Screening of the Candidate DNA Barcodes for Three Important Amorphophallus Species Identification
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Strategy
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Data Analysis
3. Results
3.1. Barcode Universality
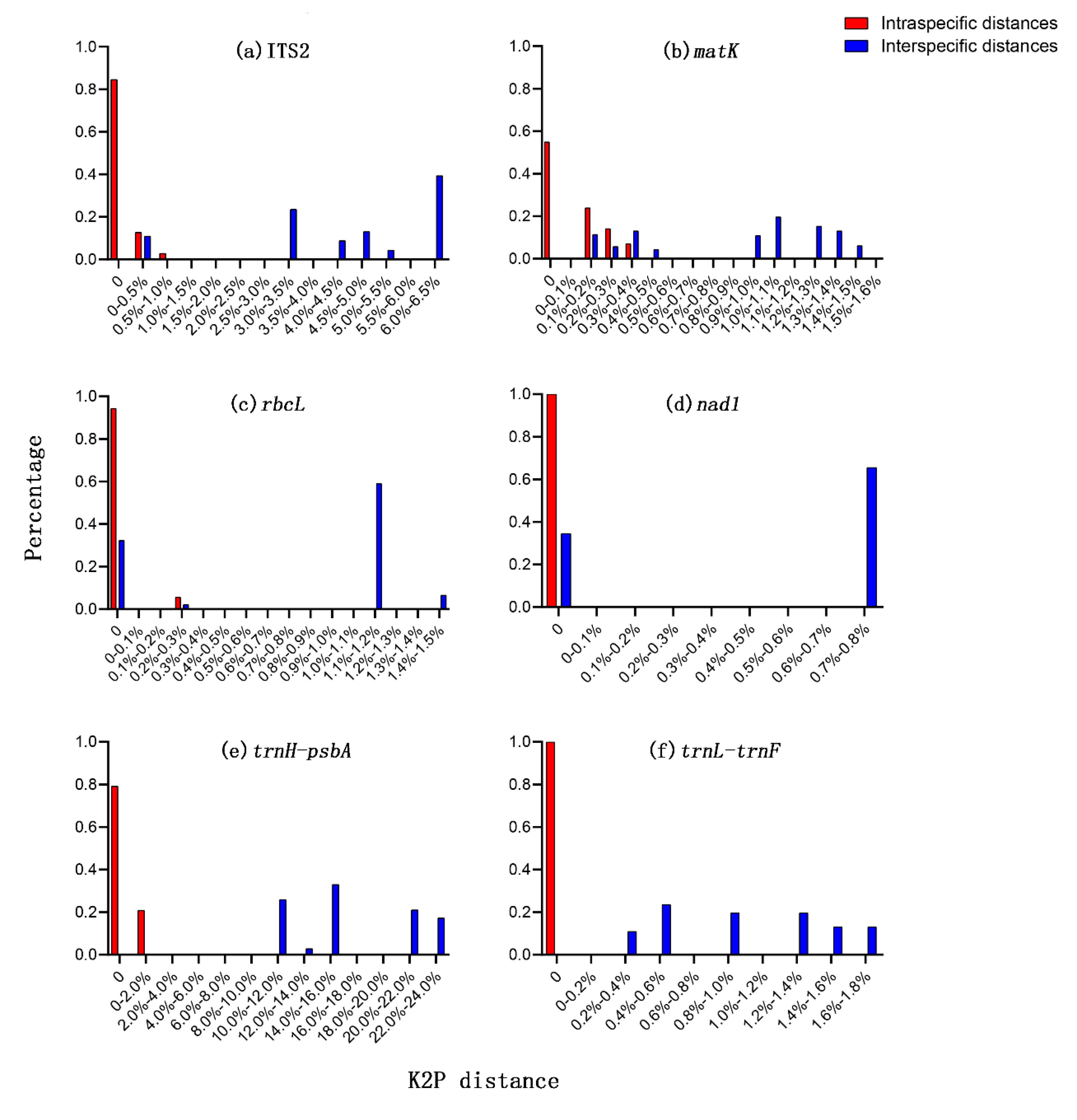
