-loop |
| PPTG_02974 | −1.4004705 | 0.03655436 | Major Facilitator Superfamily (MFS) proteins |
| PPTG_02595 | −1.49923 | 0.03655436 | NAD(P)+-dependent aldehyde dehydrogenase superfamily |
| PPTG_11386 | −1.5047674 | 0.02633833 | NA |
| PPTG_07126 | −1.5248213 | 0.04304619 | Short chain dehydrogenase |
| PPTG_12006 | −1.5957463 | 0.01170615 | Glycosyl hydrolase family 1 |
| PPTG_08585 | −1.6180257 | 0.00106403 | Old yellow enzyme (OYE)-like FMN binding domain |
| PPTG_18570 | −1.633993 | 0.01009357 | Zinc finger, C2H2 type |
| PPTG_23779 | −1.6347597 | 0.00427631 | NAD(P)H:FMN oxidoreductases, oxygen-insensitive nitroreductase, flavin reductase P, dihydropteridine reductase, NADH oxidase or NADH dehydrogenase |
| PPTG_05530 | −1.6929325 | 0.01258482 | NADPH oxidase (NOX) |
| PPTG_02448 | −1.7462909 | 0.00975047 | Major Facilitator Superfamily (MFS) proteins |
| PPTG_18743 | −1.7855609 | 0.03655436 | NA |
| PPTG_13181 | −2.0941462 | 0.01009357 | Major Facilitator Superfamily (MFS) proteins |
| PPTG_08485 | −2.1192135 | 0.02633833 | Major Facilitator Superfamily (MFS) proteins |
| PPTG_09275 | −2.1770096 | 0.0000271 | NADPH-dependent FMN reductase |
| PPTG_00236 | −2.2019316 | 0.02968312 | TonB receptor activity [Phytophthora megakarya] |
| PPTG_13068 | −2.3667825 | 0.03655436 | Membrane-associating domain |
| PPTG_20266 | -2.4697411 | 0.00888653 | NADPH-dependent FMN reductase |
| PPTG_04065 | −3.3756258 | 0.03655436 | ZIP Zinc transporter |
| PPTG_10399 | −3.9114676 | 0.01740912 | D-arabinose 1-dehydrogenase, Zn-dependent alcohol dehydrogenase family |
| Up-Regulated Gene | logFC | FDR | Annotation in NCBI |
| PPTG_01162 | 1.75973946 | 0.03399441 | Exonuclease-Endonuclease-Phosphatase (EEP) domain superfamily |
| PPTG_01484 | 1.66331002 | 0.01294692 | Amino_oxidase; Flavin containing amine oxidoreductase |
| PPTG_08721 | 1.36573335 | 0.03296512 | SCP-like extracellular protein domain |
| PPTG_12693 | 1.35565787 | 0.0217849 | NA |
| PPTG_15982 | 1.22889916 | 0.03661908 | NAD(P)-dependent dehydrogenase |
| PPTG_17442 | 1.14815854 | 0.03856894 | Isoprenylcysteine carboxyl methyltransferase (ICMT) family |
| PPTG_19949 | 1.09394216 | 0.00569986 | Peptidase domain in the S8 and S53 families |
| PPTG_06767 | 1.09239542 | 0.02680104 | Cytochrome P450 |
| PPTG_10666 | 1.05056269 | 0.02831652 | Rossmann-fold NAD(P)(+)-binding proteins |
| PPTG_23419 | 1.01740189 | 0.0217849 | large tegument protein UL36; |
| PPTG_04377 | 0.94043542 | 0.03296512 | Major Facilitator Superfamily (MFS) proteins |
| PPTG_05470 | 0.91187391 | 0.01702338 | SPRY domain in Ran binding proteins |
| PPTG_22853 | 0.85943994 | 0.03296512 | Putative storage protein LPV |
| PPTG_13013 | 0.8307265 | 0.0217849 | Cyst germination specific acidic repeat protein |
| PPTG_20368 | 0.82758849 | 0.02281486 | NA |
| PPTG_01588 | 0.82579718 | 0.02281486 | Tetratricopeptide repeat |
| PPTG_04341 | 0.78702506 | 0.014592 | Kazal type serine protease inhibitors |
| PPTG_00655 | 0.7658604 | 0.02490186 | Iron-enterobactin transporter ATP-binding protein |
| PPTG_08559 | 0.76309934 | 0.03296512 | NA |
| PPTG_00623 | 0.75665029 | 0.0217849 | The Phox Homology domain, a phosphoinositide binding module |
