OsNAC300 Positively Regulates Cadmium Stress Responses and Tolerance in Rice Roots
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
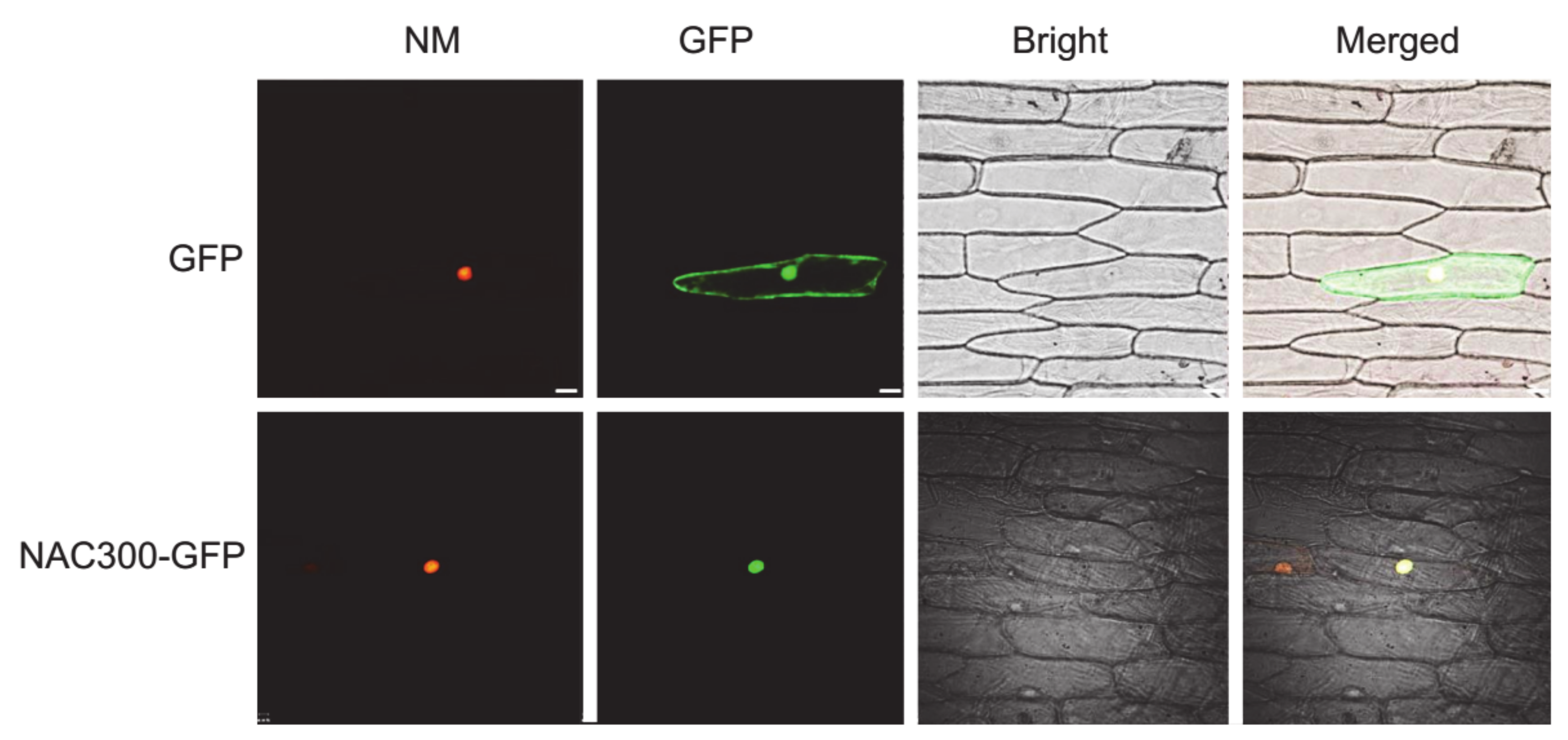
2.2. Subcellular Localization
2.3. Gene Expression Analysis
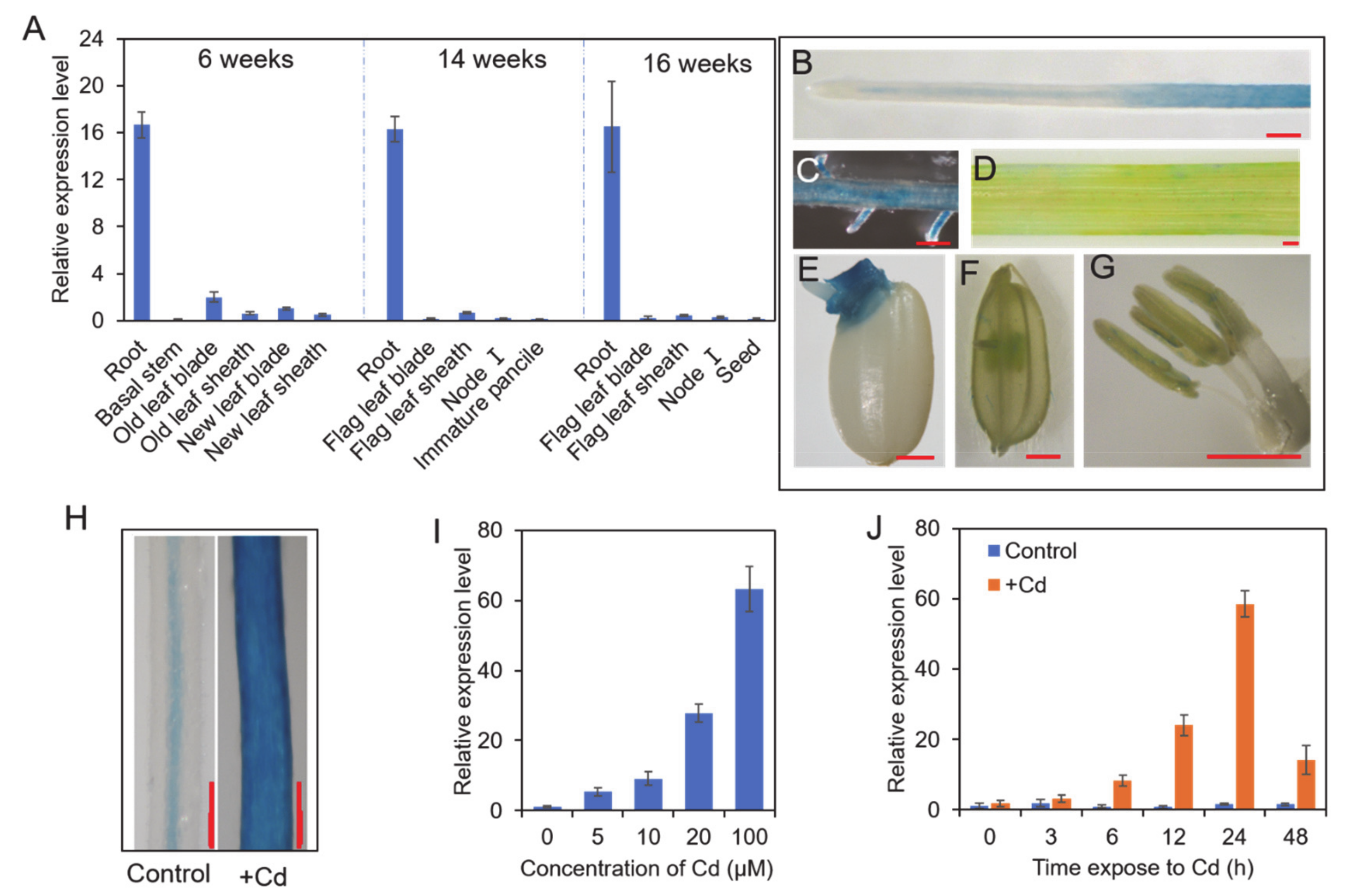
2.4. β-glucuronidase (GUS) Staining
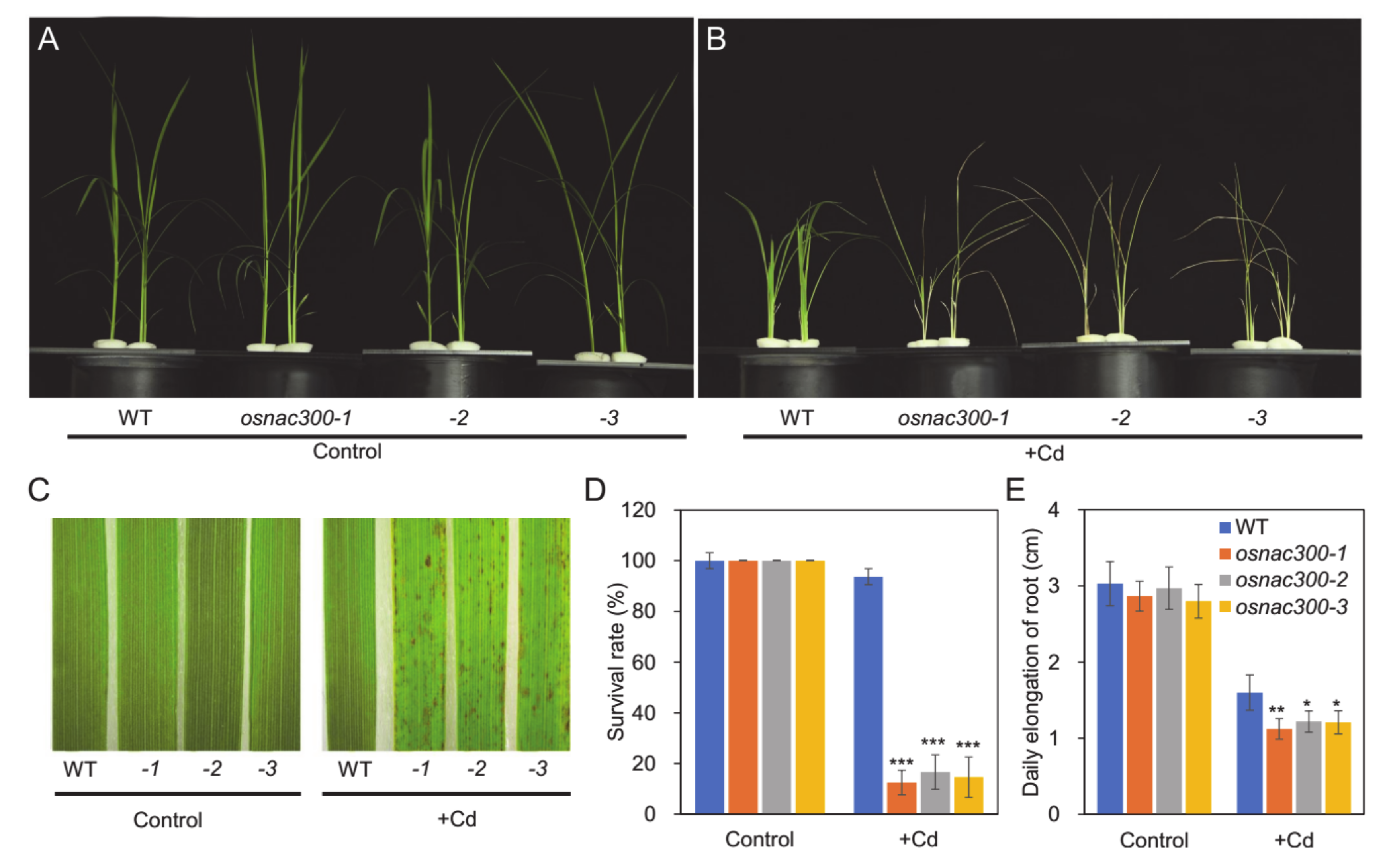
2.5. Phenotypic Analysis
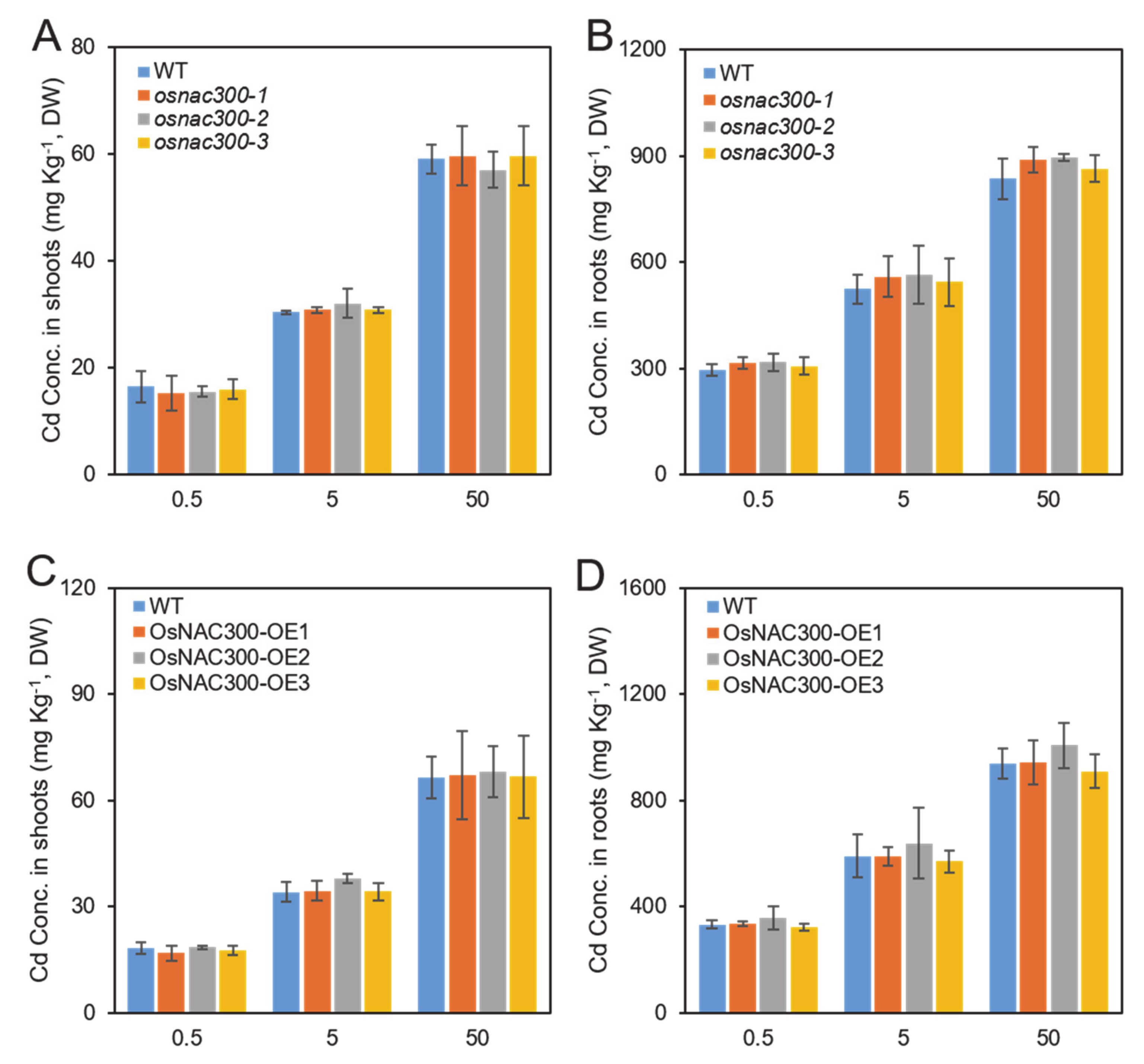
2.6. Elemental Concentration Measurement
2.7. RNA-Seq Analysis
2.8. Transient Luciferase (LUC) Reporter Assay
2.9. ChIP-qPCR Assay
2.10. Statistical Analysis
3. Results
3.1. Sequence and Subcellular Localization Analysis of OsNAC300
3.2. Expression Analysis of OsNAC300
3.3. Knock Out of OsNAC300 Enhances Sensitivity to Cd Stress
3.4. Role of OsNAC300 in the Accumulation of Cd in Rice
3.5. OsNAC300 Plays an Important Role in Changes in Global Gene Expression in Response to Cd Stress
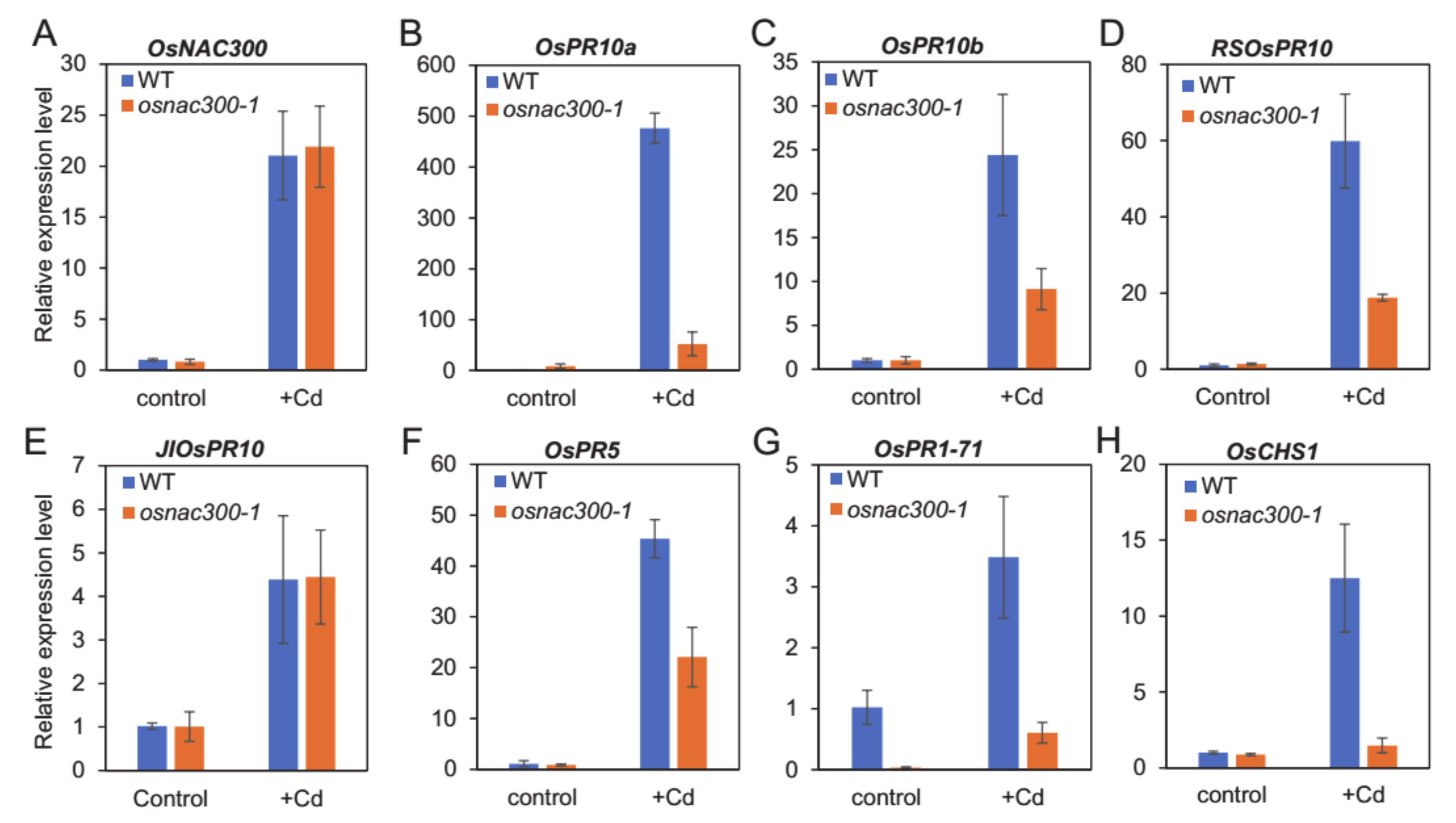
3.6. The Response of PR Genes and OsCHS1 to Cd Stress Are Dependent on OsNAC300
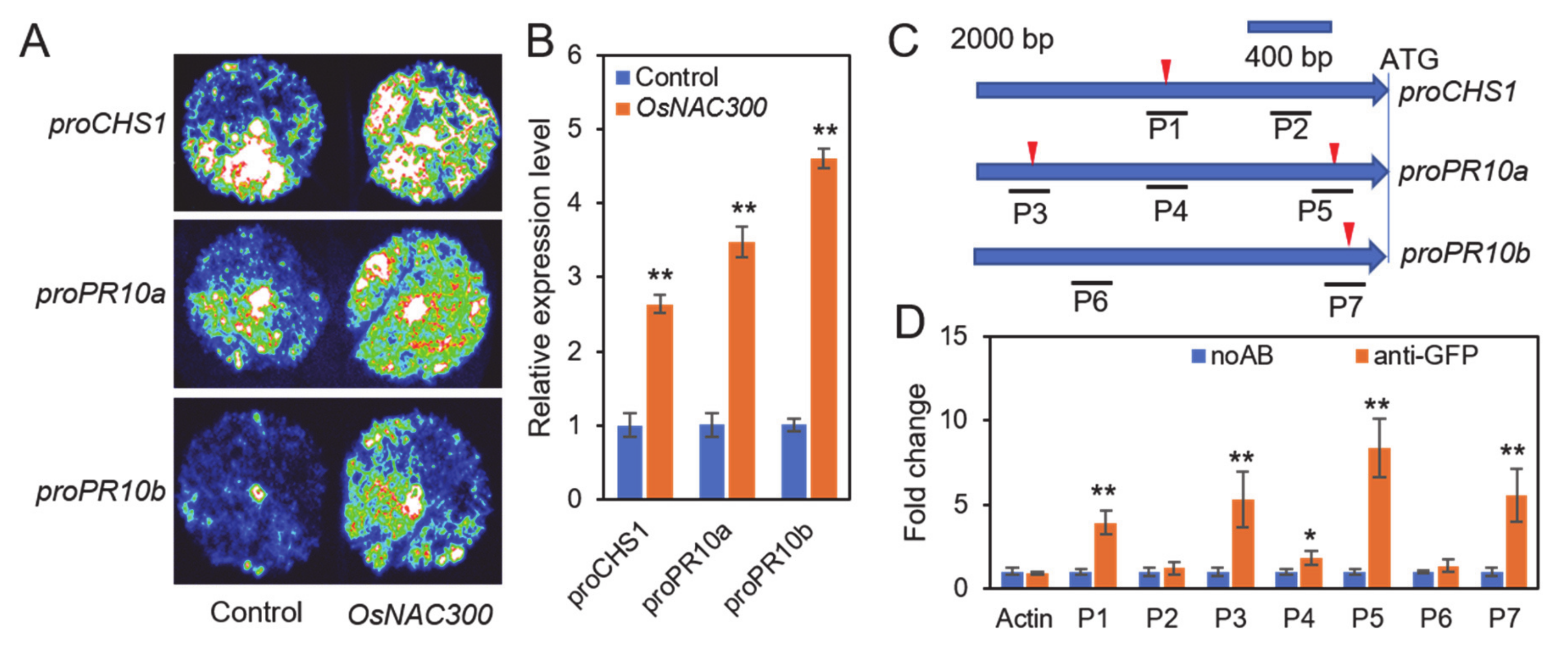
3.7. OsNAC300 Directly Binds to the Promoters of OsCHS1, OsPR10a, and OsPR10b and Enhances Their Transcription
4. Discussion
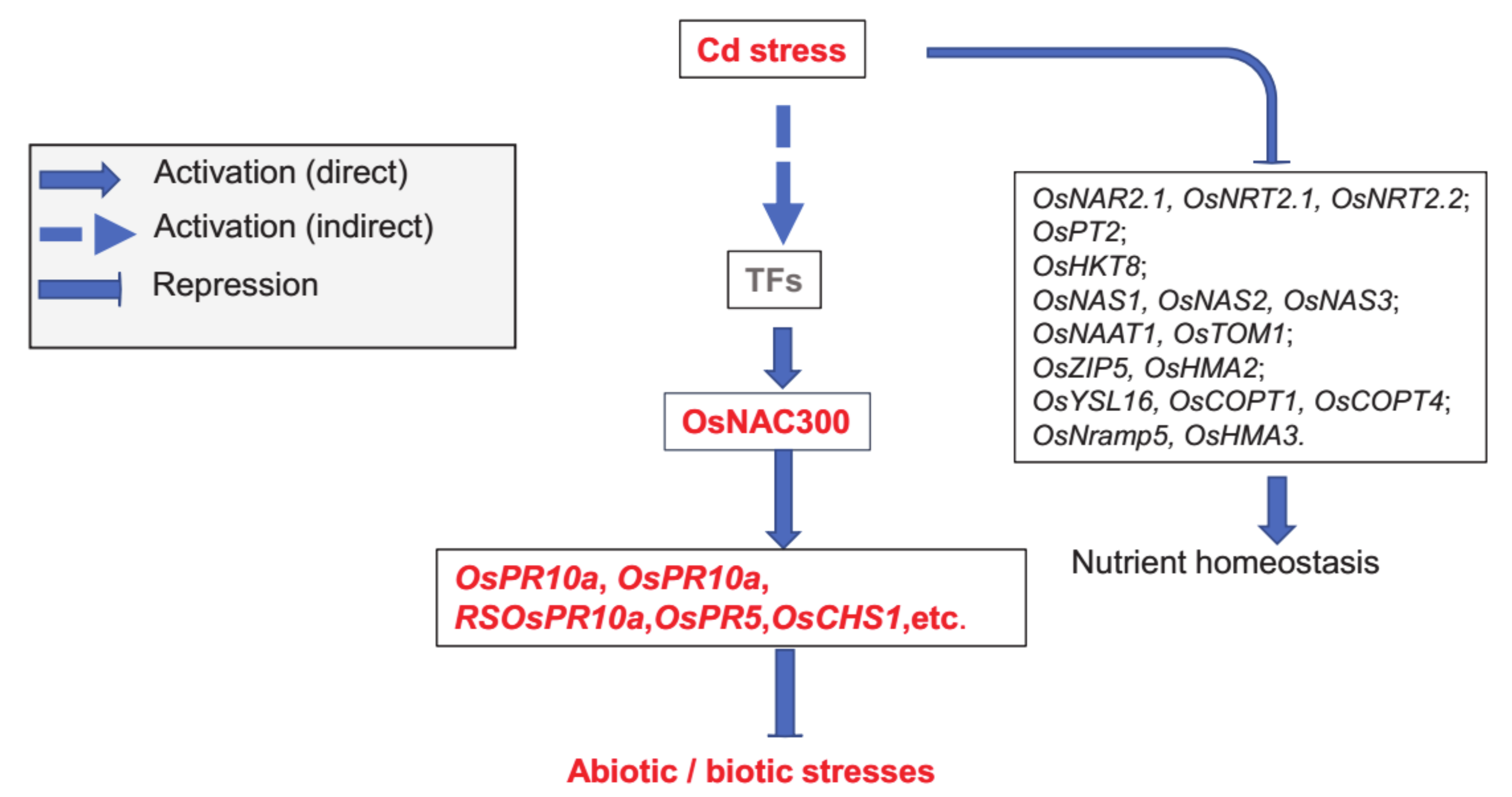
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Clemens, S.; Aarts, M.G.M.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Nawrot, T.; Plusquin, M.; Hogervorst, J.; Roels, H.A.; Celis, H.; Thijs, L.; Vangronsveld, J.; Van Hecke, E.; Staessen, J. Environmental exposure to cadmium and risk of cancer: A prospective population based study. Lancet Oncol. 2006, 7, 119–126. [Google Scholar] [CrossRef]
