Quantification of Root-Knot Nematode Infestation in Tomato Using Digital Image Analysis
Abstract
:1. Introduction
2. Materials and Methods
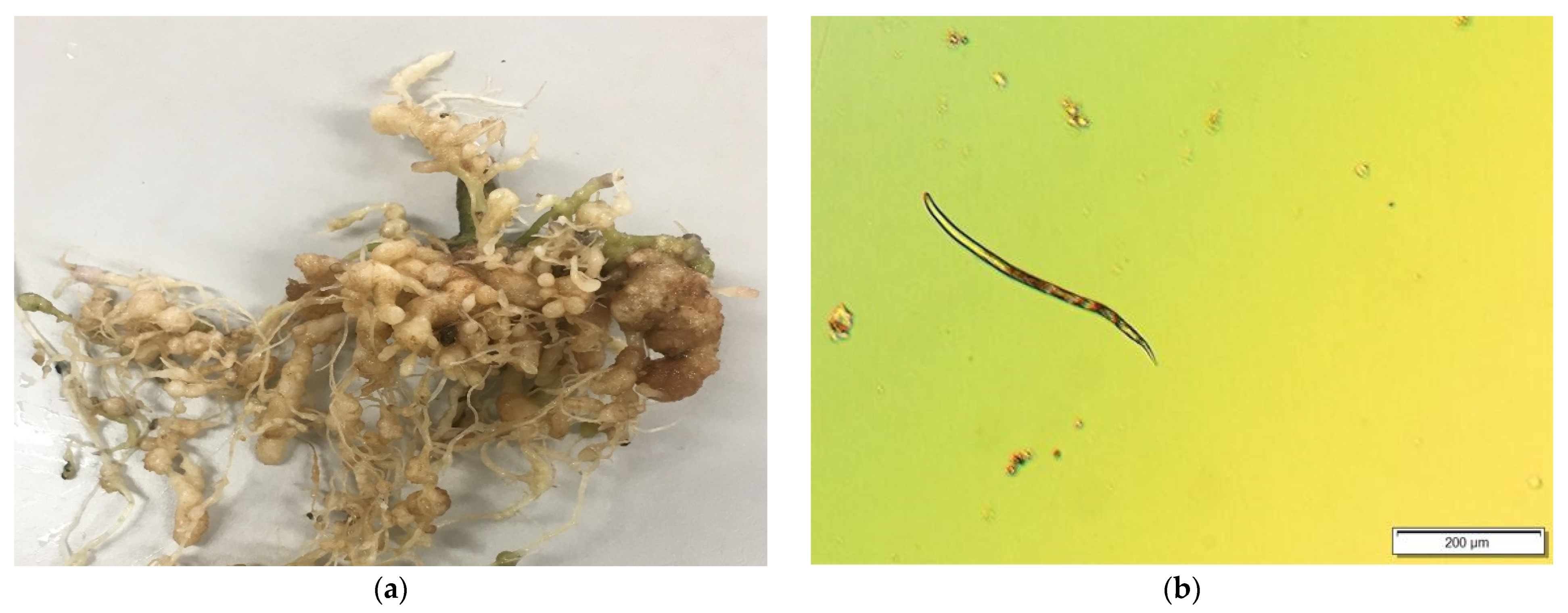
2.1. Soil Sample Preparation and Collection of Roots
2.2. Image Acquisition
2.3. Image Processing
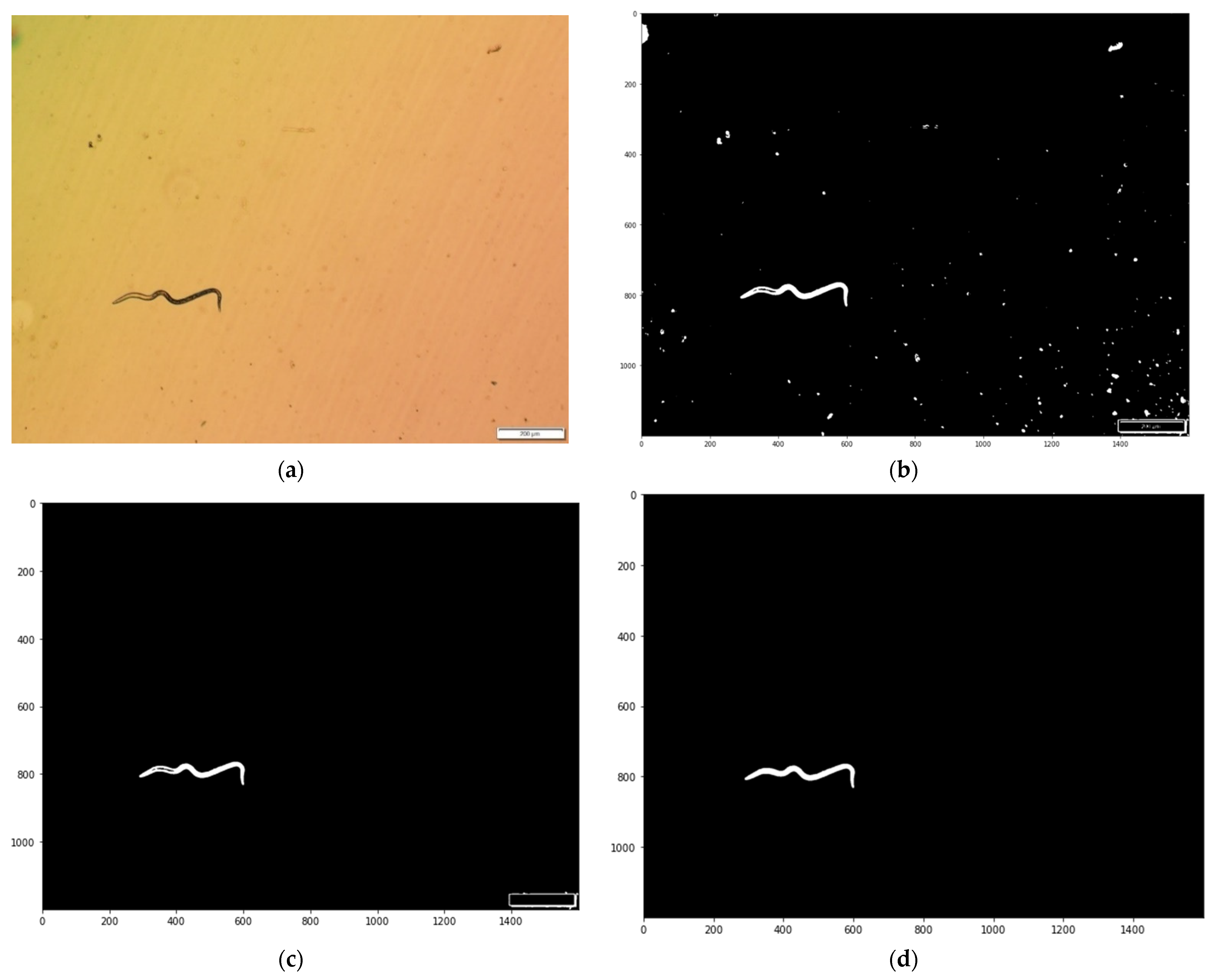
2.3.1. Image Pre-Processing
2.3.2. Image Purification
2.3.3. Morphometric Measurement
3. Evaluation Criteria
3.1. Measurement Evaluation
3.2. Coefficient of Determination (R2)
3.3. Root Mean Square Error (RMSE)
3.4. One-Way Analysis of Variance (ANOVA)
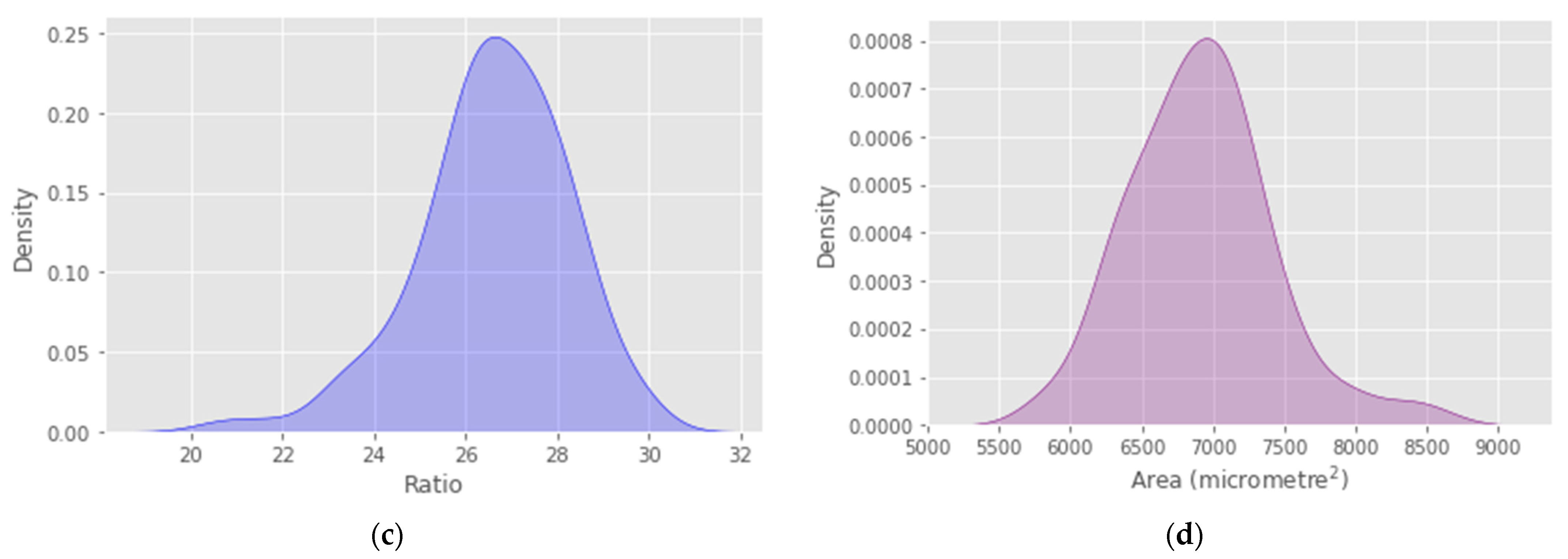
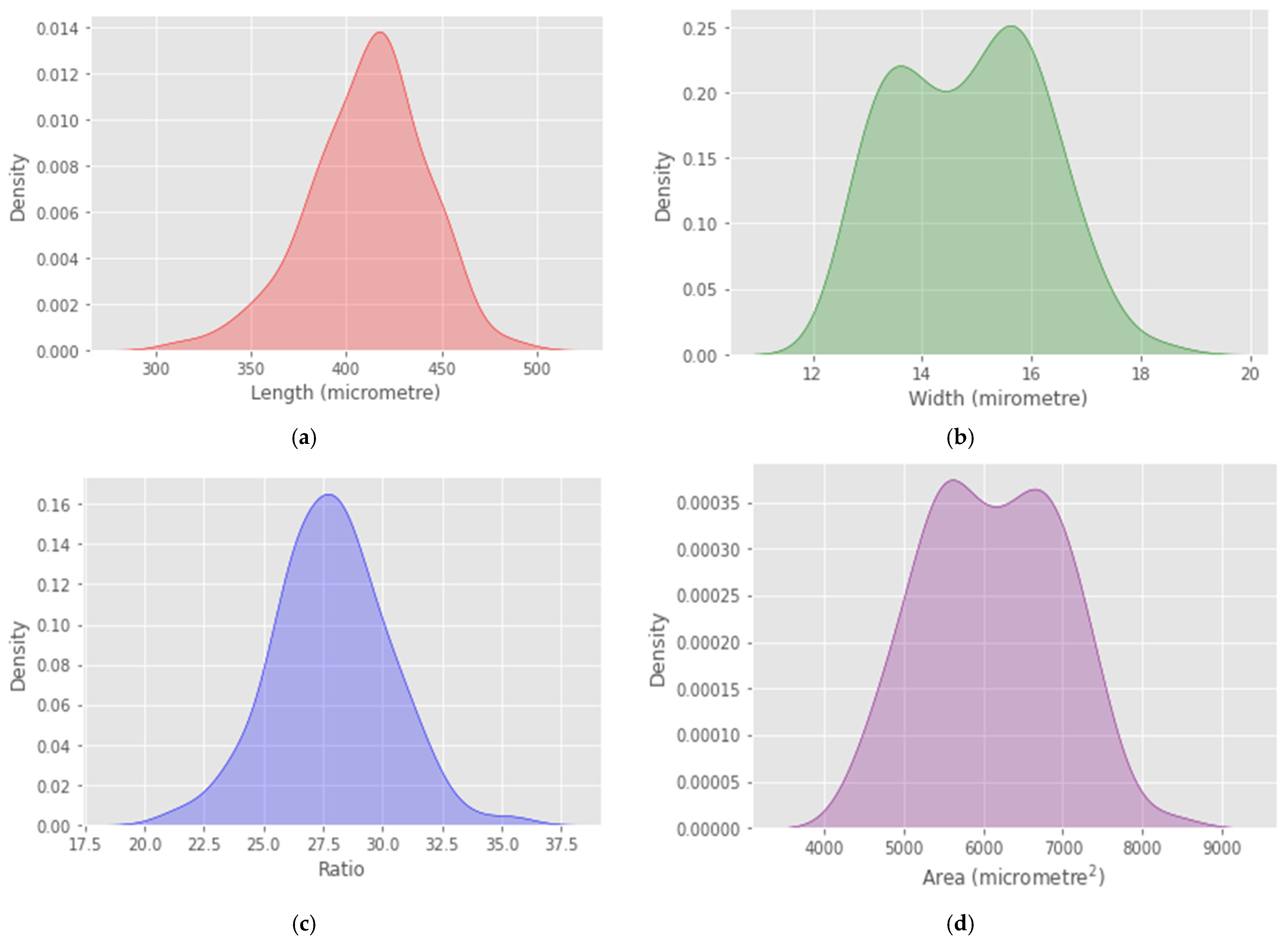
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Abbreviation | Meaning |
|---|---|
| ANOVA | Analysis of variance |
| CA | Contour arc |
| JIR | Juvenile from infected roots |