| PPTG_22560 | 0.72235522 | 0.04236865 | Ricin-type beta-trefoil |
| PPTG_07553 | 0.71836169 | 0.03296512 | HAM34-like putative membrane protein |
| PPTG_01906 | 0.69845644 | 0.03303654 | Cytochrome P450 |
| PPTG_11777 | 0.69839919 | 0.03303654 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain |
| PPTG_13016 | 0.68630319 | 0.03147853 | Glycosyltransferase (GlcNAc) |
| PPTG_17323 | 0.66267543 | 0.03303654 | tRNA binding domain |
| PPTG_15053 | 0.66097703 | 0.04910001 | NA |
| PPTG_03113 | 0.65586116 | 0.03147853 | NA |
| PPTG_00340 | 0.54904534 | 0.04815318 | Dynein heavy chain and region D6 of dynein motor |
| Down-Regulated Gene | logFC | FDR | Annotation in NCBI |
| PPTG_17561 | −0.5858454 | 0.04815318 | Biotinyl_lipoyl_domains |
| PPTG_12754 | −0.6407469 | 0.03613858 | 60S ribosomal protein L38 |
| PPTG_02651 | −0.8343073 | 0.03303654 | TLR4 regulator and MIR-interacting MSAP |
| PPTG_00501 | −0.8933285 | 0.03147853 | Abhydrolase |
| PPTG_21942 | −0.9296731 | 0.03842764 | Phosphate acetyltransferase |
| PPTG_11197 | −1.0358966 | 0.03303654 | 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase (PAPS reductase)/FAD synthetase or related enzyme |
| PPTG_08585 | −1.0629125 | 0.02281486 | Old yellow enzyme (OYE)-like FMN binding domain |
| PPTG_21974 | −1.1383951 | 0.02281486 | NA |
| PPTG_13205 | −1.2268529 | 0.04815318 | Elicitin |
| PPTG_21937 | −1.2492039 | 0.03296512 | START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC (SRPBCC) ligand-binding domain superfamily |
| PPTG_05327 | −1.2837131 | 0.03147853 | NA |
| PPTG_15084 | −1.5159809 | 0.04674932 | TKL/DRK protein kinase |
| PPTG_02595 | −1.5567119 | 0.03303654 | NAD(P)+-dependent aldehyde dehydrogenase superfamily |
| PPTG_09275 | −1.6934963 | 0.00080536 | NADPH-dependent FMN reductase |
| PPTG_18743 | −1.7231432 | 0.03661908 | NA |
| PPTG_15596 | −1.7614863 | 0.02490186 | START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC (SRPBCC) ligand-binding domain superfamily |
| PPTG_23779 | −1.7704877 | 0.00245828 | NAD(P)H:FMN oxidoreductases, oxygen-insensitive nitroreductase, flavin reductase P, dihydropteridine reductase, NADH oxidase or NADH dehydrogenase. |
| PPTG_05968 | −2.0360954 | 0.02831652 | Redox-sensitive bicupin YhaK, pirin superfamily |
| PPTG_20266 | −2.1555007 | 0.0217849 | NADPH-dependent FMN reductase |
| PPTG_05967 | −2.4675984 | 0.00569986 | Redox-sensitive bicupin YhaK, pirin superfamily |
| PPTG_16697 | −3.3993324 | 0.04099675 | Solute carrier families 5 and 6-like |
| Isolate | DEG Regulation | Term | Rich-Factor | Corrected p-Value |
|---|---|---|---|---|
| Wz-Wz | up-regulated | Spliceosome | 0.018 | 0.049 |
| down-regulated | Ubiquinone and other terpenoid-quinone biosynthesis | 0.118 | 0.008 | |
| Biosynthesis of secondary metabolites | 0.010 | 0.040 | ||
| Wz-H | down-regulated | Biosynthesis of secondary metabolites | 0.015 | 0.000 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 0.118 | 0.002 | ||
| Biosynthesis of antibiotics | 0.016 | 0.002 | ||
| Glyoxylate and dicarboxylate metabolism | 0.053 | 0.005 | ||
| Glycine, serine and threonine metabolism | 0.051 | 0.006 | ||
| Metabolic pathways | 0.005 | 0.007 | ||
