Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits
Abstract
1. Introduction
2. Doubled Haploids Procedure in Cucurbits
2.1. Pollination with Irradiated Pollen and Fruit Set
2.2. Embryo Detection and Rescue
2.3. In Vitro Culture
2.4. Chromosome Doubling
2.5. DH Seed Recovery
3. Genome Editing in Cucurbit Species
3.1. Agrobacterium-Mediated Transformation
3.2. Genome Editing Efficiency
4. Haploid Inducer-Mediated Genome-Editing in Cucurbit Species
5. Regulatory Landscape for the New Generation of Doubled Haploids
Author Contributions
Funding
Conflicts of Interest
References
- FAO. 2018. Available online: www.fao.org (accessed on 25 January 2020).
- Robinson, R.W. Rationale and methods for producing hybrid cucurbit seed. J. New Seeds 2000, 1, 1–47. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Torrico, O.; Hooghvorst, S.; Nogués, S. In situ parthenogenetic doubled haploid production in melon ‘Piel de Sapo’ for breeding purposes. Front. Plant Sci. 2020, 11, 378. [Google Scholar] [CrossRef] [PubMed]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends Plant. Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.-Q.; Zhao, W.-X.; Li, X.-H.; Liu, X.-C.; Gao, N.-N.; Huang, J.-H.; Wang, W.-Y.; Xu, X.-L.; Tang, Z.-H. Androgenesis, gynogenesis, and parthenogenesis haploids in cucurbit species. Plant. Cell Rep. 2016, 35, 1991–2019. [Google Scholar] [CrossRef]
- Lim, W.; Earle, E.D. Effect of in vitro and in vivo colchicine treatments on pollen production and fruit set of melon plants obtained by pollination with irradiated pollen. Plant. Cell. Tissue Organ. Cult. 2008, 95, 115–124. [Google Scholar] [CrossRef]
- Košmrlj, K.; Murovec, J.; Bohanec, B. Haploid Induction in Hull-less Seed Pumpkin through Parthenogenesis Induced by X-ray-irradiated Pollen. J. Am. Soc. Hortic. Sci. 2013, 138, 310–316. [Google Scholar] [CrossRef]
- Smiech, M.; Sztangret-Wisniewska, J.; Galecka, T.; Korzeniewska, A.; Marzec, L.; Kolakowska, G.; Piskurewicz, U.; Niemirowicz-Szczytt, K. Potential use of RAPD markers in characteristics of cucumber (Cucumis sativus L.) haploids and double-haploids. Acta Soc. Bot. Pol. 2008, 77, 29–34. [Google Scholar]
- Claveria, E.; Garcia-Mas, J.; Dolcet-Sanjuan, R. Optimization of cucumber doubled haploid line production using in vitro rescue of in vivo induced parthenogenic embryos. J. Am. Soc. Hortic. Sci. 2005, 130, 555–560. [Google Scholar] [CrossRef]
- Kurtar, E.S.; Balkaya, A. Production of in vitro haploid plants from in situ induced haploid embryos in winter squash (Cucurbita maxima Duchesne ex Lam.) via irradiated pollen. Plant. Cell Tissue Organ. Cult. 2010, 102, 267–277. [Google Scholar] [CrossRef]
- Kurtar, E.S.; Sarı, N.; Abak, K. Obtention of haploid embryos and plants through irradiated pollen technique in squash (Cucurbita pepo L.). Euphytica 2002, 127, 335–344. [Google Scholar] [CrossRef]
- Kurtar, E.S.; Balkaya, A.; Ozbakir, M.; Ofluoglu, T. Induction of haploid embryo and plant regeneration via irradiated pollen technique in pumpkin (Cucurbita moschata Duchesne ex. Poir). Afr. J. Biotechnol. 2009, 8, 5944–5951. [Google Scholar]
- Baktemur, G.; Yücel, N.K.; Taskin, H.; Çömlekçioğlu, S.; Büyükalaca, S. Effects of different genotypes and gamma ray doses on haploidization using irradiated pollen technique in squash. Turk. J. Biol. 2014, 38, 318–327. [Google Scholar] [CrossRef]
- Taşkın, H.; Kemal Yücel, N.; Baktemur, G.; Çömlekçioğlu, S.; Büyükalaca, S. Effects of different genotypes and gamma ray doses on haploidization with irradiated pollen technique in watermelon (Citrullus lanatus L.). Can. J. Plant Sci. 2013, 93, 1165–1168. [Google Scholar] [CrossRef]