| JEM | Juvenile from egg mass |
| J1 | First-stage juvenile |
| J2 | Second-stage juvenile |
| R2 | Coefficient of determination |
| RGB | Red, green, and blue |
| RKN | Root-knot nematode |
| RMSE | Root mean square error |
| SCN | Soybean cyst nematode |
| SG | Skeleton graph |
| TS | Thin structure |
| µm | Micrometre |
| Algorithm |
|---|
| 1. Load image (Img (x, y)) 2. Apply Gaussian filter to remove noise, Gaussian (Img (x, y), σ = 1) 3. Convert RGB image to gray, f (x, y) = Gray (Img (x, y)) 4. Apply triangle thresholding (T) to gray image 6. Apply morphological closing operation to close boundary, morphology. Closing (Img (x, y), size = 5) 7. Fill holes in the image, binary_fill_holes Img (x, y)) 8. Find all contours(c) in the image Img (x, y)) 9. For each contour(c): if (Minimum Area < contourArea(c) < Maximum Area) a. Create mask of each contour b. Compute Width, Length, Ratio and Area c. if (Minimum Ratio < ratio < Maximum Ratio): • contourArea(c) = RKN • Save mask image and measurement d. else • contourArea(c) = Dust Particle or Rubbish 10. Save Image |
Thinning Algorithm
| P9 (i − 1, j − 1) | P2 (i − 1, j) | P3 (i − 1, j + 1) |
| P8 (i, j − 1) | P1 (i, j) | P4 (i, j + 1) |
| P7 (i + 1, j − 1) | P6 (i + 1, j) | P5 (i + 1, j + 1) |
- 2 ≤ B(P1) ≤ 6
- A(P1) = 1
- P2 × P4 × P6 = 0
- P4 × P6 × P8 = 0
- 2 ≤ B(P1) ≤ 6
- A(P1) = 1
- P2 × P4 × P8 = 0
- P2 × P6 × P8 = 0
References
- Department of Agriculture and Fisheries. Root-Knot Nematode. Available online: https://www.daf.qld.gov.au/business-priorities/agriculture/plants/fruit-vegetable/insect-pests/root-knot-nematode (accessed on 20 October 2021).