| Carbon metabolism | 0.014 | 0.043 | ||
| Ascorbate and aldarate metabolism | 0.067 | 0.049 | ||
| Sulfur metabolism | 0.059 | 0.049 | ||
| Histidine metabolism | 0.056 | 0.049 |
| CHROM | POS | Gene | Annotation | Wz-Wz | Wz-H |
|---|---|---|---|---|---|
| 7000000185249081 | 1188712 | PPTG_07972 | Hypothetical protein | C/C | C/T |
| 7000000185249382 | 599392 | PPTG_05817 | Transcription factor S | C/C | C/T |
| 7000000185249084 | 546281 | PPTG_05165 | Hypothetical protein | T/C | T/T |
| 7000000185249172 | 182510 | PPTG_17734 | Crinkler family | G/A | G/G |
| 7000000185249344 | 2066387 | PPTG_03590 | Hypothetical protein | G/A | G/G |
| 7000000185249061 | 1073008 | NA | NA | A/T | T/T |
| 7000000185249344 | 2066377 | PPTG_03590 | Hypothetical protein | A/G | G/G |
| 7000000185249344 | 2066882 | PPTG_03590 | Hypothetical protein | C/T | T/T |
| Tobacco Genotype for DEG Identification | Isolate for Inoculation | Total No. of DEGs | Up-Regulated DEGs | Down-Regulated DEGs |
|---|---|---|---|---|
| K 326 Wz/Wz | Wz-Wz | 608 | 305 | 303 |
| K 326 Wz/Wz | Wz-H | 567 | 174 | 393 |
| Shared DEGs | 257 | 94 | 163 |
| Up-Regulated DEGs in K326 Wz/Wz | ||||
| GO Accession | Term | Term Type | p Value | FDR |
| GO: 0044710 | single-organism metabolic process | P | 3.40 × 10−6 | 0.0019 |
| GO: 0044281 | small molecule metabolic process | P | 0.00015 | 0.0063 |
| GO: 0044712 | single-organism catabolic process | P | 0.00015 | 0.0063 |
| GO: 0046031 | ADP metabolic process | P | 0.00018 | 0.0063 |
| GO: 0009179 | purine ribonucleoside diphosphate metabolic process | P | 0.00018 | 0.0063 |
| GO: 0046496 | nicotinamide nucleotide metabolic process | P | 0.00015 | 0.0063 |
| GO: 1901575 | organic substance catabolic process | P | 0.00013 | 0.0063 |
| GO: 0051186 | cofactor metabolic process | P | 6.50 × 10−5 | 0.0063 |
| GO: 0044723 | single-organism carbohydrate metabolic process | P | 0.00016 | 0.0063 |
| GO: 0006757 | ATP generation from ADP | P | 0.00018 | 0.0063 |
| GO: 0006732 | coenzyme metabolic process | P | 7.40 × 10−5 | 0.0063 |
| GO: 0009135 | purine nucleoside diphosphate metabolic process | P | 0.00018 | 0.0063 |
| GO: 0009185 | ribonucleoside diphosphate metabolic process | P | 0.00018 | 0.0063 |
| GO: 0072524 | pyridine-containing compound metabolic process | P | 0.00017 | 0.0063 |
| GO: 0006096 | glycolytic process | P | 0.00018 | 0.0063 |
| GO: 0019362 | pyridine nucleotide metabolic process | P | 0.00015 | 0.0063 |
| GO: 0006733 | oxidoreduction coenzyme metabolic process | P | 0.00019 | 0.0063 |
| GO: 0044724 | single-organism carbohydrate catabolic process | P | 0.00026 | 0.0075 |
| GO: 0009056 | catabolic process | P | 0.00025 | 0.0075 |
| GO: 0006090 | pyruvate metabolic process | P | 0.00029 | 0.0082 |
| GO: 0006165 | nucleoside diphosphate phosphorylation | P | 0.00032 | 0.0086 |
| GO: 0009132 | nucleoside diphosphate metabolic process | P | 0.00042 | 0.011 |
| GO: 0016209 | antioxidant activity | F | 8.40 × 10−5 | 0.011 |
| GO: 0003824 | catalytic activity | F | 8.50 × 10−5 | 0.011 |
| GO: 0046939 | nucleotide phosphorylation | P | 0.00053 | 0.012 |
| GO: 0005975 | carbohydrate metabolic process | P | 0.00053 | 0.012 |
