Efficient Biocontrol of Gaeumannomyces graminis var. Tritici in Wheat: Using Bacteria Isolated from Suppressive Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Traits of Selected Bacteria
2.1.1. Plant Growth Promoting Traits
2.1.2. Determination of Phenazine (Phz) Presence in Bacterial Strains
2.1.3. Chitinase Activity
2.2. Inoculum Preparation
Bacterial Inoculation under Greenhouse Conditions
2.3. Greenhouse Experiment
2.4. Gaeumannomyces graminis var. Tritici Detection and Quantification in Wheat Roots
2.5. Lipid Peroxidation
2.6. Superoxide Dismutase (SOD) Activity
2.7. Field Assay
2.7.1. Bacteria Inoculation under Field Conditions
2.7.2. Gaeumannomyces graminis Inoculum (Powder Inoculum)
2.7.3. Treatments and Plant Samples Collection and Analysis
2.8. Statistical Analysis
3. Results
3.1. Microbial Traits
3.2. Greenhouse Experiment
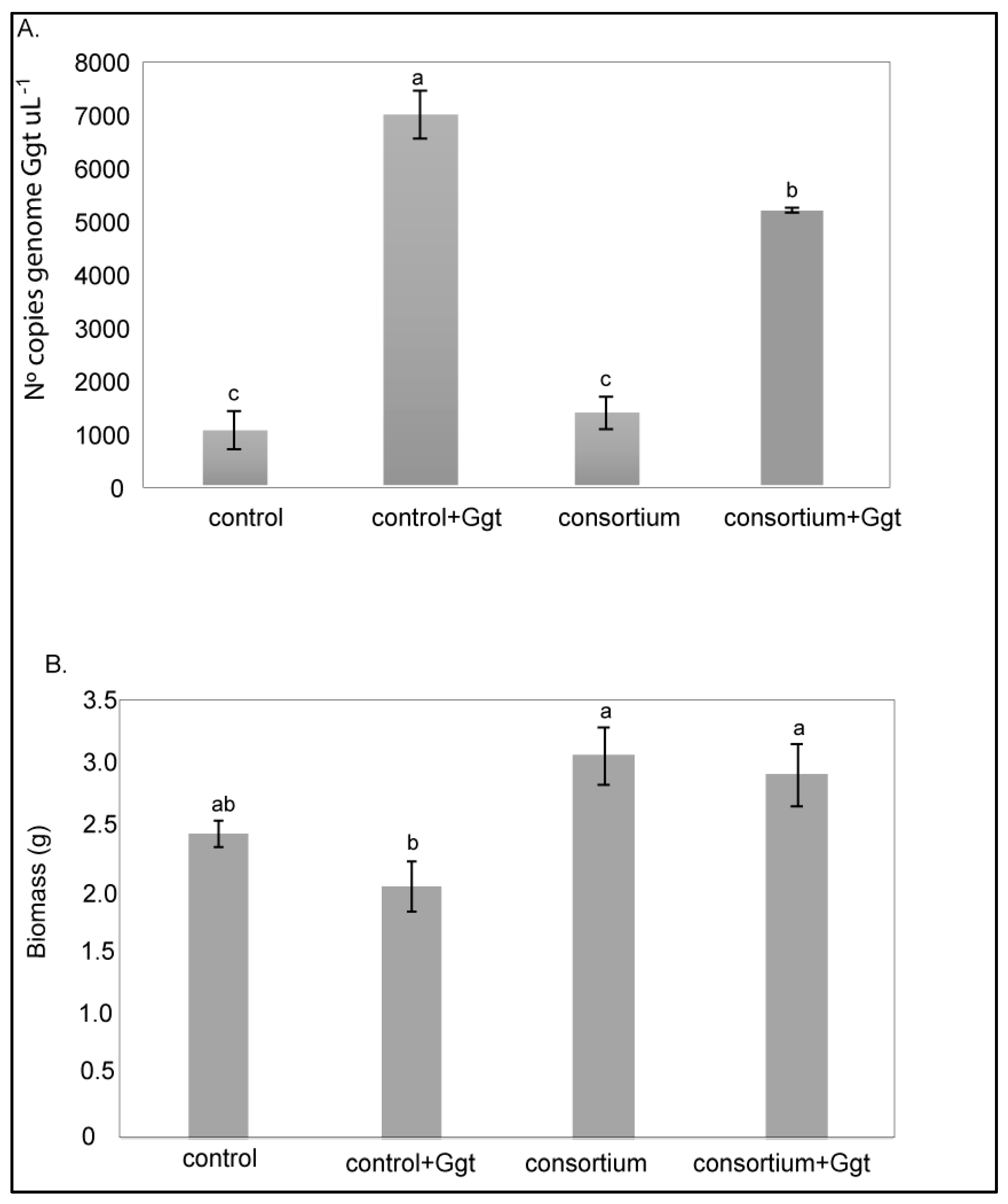
3.2.1. Gaeumannomyces graminis var. Tritici Detection and Quantification in Wheat Roots
3.2.2. Lipid Peroxidation
3.2.3. Superoxide Dismutase (SOD) Activity
3.3. Field Assay
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, P.; Benavente, E.; Manzano-Agugliaro, F.; Gimenez, E. Worldwide research trends on wheat and barley: A bibliometric comparative analysis. Agronomy 2019, 9, 352. [Google Scholar] [CrossRef]
- Duran, P.; Mora, M.D.L.L. Plant–Soil–Microorganism Interaction Involved in Natural Suppression of Take-All Disease; Kaushal, M., Prasad, R., Eds.; Springer Nature: Singapore, 2021; ISBN 9781461479789. [Google Scholar]
- Kwak, Y.S.; Weller, D.M. Take-all of wheat and natural disease suppression: A review. Plant Pathol. J. 2013, 29, 125–135. [Google Scholar] [CrossRef]
- Freeman, J.; Ward, E. Gaeumannomyces graminis, the take-all fungus and its relatives. Mol. Plant Pathol. 2004, 5, 235–252. [Google Scholar] [CrossRef] [PubMed]
- Durán, P.; Jorquera, M.; Viscardi, S.; Carrion, V.J. Screening and Characterization of Potentially Suppressive Soils against Gaeumannomyces graminis under Extensive Wheat Cropping by Chilean Indigenous Communities. Front. Microbiol. 2017, 8, 1552. [Google Scholar] [CrossRef] [PubMed]
- Duran, P.; Tortella, G.; Viscardi, S.; Barra, P.J.; Carrion, V.J.; Mora, M.D.L.L.; Pozo, M.J. Microbial Community Composition in Take-All Suppressive Soils. Front. Microbiol. 2018, 9, 2198. [Google Scholar] [CrossRef]
- Wang, G.H.; Berdy, B.M.; Velasquez, O.; Jovanovic, N.; Alkhalifa, S.; Minbiole, K.P.C.; Brucker, R.M. Changes in Microbiome Confer Multigenerational Host Resistance after Sub-toxic Pesticide Exposure. Cell Host Microbe 2020, 27, 213–224.e7. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.J. Take-all of wheat. Physiol. Mol. Plant Pathol. 2003, 62, 73–86. [Google Scholar] [CrossRef]
- Cunfer, B.M.; Buntin, G.D.; Phillips, D.V. Effect of crop rotation on take-all of wheat in double-cropping systems. Plant Dis. 2006, 90, 1161–1166. [Google Scholar] [CrossRef]