- Nicol, J.; Turner, S.; Coyne, D.; den Nijs, L.; Hockland, S.; Maafi, Z.T. Current nematode threats to world agriculture. In Genomics and Molecular Genetics of Plant-Nssematode Interactions; Jones, J., Gheysen, G., Fenoll, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 21–43. [Google Scholar]
- Elling, A.A. Major emerging problems with minor Meloidogyne species. Phytopathology 2013, 103, 1092–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Seid, A.; Fininsa, C.; Mekete, T.; Decraemer, W.; Wesemael, W.M.L. Tomato (Solanum lycopersicum) and root-knot nematodes (Meloidogyne spp.)—A century-old battle. Nematology 2015, 17, 995–1009. [Google Scholar] [CrossRef] [Green Version]
- Jagdale, G.; Arnold-Smith, L.C. Sampling for Plant-Parasitc Nematodes Identification and Diagnosis. Available online: https://athenaeum.libs.uga.edu/bitstream/handle/10724/34728/NemaSamplingGuideApr2011.pdf?sequence=1 (accessed on 7 April 2021).
- Vezina, A. Root-Knot Nematodes. Available online: https://www.promusa.org/Root-knot+nematodes (accessed on 28 August 2021).
- Hunt, D.J.; Handoo, Z.A. Root-Knot Nematodes; Perry, R.N., Moens, M., Starr, J.L., Eds.; CABI: Wallingford, UK, 2009. [Google Scholar]
- Moens, M.; Perry, R.N.; Starr, J.L. Meloidogyne species—A diverse group of novel and important plant parasites. In Root-Knot Nematodes; Perry, R.N., Moens, M., Starr, J.L., Eds.; CABI: Wallingford, UK, 2009; Volume 1, p. 483. [Google Scholar]
- Mitkowski, N.A.; Adawi, G.S. Root-knot Nematode. Plant Health Instr. 2003. [Google Scholar] [CrossRef]
- Eisenback, J.D.; Hunt, D.J. General Morphology In Root-Knot Nematodes; Starr, J.L., Moens, M., Perry, R.N., Eds.; CABI: Wallingford, UK, 2009. [Google Scholar]
- De Man, J. Die Einheimischen, Frei in der Reinen Erde und im Süssen Wasser Lebende Nematoden; Brill: Leiden, The Netherlands, 1880; Volume 5, pp. 1–104. [Google Scholar]
- De Man, J.G. Onderzoekingen over Vrij in de Aarde Levende Nematoden; Gravenhage: The Hague, The Netherlands, 1877. [Google Scholar]
- Chiu, M.T.; Xu, X.; Wei, Y.; Huang, Z.; Schwing, A.; Brunner, R.; Khachatrian, H.; Karapetyan, H.; Dozier, I.; Rose, G.; et al. Agriculture-Vision: A Large Aerial Image Database for Agricultural Pattern Analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 2828–2838. [Google Scholar]
- Gomes, J.F.S.; Leta, F.R. Applications of computer vision techniques in the agriculture and food industry: A review. Eur. Food Res. Technol. 2012, 235, 989–1000. [Google Scholar] [CrossRef]
- Tian, H.; Wang, T.; Liu, Y.; Qiao, X.; Li, Y. Computer vision technology in agricultural automation—A review. Inf. Process. Agric. 2020, 7, 1–19. [Google Scholar] [CrossRef]
- Mazurkiewicz, M.; Górska, B.; Jankowska, E.; Włodarska-Kowalczuk, M. Assessment of nematode biomass in marine sediments: A semi-automated image analysis method. Limnol. Oceanogr. Methods 2016, 14, 816–827. [Google Scholar] [CrossRef]
- Moore, B.T.; Jordan, J.M.; Baugh, L.R. WormSizer: High-throughput analysis of nematode size and shape. PLoS ONE 2013, 8, e57142. [Google Scholar] [CrossRef] [Green Version]
- Kurtulmuş, F.; Ulu, T.C. Detection of dead entomopathogenic nematodes in microscope images using computer vision. Biosyst. Eng. 2014, 118, 29–38. [Google Scholar] [CrossRef]
- Puckering, T.; Thompson, J.; Sathyamurthy, S.; Sukumar, S.; Shapira, T.; Ebert, P. Automated Wormscan. F1000Res 2017, 6, 192. [Google Scholar] [CrossRef]
- Wählby, C.; Kamentsky, L.; Liu, Z.H.; Riklin-Raviv, T.; Conery, A.L.; O’Rourke, E.J.; Sokolnicki, K.L.; Visvikis, O.; Ljosa, V.; Irazoqui, J.E.; et al. An image analysis toolbox for high-throughput C. elegans assays. Nat. Methods 2012, 9, 714–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.; Yeckel, G.; Heinz, R.; Clark, K.; Sleper, D.; Mitchum, M.G. A High-Throughput Automated Technique for Counting Females of Heterodera glycines using a Fluorescence-Based Imaging System. J. Nematol. 2010, 42, 201–206. [Google Scholar]
- Oscar, G.; Maria, F.A.; Moreno-Vazquez, S. Quantitative evaluation of Heterodera avenae females in soil and root extracts by digital image analysis. Crop. Prot. 2016, 81, 85–91. [Google Scholar] [CrossRef]
- Vagelas, I.; Pembroke, B.; Gowen, S.R. Techniques for image analysis of movement of juveniles of root-knot nematodes encumbered with Pasteuria penetransspores. Biocontrol. Sci. Technol. 2011, 21, 239–250. [Google Scholar] [CrossRef]