- Niemirowicz-Szczytt, K.; Faris, N.M.; Nikolova, V.; Rakoczy-Trojanowska, M.; Malepszy, S. Optimization of cucumber (Cucumis sativus L.) haploid production and doubling. In Proceedings of the Cucurbitaceae, Warsaw, Poland, 24–28 July 1995; Volume 94, pp. 169–171. [Google Scholar]
- Sauton, A. Effect of season and genotype on gynogenetic haploid production in muskemlon, Cucumis melo L. Sci. Hortic. Amst. 1988, 35, 71–75. [Google Scholar] [CrossRef]
- Baktemur, G.; Taşkın, H.; Büyükalaca, S. Comparison of Different Methods for Separation of Haploid Embryo Induced through Irradiated Pollen and Their Economic Analysis in Melon (Cucumis melo var. inodorus). Sci. World J. 2013, 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, M.J.; Claveria, E.; Monforte, A.J.; Dolcet-Sanjuan, R. Parthenogenic haploids in melon: Generation and molecular characterization of a doubled haploid line population. J. Am. Soc. Hortic. Sci. 2011, 136, 145–154. [Google Scholar] [CrossRef]
- Solmaz, İ.; Sarı, N.; Gürsoy, I.; Kasapoğlu, S. Comparison of in vivo and in vitro Colchicine Application for Production of Dihaploid ‘Kirkagac’and ‘Yuva Hasanbey’Melons. Afr. J. Biotechnol. 2011, 10, 15717–15724. [Google Scholar] [CrossRef]
- Sauton, A.; Dumas de Vaulx, R. Production of haploid plants in melon (Cucumis melo L.) as a result of gynogenesis induced by irradiated pollen. Agronomie 1987, 7, 141–147. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Chu, C.-C. Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Sci. Sin. 1975, 18, 659–668. [Google Scholar]
- Chée, R.P.; Leskovar, D.I.; Cantliffe, D.J. Optimizing Embryogenic Callus and Embryo Growth of a Synthetic Seed System for Sweetpotato by Varying Media Nutrient Concentrations. J. Am. Soc. Hortic. Sci. Jashs 1992, 117, 663–667. [Google Scholar] [CrossRef]
- Ebrahimzadeh, H.; Soltanloo, H.; Shariatpanahi, M.E.; Eskandari, A.; Ramezanpour, S.S. Improved chromosome doubling of parthenogenetic haploid plants of cucumber (Cucumis sativus L.) using colchicine, trifluralin, and oryzalin. Plant Cell Tissue Organ. Cult. 2018, 135, 407–417. [Google Scholar] [CrossRef]
- Lim, W.; Earle, E.D. Enhanced recovery of doubled haploid lines from parthenogenetic plants of melon (Cucumis melo L.). Plant Cell Tissue Organ. Cult. 2009, 98, 351–356. [Google Scholar] [CrossRef]
- Hu, B.; Li, D.; Liu, X.; Qi, J.; Gao, D.; Zhao, S.; Huang, S.; Sun, J.; Yang, L. Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 2017, 10, 1575–1578. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Q.; Yang, X.; Xu, J.; Liu, G.; Yao, X.; Ren, R.; Xu, J.; Lou, L. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 2020, 37, 1353–1356. [Google Scholar] [CrossRef]
- Hooghvorst, I.; López-Cristoffanini, C.; Nogués, S. Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 2019, 9, 17077. [Google Scholar] [CrossRef]
- CuGenDB. Available online: http://cucurbitgenomics.org (accessed on 1 February 2020).
- Castelblanque, L.; Marfa, V.; Claveria, E.; Martinez, I.; Perez-Grau, L.; Dolcet-Sanjuan, R.; Pitrat, M. Improving the Genetic Transformation Efficiency of Cucumis Melo Subsp. Melo ‘Piel de Sapo’via Agrobacterium. 2008. Available online: https://www.semanticscholar.org/paper/Improving-the-genetic-transformationefficiencyofCastelblanqueMarf%C3%A2/95365ce97d898fdcf36e82bf7d3dcfb52bb8f175?p2df (accessed on 1 February 2020).