- Campillo, R.; Andrade, O.; Contreras, E. Variaciones del contenido de Mn de dos suelos sometidos a esterilización y su efecto sobre la pudrición radical del trigo o “mal de pie”. Agric. Técnica 2001, 61, 339–351. [Google Scholar] [CrossRef]
- González-Castillo, J.A.; Quezada-D’angelo, T.P.; Silva-Aguayo, G.I.; Moya-Elizondo, E.A. Effect of saponins of Quillaja saponaria extracts in combination with Pseudomonas protegens to control Gaeumannomyces graminis var. tritici in wheat. Chil. J. Agric. Res. 2018, 78, 378–390. [Google Scholar] [CrossRef]
- Paz, C.; Viscardi, S.; Iturra, A.; Marin, V.; Miranda, F.; Barra, P.J.; Mendez, I.; Duran, P. Antifungal effects of drimane sesquiterpenoids isolated from Drimys winteri against Gaeumannomyces graminis var. tritici. Appl. Environ. Microbiol. 2020, 86, e01834-20. [Google Scholar] [CrossRef]
- Yuan, J.; Zhao, J.; Wen, T.; Zhao, M.; Li, R.; Goossens, P.; Huang, Q.; Bai, Y.; Vivanco, J.M.; Kowalchuk, G.A.; et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 2018, 6, 156. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Ranamukhaarachchi, S.L. Soil-borne antagonists for biological control of bacterial wilt disease caused by Ralstonia solanacearum in tomato and pepper. J. Plant Pathol. 2010, 721, 395–406. [Google Scholar]
- Barbey, C.; Crépin, A.; Bergeau, D.; Ouchiha, A.; Mijouin, L.; Taupin, L.; Orange, N.; Feuilloley, M.; Dufour, A.; Burini, J.F.; et al. In Planta Biocontrol of Pectobacterium atrosepticum by Rhodococcus erythropolis Involves Silencing of Pathogen Communication by the Rhodococcal Gamma-Lactone Catabolic Pathway. PLoS ONE 2013, 8, e66642. [Google Scholar] [CrossRef]
- Konappa, N.M.; Maria, M.; Uzma, F.; Krishnamurthy, S.; Nayaka, S.C.; Niranjana, S.R.; Chowdappa, S. Lactic acid bacteria mediated induction of defense enzymes to enhance the resistance in tomato against Ralstonia solanacearum causing bacterial wilt. Sci. Hortic. 2016, 207, 183–192. [Google Scholar] [CrossRef]
- Schreiter, S.; Babin, D.; Smalla, K.; Grosch, R. Rhizosphere competence and biocontrol effect of pseudomonas sp. RU47 independent from plant species and soil type at the field scale. Front. Microbiol. 2018, 9, 97. [Google Scholar] [CrossRef]
- Bolívar-Anillo, H.J.; Garrido, C.; Collado, I.G. Endophytic microorganisms for biocontrol of the phytopathogenic fungus Botrytis cinerea. Phytochem. Rev. 2020, 19, 721–740. [Google Scholar] [CrossRef]
- Durán, P.; Acuña, J.J.; Jorquera, M.; Azcón, R.; Paredes, C.; Rengel, Z.; de la Luz Mora, M. Endophytic bacteria from selenium-supplemented wheat plants could be useful for plant-growth promotion, biofortification and Gaeumannomyces graminis biocontrol in wheat production. Biol. Fertil. Soils 2014, 50, 983–990. [Google Scholar] [CrossRef]
- Vitti, A.; Pellegrini, E.; Nali, C.; Lovelli, S.; Sofo, A.; Valerio, M.; Scopa, A.; Nuzzaci, M. Trichoderma harzianum T-22 Induces Systemic Resistance in Tomato Infected by Cucumber mosaic virus. Front. Plant Sci. 2016, 7, 1520. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.K.; Sridhar, J.; Murali-Baskaran, R.K.; Senthil-Nathan, S.; Kaushal, P.; Dara, S.K.; Arthurs, S. Microbial biopesticides for insect pest management in India: Current status and future prospects. J. Invertebr. Pathol. 2019, 165, 74–81. [Google Scholar] [CrossRef]