- Qazi, F.; Khalid, A.; Poddar, A.; Tetienne, J.-P.; Nadarajah, A.; Aburto-Medina, A.; Shahsavari, E.; Shukla, R.; Prawer, S.; Ball, A.S. Real-time detection and identification of nematode eggs genus and species through optical imaging. Sci. Rep. 2020, 10, 7219. [Google Scholar] [CrossRef]
- Akintayo, A.; Tylka, G.L.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. A deep learning framework to discern and count microscopic nematode eggs. Sci. Rep. 2018, 8, 9145. [Google Scholar] [CrossRef] [PubMed]
- Rauf, H.T.; Saleem, B.A.; Lali, M.I.U.; Khan, M.A.; Sharif, M.; Bukhari, S.A.C. A citrus fruits and leaves dataset for detection and classification of citrus diseases through machine learning. Data Brief 2019, 26, 104340. [Google Scholar] [CrossRef] [PubMed]
- Almadhor, A.; Rauf, H.T.; Lali, M.I.U.; Damaševičius, R.; Alouffi, B.; Alharbi, A. AI-Driven Framework for Recognition of Guava Plant Diseases through Machine Learning from DSLR Camera Sensor Based High Resolution Imagery. Sensors 2021, 21, 3830. [Google Scholar] [CrossRef] [PubMed]
- Khan, W. Image Segmentation Techniques: A Survey. J. Image Graph. 2014, 166–170. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, P.K.; Soltani, S.; Wong, A.K.C. A survey of thresholding techniques. Comput. Vis. Graph. Image Process. 1988, 41, 233–260. [Google Scholar] [CrossRef]
- Netto, A.F.A.A.; Martins, R.N.; de Souza, G.S.A.; Araújo, G.D.M.; de Almeida, S.L.H.; Capelini, V.A. Segmentation of Rgb Images Using Different Vegetation Indices and Thresholding Methods. Nativa 2018, 6, 389–394. [Google Scholar] [CrossRef]
- Hakim, A.; Mor, Y.; Toker, I.A.; Levine, A.; Neuhof, M.; Markovitz, Y.; Rechavi, O. WorMachine: Machine learning-based phenotypic analysis tool for worms. BMC Biol. 2018, 16, 8. [Google Scholar] [CrossRef] [Green Version]
- Holladay, B.H.; Willett, D.S.; Stelinski, L.L. High throughput nematode counting with automated image processing. BioControl 2015, 61, 177–183. [Google Scholar] [CrossRef]
- Tajima, R.; Kato, Y. Comparison of threshold algorithms for automatic image processing of rice roots using freeware ImageJ. Field Crop Res. 2011, 121, 460–463. [Google Scholar] [CrossRef]
- Patil, S.B.; Bodhe, S.K. Leaf disease severity measurement using image processing. Int. J. Eng. Technol. 2011, 3, 297–301. [Google Scholar]
- Hussey, R. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis. Rep. 1973, 57, 1025–1028. [Google Scholar]
- Van Bezooijen, J. Methods and Techniques for Nematology; Wageningen University: Wageningen, The Netherlands, 2006. [Google Scholar]
- Fu, D.; Oh, S.; Choi, W.; Yamauchi, T.; Dorn, A.; Yaqoob, Z.; Dasari, R.R.; Feld, M.S. Quantitative DIC microscopy using an off-axis self-interference approach. Opt. Lett. 2010, 35, 2370–2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zheng, S.; Li, X.; Qin, X. A new image denoising method based on Gaussian filter. In Proceedings of the 2014 International Conference on Information Science, Electronics and Electrical Engineering, Sapporo, Japan, 26–28 April 2014; pp. 163–167. [Google Scholar]
- Deng, G.; Cahill, L. An adaptive Gaussian filter for noise reduction and edge detection. In Proceedings of the 1993 IEEE Conference Record Nuclear Science Symposium and Medical Imaging Conference, San Francisco, CA, USA, 30 October–6 November 1993; pp. 1615–1619. [Google Scholar]
- Kanan, C.; Cottrell, G.W. Color-to-grayscale: Does the method matter in image recognition? PLoS ONE 2012, 7, e29740. [Google Scholar]
- Zack, G.W.; Rogers, W.E.; Latt, S.A. Automatic measurement of sister chromatid exchange frequency. J. Histochem. Cytochem. 1977, 25, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Al-Amri, S.S.; Kalyankar, N.V. Image segmentation by using threshold techniques. arXiv 2010, arXiv:1005.4020. [Google Scholar]
- Maragos, P.; Schafer, R. Morphological filters—Part I: Their set-theoretic analysis and relations to linear shift-invariant filters. IEEE Trans. Acoust. Speech Signal Process. 1987, 35, 1153–1169. [Google Scholar] [CrossRef]
- Maragos, P. Morphological filtering for image enhancement and feature detection. In The Image and Video Processing Handbook; Academic Press: Cambridge, MA, USA, 2005; pp. 135–156. [Google Scholar]
- Zhang, T.Y.; Suen, C.Y. A fast parallel algorithm for thinning digital patterns. Commun. ACM 1984, 27, 236–239. [Google Scholar] [CrossRef]
- Choi, H.I.; Choi, S.W.; Moon, H.P. Mathematical theory of medial axis transform. Pac. J. Math. 1997, 181, 57–88. [Google Scholar] [CrossRef]
- Olympus. cellSens Life Science Imaging Software. Available online: https://investigacion.us.es/docs/web/files/manual_cellsens_en.pdf (accessed on 6 November 2020).