- Wang, J.; Yu, N.; Mu, G.; Shinwari, K.I.; Shen, Z.; Zheng, L. Screening for Cd-safe cultivars of Chinese cabbage and a preliminary study on the mechanisms of Cd accumulation. Int. J. Environ. Res. Public Health 2017, 14, 395. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, T.; Ezaki, T.; Moriguchi, J.; Furuki, K.; Shimbo, S.; Matsuda, I.N.; Ikeda, M. Rice as the most influential source of cadmium intake among general Japanese population. Sci. Total Environ. 2003, 305, 41–51. [Google Scholar] [CrossRef]
- Dalcorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Signal. Behav. 2010, 5, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Cobbett, C.S. Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 2000, 123, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S. Molecular mechanisms of plant metal tolerance and homeostasis. Planta 2001, 212, 475–486. [Google Scholar] [CrossRef]
- Luo, J.; Huang, J.; Zeng, D.; Peng, J.; Zhang, G.; Ma, H.; Guan, Y.; Yi, H.; Fu, Y.; Han, B.; et al. A defensin-like protein drives cadmium eux and allocation in rice. Nat. Commun. 2018, 9, 645. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Koes, R.E.; Spelt, C.E.; Mol, J.N.M. The chalcone synthase multigene family of Petunia hybrida (V30): Differential, light-regulated expression during flower development and UV light induction. Plant Mol. Biol. 1989, 12, 213–225. [Google Scholar] [CrossRef]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kim, S.G.; Kang, K.Y.; Kim, J.G.; Park, S.R.; Gupta, R.; Kim, Y.H.; Wang, Y.; Kim, S.T. Overexpression of a pathogenesis-related protein 10 enhances biotic and abiotic stress tolerance in rice. Plant Pathol. J. 2016, 32, 552–562. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ekramoddoullah, A.K.M. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 2006, 68, 3–13. [Google Scholar] [CrossRef]
- Lee, E.J.; Facchini, P. Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 2010, 22, 3489–3503. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, Z.; Brown, R.L.; Bhatnagar, D. Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J. Plant Physiol. 2010, 167, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Gyohda, A.; Tominaga, M.; Kawakatsu, M.; Hatakeyama, A.; Ishii, N.; Shimaya, K.; Nishimura, T.; Riemann, M.; Nick, P.; et al. RSOSPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol. 2011, 52, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- McGee, J.D.; Hamer, J.E.; Hodges, T.K. Characterization of a PR-10 pathogenesis related gene family induced in rice during infection with Magnaporthe grisea. Mol. Plant-Microbe Interact. 2001, 14, 877–886. [Google Scholar] [CrossRef]
- Jwa, N.S.; Kumar, A.G.; Rakwal, R.; Park, C.H.; Prasad, A.V. Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem. Biophys. Res. Commun. 2001, 286, 973–983. [Google Scholar] [CrossRef]
- Hashimoto, M.; Kisseleva, L.; Sawa, S.; Furukawa, T.; Komatsu, S.; Koshiba, T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 2004, 45, 550–559. [Google Scholar] [CrossRef]
- Midoh, N.; Iwata, M. Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol. 1996, 37, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.S.; Han, M.; Lee, S.K.; Cho, J.I.; Ryoo, N.; Heu, S.; Lee, Y.H.; Bhoo, S.H.; Wang, G.L.; Hahn, T.R.; et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006, 25, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Rakwal, R.; Agrawal, G.K.; Agrawal, V.P. Jasmonate, salicylate, protein phosphatase 2A inhibitors and kinetin up-regulate OsPR5 expression in cut responsive rice (Oryza sativa). J. Plant Physiol. 2001, 158, 1357–1362. [Google Scholar] [CrossRef]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef]
- Yoon, Y.; Deok, H.S.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The role of stress responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Greve, K.; La Cour, T.; Jensen, M.K.; Poulsen, F.M.; Skriver, K. Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem. J. 2003, 371, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Moumeni, A.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Attia, K.; Kikuchi, S. Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday Selection (drought tolerant) and IR64. Mol. Genet. Genom. 2012, 287, 389–410. [Google Scholar] [CrossRef]
- Sun, L.; Huang, L.; Hong, Y.; Zhang, H.; Song, F.; Li, D. Comprehensive analysis suggests overlapping expression of rice ONAC transcription factors in abiotic and biotic stress responses. Int. J. Mol. Sci. 2015, 16, 4306–4326. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.P.; Leach, J.E. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019, 9, 6273. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yu, Y.; Chen, Q.; Mu, G.; Shen, Z.; Zheng, L. OsMYB45 plays an important role in rice resistance to cadmium stress. Plant Sci. 2017, 264, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chmielowska-Bąk, J.; Gzyl, J.; Rucińska-Sobkowiak, R.; Arasimowicz-Jelonek, M.; Deckert, J. The new insights into cadmium sensing. Front. Plant Sci. 2014, 5, 245. [Google Scholar] [PubMed]