| GO: 0016052 | carbohydrate catabolic process | P | 0.00056 | 0.012 |
| GO: 0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors | F | 0.00017 | 0.014 |
| GO: 0006082 | organic acid metabolic process | P | 0.0014 | 0.03 |
| GO: 0043436 | oxoacid metabolic process | P | 0.0016 | 0.033 |
| GO: 0019752 | carboxylic acid metabolic process | P | 0.0016 | 0.033 |
| GO: 0043168 | anion binding | F | 0.00057 | 0.035 |
| GO: 1901135 | carbohydrate derivative metabolic process | P | 0.0025 | 0.048 |
| Down-Regulated DEGs in K326 Wz/Wz | ||||
| GO Accession | Term | Term Type | p Value | FDR |
| GO: 0000786 | nucleosome | C | 5.10 × 10−27 | 2.10 × 10−25 |
| GO: 0032993 | protein-DNA complex | C | 5.10 × 10−27 | 2.10 × 10−25 |
| GO: 0044815 | DNA packaging complex | C | 8.30 × 10−27 | 2.20 × 10−25 |
| GO: 0000785 | chromatin | C | 3.40 × 10−26 | 6.90 × 10−25 |
| GO: 0046982 | protein heterodimerization activity | F | 1.60 × 10−26 | 2.40 × 10−24 |
| GO: 0031497 | chromatin assembly | P | 5.30 × 10−26 | 7.70 × 10−24 |
| GO: 0065004 | protein-DNA complex assembly | P | 8.30 × 10−26 | 7.70 × 10−24 |
| GO: 0034728 | nucleosome organization | P | 5.30 × 10−26 | 7.70 × 10−24 |
| GO: 0006334 | nucleosome assembly | P | 5.30 × 10−26 | 7.70 × 10−24 |
| GO: 0071824 | protein-DNA complex subunit organization | P | 8.30 × 10−26 | 7.70 × 10−24 |
| GO: 0006333 | chromatin assembly or disassembly | P | 1.10 × 10−25 | 8.80 × 10−24 |
| GO: 0006323 | DNA packaging | P | 1.70 × 10−25 | 1.20 × 10−23 |
| GO: 0071103 | DNA conformation change | P | 2.60 × 10−24 | 1.50 × 10−22 |
| GO: 0044427 | chromosomal part | C | 1.00 × 10−23 | 1.70 × 10−22 |
| GO: 0005694 | chromosome | C | 1.90 × 10−22 | 2.50 × 10−21 |
| GO: 0006325 | chromatin organization | P | 7.90 × 10−23 | 4.10 × 10−21 |
| GO: 0034622 | cellular macromolecular complex assembly | P | 4.30 × 10−21 | 2.00 × 10−19 |
| GO: 0051276 | chromosome organization | P | 1.00 × 10−20 | 4.20 × 10−19 |
| GO: 0065003 | macromolecular complex assembly | P | 1.30 × 10−20 | 5.00 × 10−19 |
| GO: 0070271 | protein complex biogenesis | P | 8.40 × 10−20 | 2.80 × 10−18 |
| GO: 0006461 | protein complex assembly | P | 8.40 × 10−20 | 2.80 × 10−18 |
| GO: 0022607 | cellular component assembly | P | 4.90 × 10−19 | 1.50 × 10−17 |
| GO: 0071822 | protein complex subunit organization | P | 1.40 × 10−18 | 3.90 × 10−17 |
| GO: 0043933 | macromolecular complex subunit organization | P | 4.60 × 10−18 | 1.20 × 10−16 |
| GO: 0044085 | cellular component biogenesis | P | 3.80 × 10−17 | 9.70 × 10−16 |
| GO: 0006996 | organelle organization | P | 5.30 × 10−17 | 1.30 × 10−15 |
| GO: 0046983 | protein dimerization activity | F | 1.70 × 10−13 | 1.30 × 10−11 |
| GO: 0016043 | cellular component organization | P | 5.30 × 10−12 | 1.20 × 10−10 |
| GO: 0071840 | cellular component organization or biogenesis | P | 3.10 × 10−11 | 6.90 × 10−10 |
| GO: 0044422 | organelle part | C | 2.30 × 10−10 | 2.30 × 10−9 |
| GO: 0044446 | intracellular organelle part | C | 2.30 × 10−10 | 2.30 × 10−9 |
| GO: 0003677 | DNA binding | F | 5.50 × 10−11 | 2.80 × 10−9 |
| GO: 0043234 | protein complex | C | 4.90 × 10−9 | 4.50 × 10−8 |
| GO: 0043232 | intracellular non-membrane-bounded organelle | C | 1.30 × 10−8 | 9.40 × 10−8 |