- Thangavel, P.; Sridevi, G. Environmental Sustainability: Role of Green Technologies; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–324. [Google Scholar] [CrossRef]
- Revathi, K.; Chandrasekaran, R.; Thanigaivel, A.; Arunachalam Kirubakaran, S.; Senthil-Nathan, S. Biocontrol efficacy of protoplast fusants between Bacillus thuringiensis and Bacillus subtilis against Spodoptera litura Fabr. Arch. Phytopathol. Plant Prot. 2014, 47, 1365–1375. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Shi, X.-C.; Chen, X.; Laborda, P.; Zhao, Y.-Y.; Liu, F.-Q.; Laborda, P. Biocontrol ability of phenazine-producing strains for the management of fungal plant pathogens: A review. Biol. Control 2021, 155, 104548. [Google Scholar] [CrossRef]
- Wall, D.H.; Bardgett, R.D.; Behan-Pelletier, V.; Herrick, J.E.; Jones, T.H.; Ritz, K.; Six, J.; Strong, D.R.; van der Putten, W.H. Soil Ecology and Ecosystem Services, 1st ed.; Wall, D., Ed.; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease Suppressive Soils: New Insights from the Soil Microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef]
- Sheth, R.U.; Cabral, V.; Chen, S.P.; Wang, H.H. Manipulating Bacterial Communities by in situ Microbiome Engineering. Trends Genet. 2016, 32, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef] [PubMed]
- Toharisman, A.; Suhartono, M.T.; Spindler-Barth, M.; Hwang, J.-K.; Pyun, Y.-R. Purification and characterization of a thermostable chitinase from Bacillus licheniformis Mb-2. World J. Microbiol. Biotechnol. 2005, 21, 733–738. [Google Scholar] [CrossRef]
- Kuddus, M.; Ahmad, I.Z. Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J. Genet. Eng. Biotechnol. 2013, 11, 39–46. [Google Scholar] [CrossRef]
- Taylor, G.; Foyd, C. Mechanisms of Aluminum Tolerance In Triticum aestivum L. (Wheat) Iii. Long-Term Ph Changes Induced In Nutrient Solutions By Winter Cultivars Differing In Tolerance To Aluminum. J. Plant Nutr. 1985, 8, 613–628. [Google Scholar] [CrossRef]
- Bithell, S.L.; McKay, A.; Butler, R.C.; Herdina; Ophel-Keller, K.; Hartley, D.; Cromey, M.G. Predicting take-all severity in second-year wheat using soil DNA concentrations of Gaeumannomyces graminis var. tritici determined with qPCR. Plant Dis. 2012, 96, 443–451. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Durán, P.; Viscardi, S.; Acuña, J.J.; Cornejo, P.; Azcón, R.; Mora, M.D.L.L. Endophytic selenobacteria and arbuscular mycorrhizal fungus for Selenium biofortification and Gaeumannomyces graminis biocontrol. J. Soil Sci. Plant Nutr. 2018, 18, 1021–1035. [Google Scholar] [CrossRef]
- Barra, P.J.; Inostroza, N.G.; Mora, M.L.; Crowley, D.E.; Jorquera, M.A. Bacterial consortia inoculation mitigates the water shortage and salt stress in an avocado (Persea americana Mill.) nursery. Appl. Soil Ecol. 2016, 111, 39–47. [Google Scholar] [CrossRef]