- Chen, J.; Yang, L.; Yan, X.; Liu, Y.; Wang, R.; Fan, T.; Ren, Y.; Tang, X.; Xiao, F.; Liu, Y.; et al. Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol. 2016, 171, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, S.; Jozefczak, M.; Wójcik, M.; Deckers, J.; Vangronsveld, J.; Cuypers, A. Glutathione: A key player in metal chelation, nutrient homeostasis, cell cycle regulation and the DNA damage response in cadmium-exposed Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 154, 498–507. [Google Scholar] [CrossRef]
- He, F.; Liu, Q.; Zheng, L.; Cui, Y.; Shen, Z.; Zheng, L. RNA-Seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front. Plant Sci. 2015, 6, 1136. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, H.; Reinfelder, J.R.; Liang, X.; Sun, C.; Liu, C.; Li, F.; Yi, J. A transcriptomic (RNA-seq) analysis of genes responsive to both cadmium and arsenic stress in rice root. Sci. Total Environ. 2019, 666, 445–460. [Google Scholar] [CrossRef]
- Oono, Y.; Yazawa, T.; Kanamori, H.; Sasaki, H.; Mori, S.; Handa, H.; Matsumoto, T. Genome-wide transcriptome analysis of cadmium stress in rice. BioMed Res. Int. 2016, 9739505. [Google Scholar] [CrossRef]