| GO: 0043228 | non-membrane-bounded organelle | C | 1.30 × 10−8 | 9.40 × 10−8 |
| GO: 0005634 | nucleus | C | 3.00 × 10−6 | 2.00 × 10−5 |
| GO: 0009987 | cellular process | P | 2.10 × 10−6 | 4.30 × 10−5 |
| GO: 0003676 | nucleic acid binding | F | 1.30 × 10−6 | 5.10 × 10−5 |
| GO: 0032991 | macromolecular complex | C | 8.20 × 10−6 | 5.10 × 10−5 |
| GO: 0006352 | DNA-templated transcription, initiation | P | 6.00 × 10−6 | 0.00012 |
| GO: 0043231 | intracellular membrane-bounded organelle | C | 0.0002 | 0.0011 |
| GO: 0043227 | membrane-bounded organelle | C | 0.0002 | 0.0011 |
| GO: 0043229 | intracellular organelle | C | 0.00071 | 0.0034 |
| GO: 0043226 | organelle | C | 0.00071 | 0.0034 |
| GO: 1901576 | organic substance biosynthetic process | P | 0.00025 | 0.0046 |
| GO: 0009058 | biosynthetic process | P | 0.00025 | 0.0046 |
| GO: 1901363 | heterocyclic compound binding | F | 0.00019 | 0.005 |
| GO: 0097159 | organic cyclic compound binding | F | 0.00019 | 0.005 |
| GO: 0019438 | aromatic compound biosynthetic process | P | 0.00045 | 0.008 |
| GO: 1901362 | organic cyclic compound biosynthetic process | P | 0.0006 | 0.009 |
| GO: 0097659 | nucleic acid-templated transcription | P | 0.00059 | 0.009 |
| GO: 0006351 | transcription, DNA-templated | P | 0.00059 | 0.009 |
| GO: 0032774 | RNA biosynthetic process | P | 0.0006 | 0.009 |
| GO: 0044249 | cellular biosynthetic process | P | 0.00057 | 0.009 |
| GO: 0034654 | nucleobase-containing compound biosynthetic process | P | 0.0018 | 0.027 |
| GO: 0044424 | intracellular part | C | 0.0061 | 0.028 |
| GO: 0065007 | biological regulation | P | 0.0024 | 0.034 |
| GO: 0050794 | regulation of cellular process | P | 0.0025 | 0.034 |
| GO: 0005622 | intracellular | C | 0.0081 | 0.035 |
| GO: 0044711 | single-organism biosynthetic process | P | 0.0029 | 0.039 |
| GO: 0050789 | regulation of biological process | P | 0.0031 | 0.04 |
| GO: 0065008 | regulation of biological quality | P | 0.0034 | 0.042 |
| GO: 0018130 | heterocycle biosynthetic process | P | 0.0036 | 0.043 |
| Down-Regulated DEGs in K326 Wz/Wz | ||||
|---|---|---|---|---|
| GO Accession | Term | Term Type | p Value | FDR |
| GO: 0006457 | protein folding | P | 9.70 × 10−11 | 3.60 × 10−8 |
| GO: 0051082 | unfolded protein binding | F | 1.30 × 10−9 | 2.90 × 10−7 |
| GO: 0005488 | binding | F | 2.30 × 10−5 | 0.0025 |
| GO: 0043565 | sequence-specific DNA binding | F | 0.00035 | 0.026 |
| GO: 1901363 | heterocyclic compound binding | F | 0.0011 | 0.048 |
| GO: 0001071 | nucleic acid binding transcription factor activity | F | 0.0015 | 0.048 |
| GO: 0097159 | organic cyclic compound binding | F | 0.0011 | 0.048 |
| GO: 0003700 | transcription factor activity, sequence-specific DNA binding | F | 0.0015 | 0.048 |
| GO Terms Enriched for Up-Regulated DEGs in K326 Wz/Wz | ||||
| GO Accession | Term | Term Type | p Value | FDR |
| GO: 0004867 | serine-type endopeptidase inhibitor activity | F | 2.60 × 10−11 | 4.20 × 10−9 |
| GO: 0030414 | peptidase inhibitor activity | F | 1.40 × 10−8 | 4.40 × 10−7 |
| GO: 0061134 | peptidase regulator activity | F | 1.40 × 10−8 | 4.40 × 10−7 |