- Harrison, J.; Norton, A. The Gauss-Green theorem for fractal boundaries. Duke Math. J. 1992, 67, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Lund, S.P.; Vyzasatya, R.; Vanguri, P.; Elliott, J.T.; Plant, A.L.; Lin-Gibson, S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017, 19, 1509–1521. [Google Scholar] [CrossRef] [PubMed]
- Reisinger, H. The impact of research designs on R2 in linear regression models: An exploratory meta-analysis. J. Empir. Gen. Mark. Sci. 1997, 2, 1–12. [Google Scholar]
- Chicco, D.; Warrens, M.J.; Jurman, G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef]
- Si-Mohamed, S.; Bar-Ness, D.; Sigovan, M.; Tatard-Leitman, V.; Cormode, D.P.; Naha, P.C.; Coulon, P.; Rascle, L.; Roessl, E.; Rokni, M. Multicolour imaging with spectral photon-counting CT: A phantom study. Eur. Radiol. Exp. 2018, 2, 34. [Google Scholar] [CrossRef]
- Chai, T.; Draxler, R.R. Root mean square error (RMSE) or mean absolute error (MAE)?—Arguments against avoiding RMSE in the literature. Geosci. Model Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef] [Green Version]
- Neill, S.P.; Hashemi, M.R. Ocean Modelling for Resource Characterization. In Fundamentals of Ocean Renewable Energy; Neill, S.P., Hashemi, M.R., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 193–235. [Google Scholar]
- Kim, T.K. Understanding one-way ANOVA using conceptual figures. Korean J. Anesthesiol. 2017, 70, 22. [Google Scholar] [CrossRef] [Green Version]
- Faraway, J.J. Practical Regression and ANOVA Using R; University of Bath: Bath, UK, 2002; Volume 168. [Google Scholar]
- Saha, P.K.; Borgefors, G.; di Baja, G.S. A survey on skeletonization algorithms and their applications. Pattern Recognit. Lett. 2016, 76, 3–12. [Google Scholar] [CrossRef]
- Hsu, Y.-Y.; Schuff, N.; Du, A.-T.; Mark, K.; Zhu, X.; Hardin, D.; Weiner, M.W. Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. J. Magn. Reson. Imaging 2002, 16, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.K.; Udupa, J.K. Optimum image thresholding via class uncertainty and region homogeneity. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 689–706. [Google Scholar] [CrossRef]