- Mora, L.; Demanet, R.; Acuña, J.J.; Viscardi, S.; Jorquera, M.; Rengel, Z.; Durán, P. Aluminum-tolerant bacteria improve the plant growth and phosphorus content in ryegrass grown in a volcanic soil amended with cattle dung manure. Appl. Soil Ecol. 2017, 115, 19–26. [Google Scholar] [CrossRef]
- Shen, Z.; Ruan, Y.; Chao, X.; Zhang, J.; Li, R.; Shen, Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol. Fertil. Soils 2015, 51, 553–562. [Google Scholar] [CrossRef]
- Garbeva, P.; Postma, J.; Van Veen, J.A.; Van Elsas, J.D. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ. Microbiol. 2006, 8, 233–246. [Google Scholar] [CrossRef]
- Yang, L.; Han, X.; Zhang, F.; Goodwin, P.H.; Yang, Y.; Li, J.; Xia, M.; Sun, R.; Jia, B.; Zhang, J.; et al. Screening Bacillus species as biological control agents of Gaeumannomyces graminis var. Tritici on wheat. Biol. Control 2018, 118, 1–9. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ.-Sci. 2013, 26, 1–20. [Google Scholar] [CrossRef]
- Ji, W.; Leng, X.; Jin, Z.; Li, H. Plant growth promoting bacteria increases biomass, effective constituent, and modifies rhizosphere bacterial communities of Panax ginseng. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2019, 69, 135–146. [Google Scholar] [CrossRef]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Win, T.T.; Bo, B.; Malec, P.; Fu, P. The effect of a consortium of Penicillium sp. and Bacillus spp. in suppressing banana fungal diseases caused by Fusarium sp. and Alternaria sp. J. Appl. Microbiol. 2021, 131, 1890–1908. [Google Scholar] [CrossRef]
- Andrade, O.; Campillo, R.; Peyrelongue, A. Soils suppressive against Gaeumannomyces graminis var. tritici identified under wheat crop monoculture in southern Chile. Cienc. Investig. Agrar. 2011, 38, 345–356. [Google Scholar] [CrossRef]
- Madariaga, B.; Vera, C. Fitopatología-Enfermedades en Cultivos: Mal del Pie; Ficha Técnica INIA: Santiago, Chile, 2017; pp. 1–2. [Google Scholar]
- Fones, H.; Preston, G.M. Reactive oxygen and oxidative stress tolerance in plant pathogenic Pseudomonas. FEMS Microbiol. Lett. 2012, 327, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Soydam Aydin, S.; Büyük, I.; Aras, S. Relationships among lipid peroxidation, SOD enzyme activity, and SOD gene expression profile in Lycopersicum esculentum L. exposed to cold stress. Genet. Mol. Res. 2013, 12, 3220–3229. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Vlami, M.; Souza, J.T. De Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 2002, 81, 537. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Blankenfeldt, W.; Thomashow, L.S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 2006, 44, 417–445. [Google Scholar] [CrossRef]
- Gupta, P.; Ravi, I.; Sharma, V. Induction of β-1,3-glucanase and chitinase activity in the defense response of Eruca sativa plants against the fungal pathogen Alternaria brassicicola. J. Plant Interact. 2013, 8, 155–161. [Google Scholar] [CrossRef]