- Ren, F.; Ren, C.; Zhang, Z.; Duan, W.; Lecourieux, D.; Li, S.; Liang, Z. Efficiency Optimization of CRISPR/Cas9-Mediated Targeted Mutagenesis in Grape. Front. Plant Sci. 2019, 10, 612. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, C.; Zhang, F.; Mendez, J.; Lozano, Y.; Chatpar, K.; Irish, V.F.; Jacob, Y. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2018, 93, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef] [PubMed]
- Gilles, L.M.; Khaled, A.; Laffaire, J.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef]
- Prigge, V.; Xu, X.; Li, L.; Babu, R.; Chen, S.; Atlin, G.N.; Melchinger, A.E. New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 2012, 190, 781–793. [Google Scholar] [CrossRef]
- Laurie, D.A.; Bennett, M.D. The production of haploid wheat plants from wheat x maize crosses. Theor. Appl. Genet. 1988, 76, 393–397. [Google Scholar] [CrossRef]
- Coe, E.H. A Line of Maize with High Haploid Frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Burk, L.G.; Gerstel, D.U.; Wernsman, E.A. Maternal haploids of Nicotiana tabacum L. from seed. Science 1979, 206, 585. [Google Scholar] [CrossRef]
- Kasha, K.J.; Kao, K.N. High frequency haploid production in barley (Hordeum vulgare L.). Nature 1970, 225, 874. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Britt, A.B.; Kuppu, S. Cenh3: An Emerging Player in Haploid Induction Technology. Front. Plant Sci. 2016, 7, 357. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Chan, S.W.L. Haploid plants produced by centromere-mediated genome elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef]
- Kuppu, S.; Tan, E.H.; Nguyen, H.; Rodgers, A.; Comai, L.; Chan, S.W.L.; Britt, A.B. Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance. PLoS Genet. 2015, 11, e1005494. [Google Scholar] [CrossRef]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Wang, W.; McCuiston, J.; Zhong, H.; Nuccio, M.L.; Martin, B. Maternal Haploids Are Preferentially Induced by CENH3-tailswap Transgenic Complementation in Maize. Front. Plant Sci. 2016, 7, 414. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, B.; Li, M.; Wang, D.; Jiao, Y.; Qi, X.; Wang, M.; Liu, Z.; Chen, C.; Wang, Y.; et al. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants 2020, 6, 466–472. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2019, 71, 1337–1349. [Google Scholar] [CrossRef]