- Nunez-Iglesias, J.; Blanch, A.J.; Looker, O.; Dixon, M.W.; Tilley, L. A new Python library to analyse skeleton images confirms malaria parasite remodelling of the red blood cell membrane skeleton. PeerJ 2018, 6, e4312. [Google Scholar] [CrossRef] [Green Version]
- Curtis, R.H.; Robinson, A.F.; Perry, R.N. Hatch and host location. In Root-Knot Nematodes; Perry, R.N., Moens, M., Starr, J.L., Eds.; CABI: Wallingford, UK, 2009; Volume 1, pp. 139–162. [Google Scholar]
- Caillaud, M.-C.; Dubreuil, G.; Quentin, M.; Perfus-Barbeoch, L.; Lecomte, P.; de Almeida Engler, J.; Abad, P.; Rosso, M.-N.; Favery, B. Root-knot nematodes manipulate plant cell functions during a compatible interaction. J. Plant Physiol. 2008, 165, 104–113. [Google Scholar] [CrossRef]




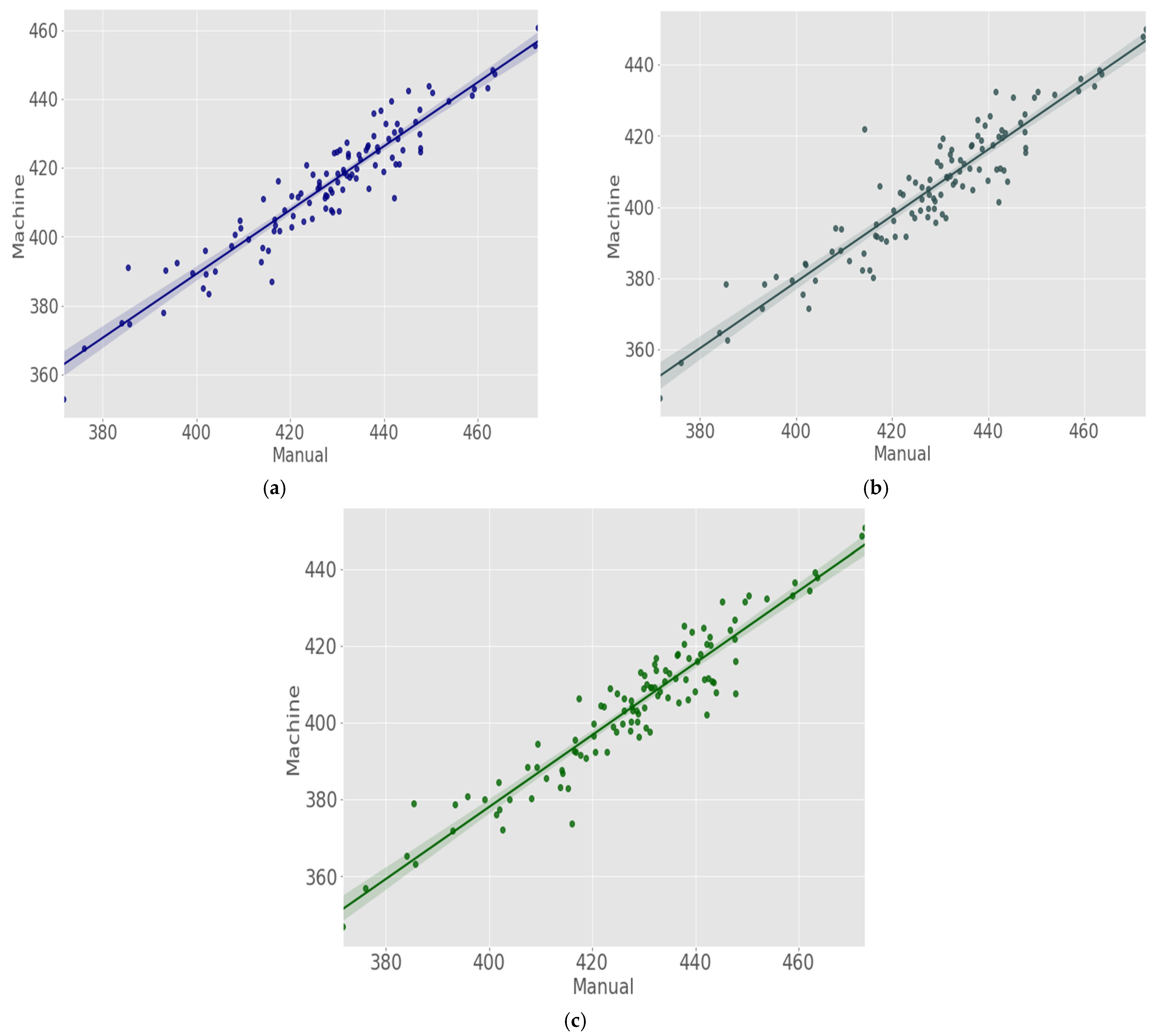
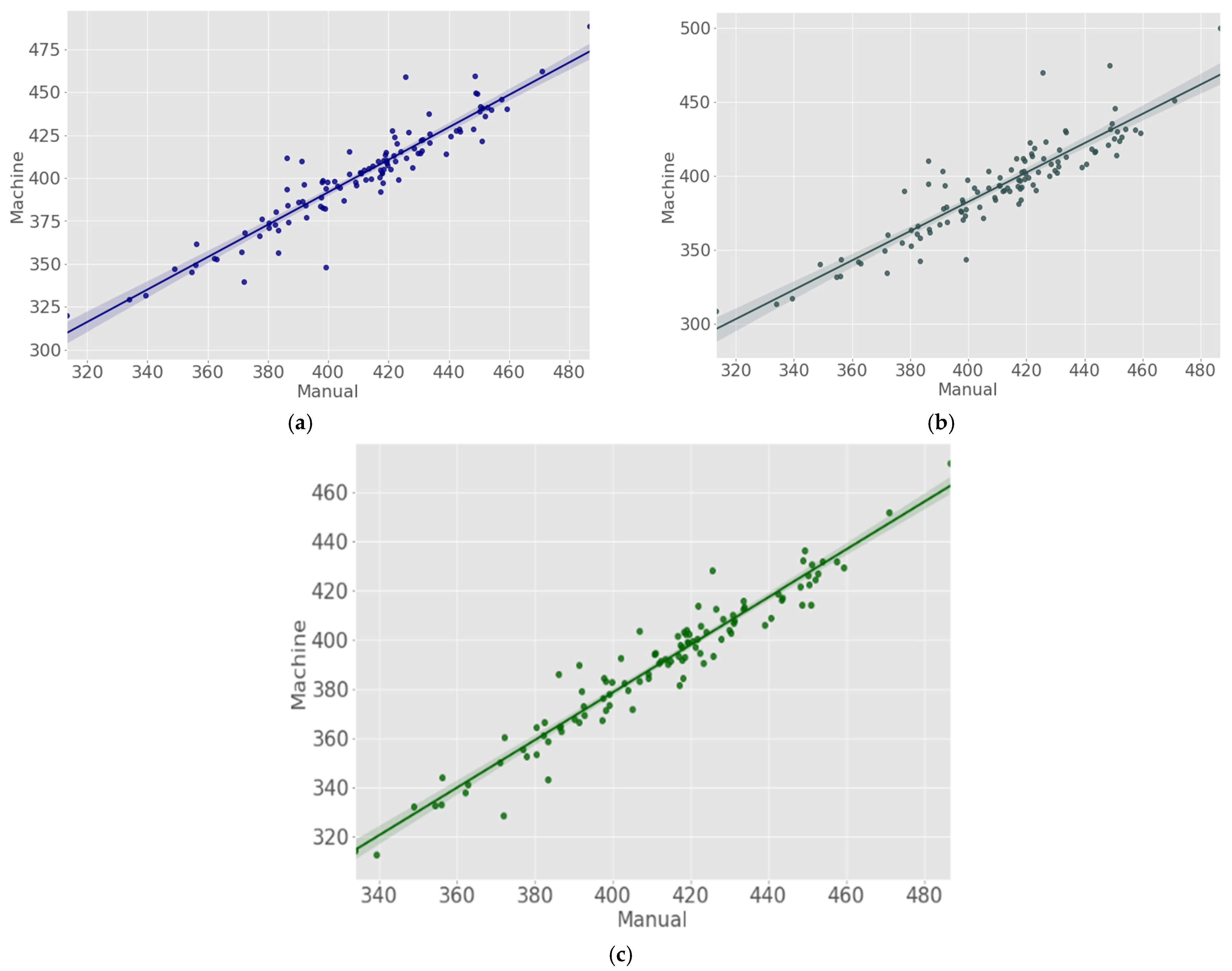
| Authors | Nematode Type | Imaging Technology | Magnification | Image Size | Image Analysis Method | Performance Metrics |
|---|---|---|---|---|---|---|
| [33] | Entomopathogenic nematodes: Steinernema diaprepesi and Heterorhabditis indica | Dino-Lite Edge AM4815ZT digital microscope and Leica M165C microscope | 30×, 20× | 2560 × 2048 | ImageJ | CV, CV(RMSE) |
| [23] | Cereal cyst nematode: Heterodera avenae | HP Scanjet 2400 | 4800 × 4800 pixel, 800 dpi | Software KS-400 V.3.0 with LDA and NBC | Accuracy, variance, correlation | |
| [19] | Caenorhabditis elegans, Heterorhabditis bacteriophora | Leica S8 Apo stereomicroscope | 2.6× | 2048 × 1536 | Python: Scipy, NumPy, Scikit-image | SEM, R2 |
| [22] | Soybean cyst nematode: Heterodera glycines | Kodak Image Station 4000 MM Pro | 15 × 15 | Fluorescence based imaging system | R2 | |
| [17] | Sea nematodes | Leica DFC450 Leica M205C Micro- scope | 10× | 2560 × 1920 | Leica Application Suite | PERNOVA test |
| [24] | Root-knot nematodes | Inverted Microscope with digital camera | 200× | 320 × 240 | Image J, GenStat | R2 |
| [32] | C. elegans | Olympus IX83 microscope | 4× | 64 × 128 | Machine Learning | Accuracy |
| [18] | C. elegans | V20 M2Bio Stereomicroscope | 10× | ImageJ | Coefficient of variation | |
| [20] | C. elegans | Epson v700 (or v800) photo scanner | 2400 dpi | Fiji | p-value | |