- Loc, N.H.; Huy, N.D.; Quang, H.T.; Lan, T.T.; Thu Ha, T.T. Characterisation and antifungal activity of extracellular chitinase from a biocontrol fungus, Trichoderma asperellum PQ34. Mycology 2020, 11, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Dukare, A.; Paul, S.; Arambam, A. Isolation and efficacy of native chitinolytic rhizobacteria for biocontrol activities against Fusarium wilt and plant growth promotion in pigeon pea (Cajanus cajan L.). Egypt. J. Biol. Pest Control 2020, 30, 56. [Google Scholar] [CrossRef]
- Barra, P.J.; Pontigo, S.; Delgado, M.; Parra–Almuna, L.; Duran, P.; Valentine, A.J.; Jorquera, M.A.; Mora, M.D.L.L. Phosphobacteria inoculation enhances the benefit of P–fertilization on Lolium perenne in soils contrasting in P–availability. Soil Biol. Biochem. 2019, 136, 107516. [Google Scholar] [CrossRef]
- Niu, B.; Wang, W.; Yuan, Z.; Sederoff, R.R.; Sederoff, H.; Chiang, V.L.; Borriss, R. Microbial Interactions Within Multiple-Strain Biological Control Agents Impact Soil-Borne Plant Disease. Front. Microbiol. 2020, 11, 585404. [Google Scholar] [CrossRef]




| Variable | Values |
|---|---|
| N (mg kg−1) | 29 |
| P (mg kg−1) | 27 |
| K (mg kg−1) | 344 |
| pH (water 1:5, w:v) | 5.75 |
| Organic matter (%) | 17 |
| K (cmol+ kg−1) | 0.88 |
| Na (cmol+ kg−1) | 0.15 |
| Ca (cmol+ kg−1) | 7.61 |
| Mg (cmol+ kg−1) | 1.77 |
| Al (cmol+ kg−1) | 0.05 |
| Al sat (%) | 0.48 |
| CICE (cmol+ kg−1) | 10.46 |
| Bas. sat (cmol+ kg−1) | 10.41 |
| Strain | PS * | Siderophores | Ggt Inhibition (%) | Phenazine | Chitinase ** (μmol mg protein−1) |
|---|---|---|---|---|---|
| Bacillus sp. | +++ | + | 40 | + | 2.1 bc ± 0.43 |
| Serratia sp. | + | + | 20 | + | 1.9 bc ± 0.35 |
| Acinetobacter sp. | - | - | 100 | + | 4.1 ab ± 0.52 |
| Consortium | +++ | + | 80 | + | 9.9 a ± 2.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez, I.; Fallard, A.; Soto, I.; Tortella, G.; de la Luz Mora, M.; Valentine, A.J.; Barra, P.J.; Duran, P. Efficient Biocontrol of Gaeumannomyces graminis var. Tritici in Wheat: Using Bacteria Isolated from Suppressive Soils. Agronomy 2021, 11, 2008. https://doi.org/10.3390/agronomy11102008
Méndez I, Fallard A, Soto I, Tortella G, de la Luz Mora M, Valentine AJ, Barra PJ, Duran P. Efficient Biocontrol of Gaeumannomyces graminis var. Tritici in Wheat: Using Bacteria Isolated from Suppressive Soils. Agronomy. 2021; 11(10):2008. https://doi.org/10.3390/agronomy11102008
Chicago/Turabian StyleMéndez, Isabel, Ana Fallard, Isabel Soto, Gonzalo Tortella, María de la Luz Mora, Alex J. Valentine, Patricio Javier Barra, and Paola Duran. 2021. "Efficient Biocontrol of Gaeumannomyces graminis var. Tritici in Wheat: Using Bacteria Isolated from Suppressive Soils" Agronomy 11, no. 10: 2008. https://doi.org/10.3390/agronomy11102008
APA StyleMéndez, I., Fallard, A., Soto, I., Tortella, G., de la Luz Mora, M., Valentine, A. J., Barra, P. J., & Duran, P. (2021). Efficient Biocontrol of Gaeumannomyces graminis var. Tritici in Wheat: Using Bacteria Isolated from Suppressive Soils. Agronomy, 11(10), 2008. https://doi.org/10.3390/agronomy11102008