- Abbott, A. Europe’s genetically edited plants stuck in legal limbo. Nat. News 2015, 528, 319. [Google Scholar] [CrossRef] [PubMed]
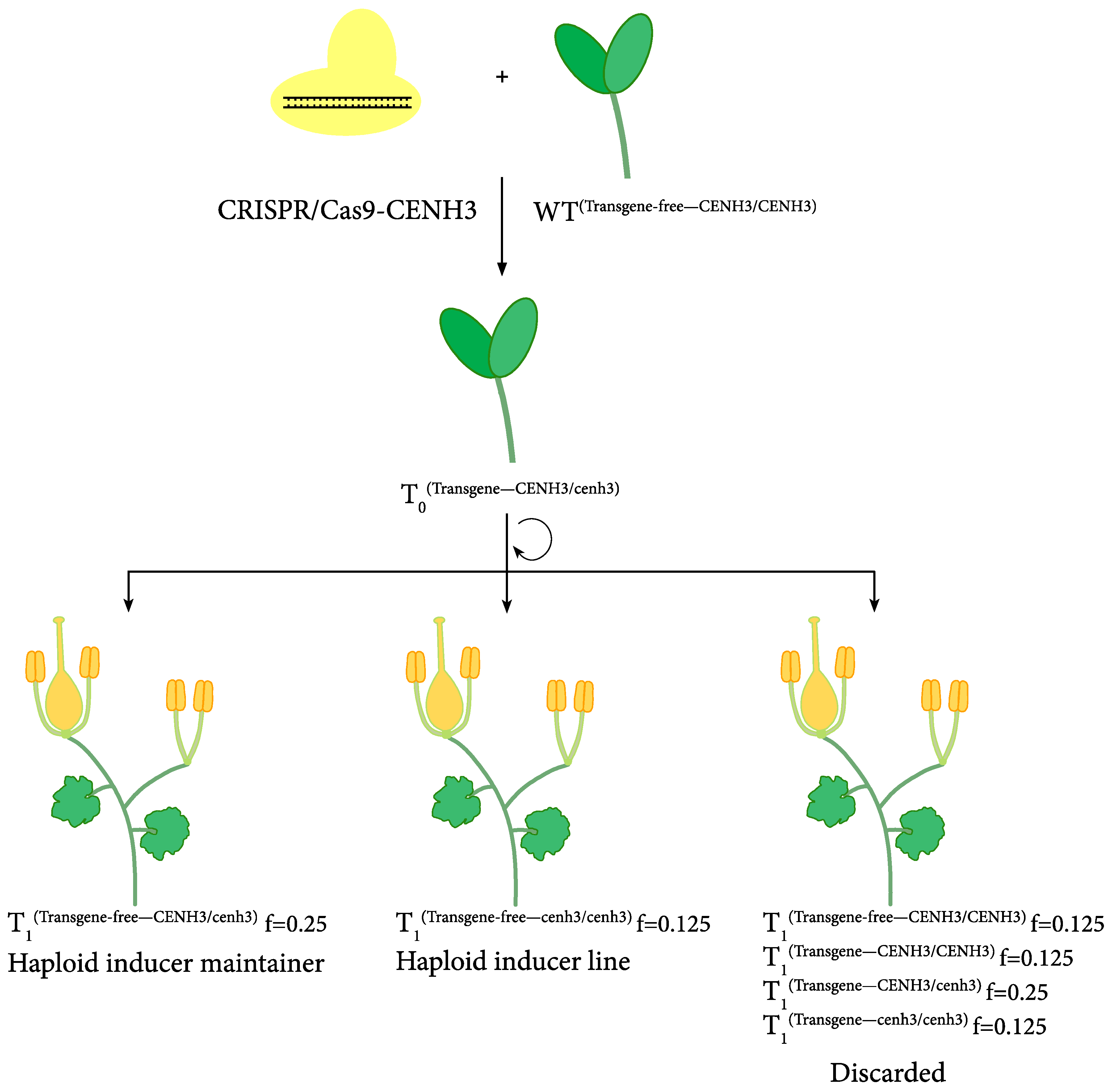
| Species | Embryo Detection Method | Embryos per Fruit | Mortality Rate In Vitro | Ploidy-Level | Chromosome Doubling | Reference | |
|---|---|---|---|---|---|---|---|
| Method | Efficiency | ||||||
| Cucumber | X-ray | 0.23 | 68.23% | 62% H 38% M | E20H8 medium suppl. 500 µM colchicine for 48 h | 55% M 30% DH | Claveria et al. (2005) [10] |
| Cucumber | One-by-one | 5.79 | 79.73% | - | - | - | Smiech et al. (2008) [9] |
| Melon | One-by-one | 6.27 | 30.85% | 73% H 27% M | In vitro solution 500 mg·L−1 colchicine for 3 h | 26% DH | Lim and Earle (2008) [7] |
| Melon | X-ray | 0.30 | 50.94% | 73.1 H 23.1% DH 3.8% M | In vivo solution 5000 mg·L−1 colchicine for 2 h | 9.38% M 20.31% DH | Hooghvorst et al. (2020) [3] |
| Melon | One-by-one | 1.97 | 34.22% | 90% H | E20H8 medium suppl. 500 µM colchicine for 48 h | - | Gonzalo et al. (2011) [19] |
| Melon | - | - | - | - | In vivo solution 0.5% colchicine for 2 h | 46.03% DH | Solmaz et al. (2011) [20] |
| Pumpkin | One-by-one | 16.38 | 84.04% | 5.9% H | - | - | Kurtar et al. (2009) [13] |
| Pumpkin | One-by-one | 69.85 | - | 0.86% H | Košmrlj et al. (2013) [8] | ||
| Squash | One-by-one | 0.2–10.5 | 71.2–80.1% | 43.7% H 56.3% D | - | - | Kurtar et al. (2002) [12] |
| Squash | One-by-one | 18.45 | - | 65.6% H | Baktemur et al. (2014) [14] | ||
| Watermelon | One-by-one | 1.40 | - | - | - | - | Taskin et al. (2013) [15] |
| Winter squash | One-by-one | 13.72 | 85% | 10.9% H | - | - | Kurtar and Balkaya (2010) [11] |
| Species | Target Gene | Plant Material | Transformation | Genome Editing Efficiency | Reference | |
|---|---|---|---|---|---|---|
| Method | Efficiency | |||||
| Cucumber | eIF4E | Cotyledonary explants | Agrobacterium-mediated | - | 20% | Chandrasekaran et al. (2016) [28] |
| Cucumber | WIP1 | Cotyledonary explants | Agrobacterium-mediated | 1.32% | 65.2% | Hu et al. (2017) [27] |
| Watermelon | PDS | Cotyledonary explants | Agrobacterium-mediated | 1.67% | 100% | Tian et al. (2017) [29] |
| Watermelon | PDS | Protoplast | Protoplast transfection | - | 42.1–51.6% | Tian et al. (2017) [29] |