| [21] | C. elegans | Discovery-1 microscope, Axioscope (Zeiss) | 2×, 2.5× | 696 × 520 pixels | Cell Profiler | Accuracy, Precision |
| Method | Sample | R2 | RMSE |
|---|---|---|---|
| CA | JIR | 0.898 | 14.237 |
| JEM | 0.881 | 13.603 | |
| TS | JIR | 0.875 | 23.975 |
| JEM | 0.823 | 22.426 | |
| SG | JIR | 0.898 | 24.501 |
| JEM | 0.924 | 23.832 |
| Ratio | R2 | RMSE |
|---|---|---|
| CA | 0.220 | 2.137 |
| TS | 0.227 | 2.528 |
| SG | 0.206 | 2.590 |
| Ratio | Method | R2 | RMSE | Misidentified | Undetected |
|---|---|---|---|---|---|
| 19–30 | CA | 0.857 | 0.481 | 3 | 97 |
| TS | 0.835 | 0.520 | 2 | 110 | |
| SG | 0.828 | 0.533 | 3 | 114 | |
| 15–35 | CA | 0.961 | 0.232 | 6 | 20 |
| TS | 0.959 | 0.236 | 8 | 17 | |
| SG | 0.961 | 0.232 | 6 | 20 | |
| 12–35 | CA | 0.966 | 0.215 | 11 | 9 |
| TS | 0.961 | 0.232 | 6 | 20 | |
| SG | 0.967 | 0.210 | 8 | 13 | |
| 10–35 | CA | 0.965 | 0.219 | 13 | 8 |
| TS | 0.958 | 0.240 | 16 | 8 | |
| SG | 0.973 | 0.191 | 7 | 10 | |
| 8–35 | CA | 0.947 | 0.271 | 22 | 8 |
| TS | 0.945 | 0.278 | 23 | 7 | |
| SG | 0.969 | 0.206 | 11 | 9 | |
| 6–35 | CA | 0.927 | 0.326 | 33 | 6 |
| TS | 0.919 | 0.346 | 39 | 7 | |
| SG | 0.961 | 0.232 | 17 | 9 | |
| 4–35 | CA | 0.872 | 0.452 | 68 | 6 |
| TS | 0.881 | 0.435 | 67 | 5 | |
| SG | 0.949 | 0.267 | 27 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pun, T.B.; Neupane, A.; Koech, R. Quantification of Root-Knot Nematode Infestation in Tomato Using Digital Image Analysis. Agronomy 2021, 11, 2372. https://doi.org/10.3390/agronomy11122372
Pun TB, Neupane A, Koech R. Quantification of Root-Knot Nematode Infestation in Tomato Using Digital Image Analysis. Agronomy. 2021; 11(12):2372. https://doi.org/10.3390/agronomy11122372
Chicago/Turabian StylePun, Top Bahadur, Arjun Neupane, and Richard Koech. 2021. "Quantification of Root-Knot Nematode Infestation in Tomato Using Digital Image Analysis" Agronomy 11, no. 12: 2372. https://doi.org/10.3390/agronomy11122372
APA StylePun, T. B., Neupane, A., & Koech, R. (2021). Quantification of Root-Knot Nematode Infestation in Tomato Using Digital Image Analysis. Agronomy, 11(12), 2372. https://doi.org/10.3390/agronomy11122372