- Kusano, H.; Asano, T.; Shimada, H.; Kadowaki, K.I. Molecular characterization of ONAC300, a novel NAC gene specifically expressed at early stages in various developing tissues of rice. Mol. Genet. Genom. 2005, 272, 616–626. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-Guided genome editing in plants using a CRISPR–cas9 system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef]
- Ozawa, K. Establishment of a high efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Plant Sci. 2009, 176, 522–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Yue, X.; Zeng, H.; Zhu, J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 2014, 26, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Alvarez, V.R.; Avramova, Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 2008, 3, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Huang, A.; Li, J.; Gao, L.; Feng, Y.; Pemberton, E.; Chen, C. OsNAC45 plays complex roles by mediating POD activity and the expression of development related genes under various abiotic stresses in rice root. Plant Growth Regul. 2018, 84, 519–531. [Google Scholar] [CrossRef]
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi, S.K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Song, S.Y.; Chen, Y.; Chen, J.; Dai, X.Y.; Zhang, W.H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N.K. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.Y.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef]
- Lee, K.; Bae, D.W.; Kim, S.H.; Han, H.J.; Liu, X.; Park, H.C.; Lim, C.O.; Lee, S.Y.; Chung, W.S. Comparative proteomic analysis of the short-term responses of rice roots and leaves to cadmium. J. Plant Physiol. 2010, 167, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signaling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [PubMed]