| Watermelon | ACS | Cotyledonary explants | Agrobacterium-mediated | - | 23% | Tian et al. (2018) [30] |
| Watermelon | PSK1 | Cotyledonary explants | 2.3% | - | Zhang et al. (2020) [31] | |
| Melon | PDS | Cotyledonary explants | Agrobacterium-mediated | 5.63% | 42–45% | Hooghvorst et al. (2019) [32] |
| Melon | PDS | Protoplast | Protoplast transfection | - | 25% | Hooghvorst et al. (2019) [32] |
| Species | Target Gene | Mutation or Disruption Method | Haploid Progeny | Reference |
|---|---|---|---|---|
| Arabidopsis | CENH3 | GFP-tailswap disruption | 4–34% | Ravi and Chan (2010) [48] |
| Arabidopsis | CENH3 | GFP-tailswap disruption | 2.50% | Kuppu et al. (2015) [49] |
| Arabidopsis | CENH3 | GFP-tailswap disruption | 4.80% | Karimi-Ashtiyani et al. (2015) [50] |
| Arabidopsis | CENH3 | GFP-tailswap disruption | 3.60% | Kelliher et al. (2016) [51] |
| Arabidopsis | DMP | CRISPR/Cas9 | 2.1% | Zhong et al. (2020) [52] |
| Cucumber | CENH3 | EMS-induced mutation | 1% | WO 2017 081,011 A1/RIJK ZWAAN |
| Maize | CENH3 | GFP-tailswap disruption | 3.6% | Kelliher et al. (2016) [51] |
| Maize | MATL | TALEN | 6.70% | Kelliher et al. (2017) [45] |
| Maize | DMP | CRISPR/Cas9 | 0.1–0.3% | Zhong et al. (2019) [53] |
| Melon | CENH3 | EMS-induced mutation | 1.50% | WO 2017 081,011 A1/RIJK ZWAAN |
| Rice | CENH3 | EMS-induced mutation | 1% | WO 2017 200386/KEYGENE |
| Rice | MATL | CRISPR/Cas9 | 2–6% | Yao et al. (2018) [46] |
| Tomato | CENH3 | GFP-tailswap disruption | 0.2–2.3% | WO 2017 200386/KEYGENE |
| Wheat | MATL | CRISPR/Cas9 | 18.9% | Liu et al. (2019) [54] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hooghvorst, I.; Nogués, S. Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits. Agronomy 2020, 10, 1441. https://doi.org/10.3390/agronomy10091441
Hooghvorst I, Nogués S. Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits. Agronomy. 2020; 10(9):1441. https://doi.org/10.3390/agronomy10091441
Chicago/Turabian StyleHooghvorst, Isidre, and Salvador Nogués. 2020. "Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits" Agronomy 10, no. 9: 1441. https://doi.org/10.3390/agronomy10091441
APA StyleHooghvorst, I., & Nogués, S. (2020). Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits. Agronomy, 10(9), 1441. https://doi.org/10.3390/agronomy10091441