| GO: 0004866 | endopeptidase inhibitor activity | F | 1.40 × 10−8 | 4.40 × 10−7 |
| GO: 0061135 | endopeptidase regulator activity | F | 1.40 × 10−8 | 4.40 × 10−7 |
| GO: 0044710 | single-organism metabolic process | P | 2.10 × 10−6 | 0.00012 |
| GO: 0004857 | enzyme inhibitor activity | F | 1.30 × 10−5 | 0.00036 |
| GO: 0050660 | flavin adenine dinucleotide binding | F | 8.80 × 10−5 | 0.0018 |
| GO: 0030234 | enzyme regulator activity | F | 7.70 × 10−5 | 0.0018 |
| GO: 0098772 | molecular function regulator | F | 0.00014 | 0.0025 |
| GO: 0044699 | single-organism process | P | 0.00015 | 0.0043 |
| GO: 0016491 | oxidoreductase activity | F | 0.00035 | 0.0057 |
| GO: 0055114 | oxidation-reduction process | P | 0.00032 | 0.0063 |
| GO: 0050662 | coenzyme binding | F | 0.00085 | 0.012 |
| GO: 0048037 | cofactor binding | F | 0.0021 | 0.029 |
| GO Terms Enriched for Down-Regulated DEGs in K326 Wz/Wz | ||||
| GO Accession | Term | Term Type | p Value | FDR |
| GO: 0016758 | transferase activity, transferring hexosyl groups | F | 3.9 × 10−5 | 0.01 |
| GO: 0016757 | transferase activity, transferring glycosyl groups | F | 0.00029 | 0.013 |
| GO: 0015405 | P-P-bond-hydrolysis-driven transmembrane transporter activity | F | 0.00043 | 0.013 |
| GO: 0042626 | ATPase activity, coupled to transmembrane movement of substances | F | 0.00021 | 0.013 |
| GO: 0015399 | primary active transmembrane transporter activity | F | 0.00043 | 0.013 |
| GO: 0043492 | ATPase activity, coupled to movement of substances | F | 0.00023 | 0.013 |
| GO: 0016491 | oxidoreductase activity | F | 0.00048 | 0.013 |
| GO: 0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | F | 0.00042 | 0.013 |
| GO: 0016667 | oxidoreductase activity, acting on a sulfur group of donors | F | 0.00036 | 0.013 |
| GO: 0016887 | ATPase activity | F | 0.00073 | 0.018 |
| GO: 0006457 | protein folding | P | 0.00031 | 0.032 |
| GO: 0042592 | homeostatic process | P | 0.0004 | 0.032 |
| GO: 0019725 | cellular homeostasis | P | 0.00022 | 0.032 |
| GO: 0045454 | cell redox homeostasis | P | 0.00017 | 0.032 |
| GO: 0042623 | ATPase activity, coupled | F | 0.0021 | 0.048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, J.; Shi, R.; Lewis, R.S.; Shew, H.D. RNAseq Reveals Differential Gene Expression Contributing to Phytophthora nicotianae Adaptation to Partial Resistance in Tobacco. Agronomy 2021, 11, 656. https://doi.org/10.3390/agronomy11040656
Jin J, Shi R, Lewis RS, Shew HD. RNAseq Reveals Differential Gene Expression Contributing to Phytophthora nicotianae Adaptation to Partial Resistance in Tobacco. Agronomy. 2021; 11(4):656. https://doi.org/10.3390/agronomy11040656
Chicago/Turabian StyleJin, Jing, Rui Shi, Ramsey Steven Lewis, and Howard David Shew. 2021. "RNAseq Reveals Differential Gene Expression Contributing to Phytophthora nicotianae Adaptation to Partial Resistance in Tobacco" Agronomy 11, no. 4: 656. https://doi.org/10.3390/agronomy11040656
APA StyleJin, J., Shi, R., Lewis, R. S., & Shew, H. D. (2021). RNAseq Reveals Differential Gene Expression Contributing to Phytophthora nicotianae Adaptation to Partial Resistance in Tobacco. Agronomy, 11(4), 656. https://doi.org/10.3390/agronomy11040656