- Matallana-Ramirez, L.P.; Rauf, M.; Farage-Barhom, S.; Dortay, H.; Xue, G.P.; Dröge-Laser, W.; Lers, A.; Balazadeh, S.; Mueller-Roeber, B. NAC transcription factor ORE1 and Senescence-Induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 2013, 6, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.H.; Chu, H.; Tang, L.K.; Sakamoto, W.; Maekawa, M.; Chu, I.K.; Wang, M.; Lo, C. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 2008, 228, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Hernández, I.; Alegre, L.; Van Breusegem, F.; Munné-Bosch, S. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 2009, 14, 125–132. [Google Scholar] [CrossRef]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef]

| WT | osnac300-1 | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | Control | +Cd | FC | Control | +Cd | FC | Symbol | Description |
| pathogenesis-related protein | ||||||||
| Os12g0555500 | 1.3 | 229.2 | 173.8 | 8.4 | 62.2 | 7.4 | OsPR10A | Probenazole-inducible protein PBZ1. |
| Os12g0555200 | 19.3 | 253.9 | 13.2 | 16.2 | 78.0 | 4.8 | OsPR10B | Similar to Probenazole-inducible protein PBZ1. |
| Os12g0555000 | 39.3 | 956.6 | 24.3 | 48.6 | 479.8 | 9.9 | --- | Similar to Probenazole-inducible protein PBZ1. |
| Os03g0663600 | 4.1 | 155.3 | 38.1 | 4.6 | 87.6 | 19.2 | --- | Similar to Pathogenesis-related thaumatin-like protein. |
| Os12g0628600 | 1.7 | 69.8 | 41.6 | 1.6 | 29.7 | 18.1 | OsPR5 | Similar to Thaumatin-like pathogenesis-related protein 3 precursor. |
| Metabolism | ||||||||
| Os11g0530600 | 0.4 | 3.9 | 9.2 | 0.4 | 0.5 | 1.3 | OsCHS1 | Similar to Chalcone synthase C2 (EC 2.3.1.74). |
| Other biology process | ||||||||
| Os11g0514500 | 7.6 | 51.5 | 6.8 | 9.3 | 29.1 | 3.1 | --- | leucine-rich repeat-containing extracellular glycoprotein precursor. |
| Os03g0661600 | 19.7 | 74.7 | 3.8 | 24.6 | 46.9 | 1.9 | --- | Similar to Alpha-amylase/trypsin inhibitor (Antifungal protein). |
| Unknown | ||||||||
| Os09g0272600 | 75.8 | 754.7 | 10.0 | 86.4 | 482.2 | 5.6 | --- | Conserved hypothetical protein. |
| Os10g0452100 | 11.1 | 50.5 | 4.5 | 15.0 | 32.3 | 2.2 | --- | Conserved hypothetical protein. |
| Os04g0653700 | 2.3 | 9.4 | 4.1 | 3.3 | 5.6 | 1.7 | --- | Conserved hypothetical protein. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Shinwari, K.I.; Song, Y.; Xia, J.; Xu, H.; Du, B.; Luo, L.; Zheng, L. OsNAC300 Positively Regulates Cadmium Stress Responses and Tolerance in Rice Roots. Agronomy 2021, 11, 95. https://doi.org/10.3390/agronomy11010095
Hu S, Shinwari KI, Song Y, Xia J, Xu H, Du B, Luo L, Zheng L. OsNAC300 Positively Regulates Cadmium Stress Responses and Tolerance in Rice Roots. Agronomy. 2021; 11(1):95. https://doi.org/10.3390/agronomy11010095
Chicago/Turabian StyleHu, Shubao, Kamran Iqbal Shinwari, Yuxinrui Song, Jixing Xia, Heng Xu, Binbin Du, Le Luo, and Luqing Zheng. 2021. "OsNAC300 Positively Regulates Cadmium Stress Responses and Tolerance in Rice Roots" Agronomy 11, no. 1: 95. https://doi.org/10.3390/agronomy11010095
APA StyleHu, S., Shinwari, K. I., Song, Y., Xia, J., Xu, H., Du, B., Luo, L., & Zheng, L. (2021). OsNAC300 Positively Regulates Cadmium Stress Responses and Tolerance in Rice Roots. Agronomy, 11(1), 95. https://doi.org/10.3390/agronomy11010095