3.2. Sequence Characteristics
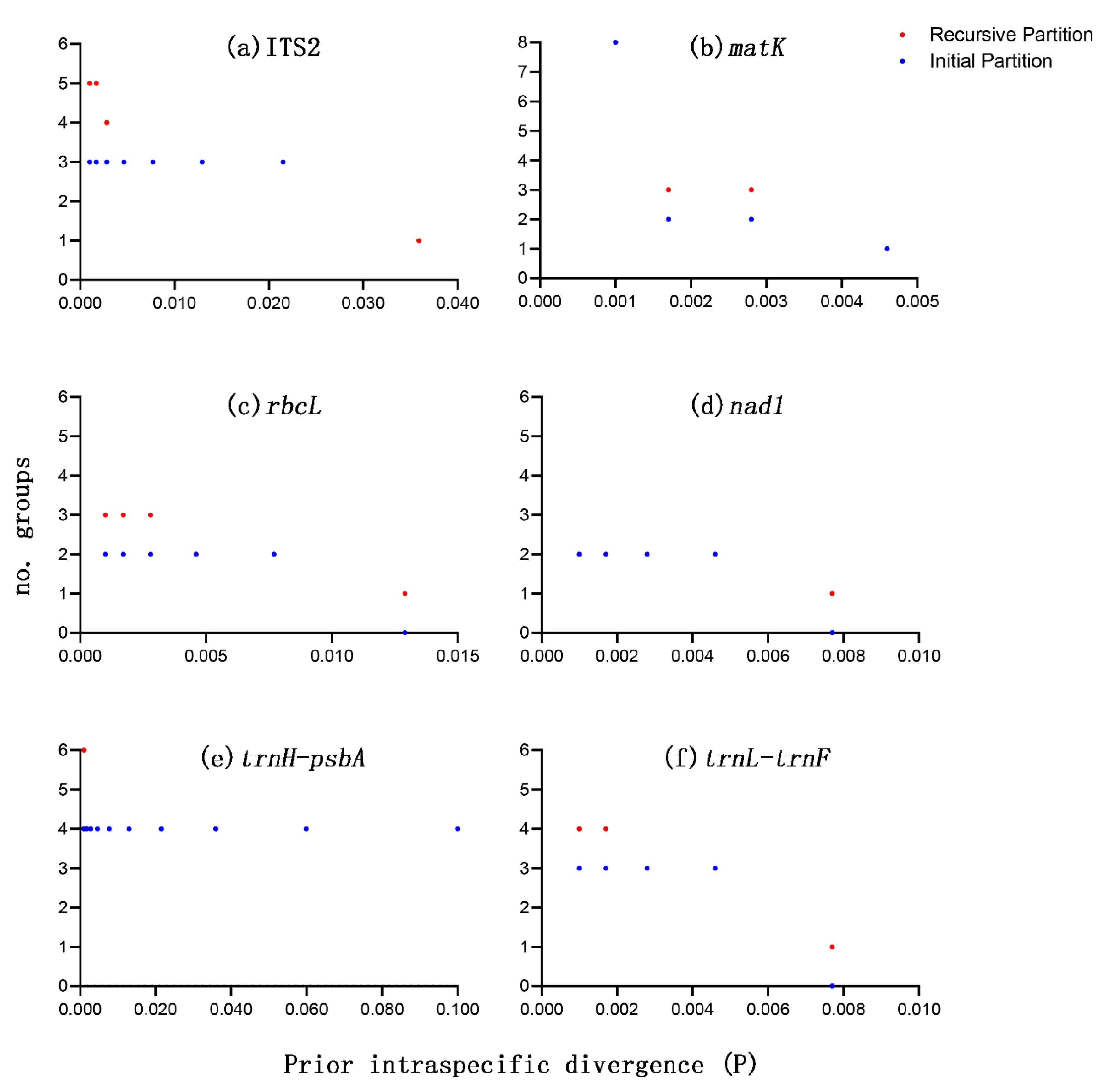
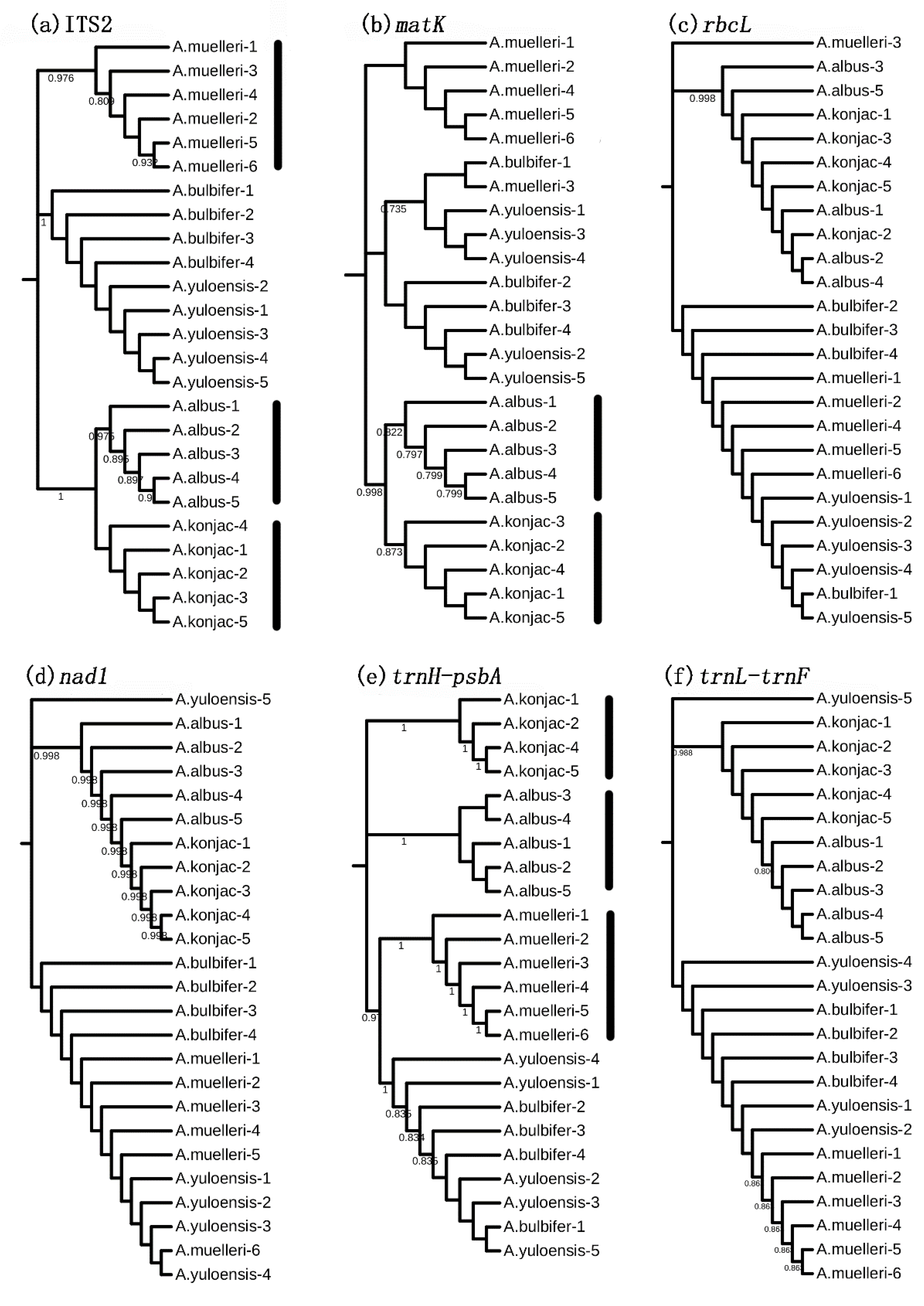
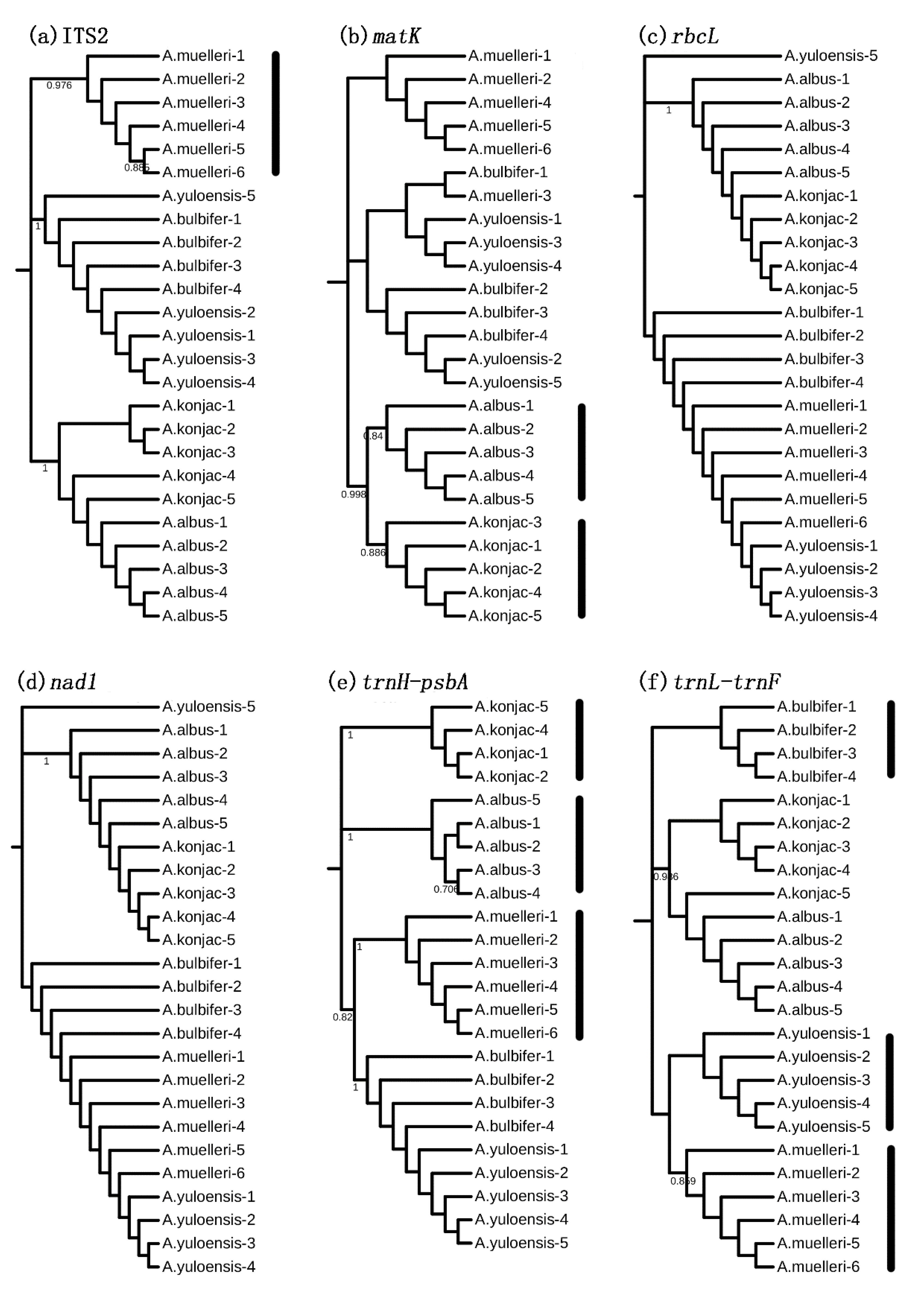
3.3. Species Discrimination Based on Different Analytical Methods
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, E.; Yang, C.; Liu, J.; Jin, S.; Harijati, N.; Hu, Z.; Diao, Y.; Zhao, L. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci. Rep. 2019, 9, 809. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, R.D.; Reddy, C.K.; Xu, B. Health-Promoting effects of konjac glucomannan and its practical applications: A critical review. Int. J. Biol. Macromol. 2019, 126, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yuan, L.-Q.; Li, L.-J.; Lv, Y.; Chen, P.F.; Pan, L. Suppression of gastric cancer by extract from the tuber of amorphophallus konjac via induction of apoptosis and autophagy. Oncol. Rep. 2017, 38, 1051–1058. [Google Scholar] [CrossRef]
- Suwannaporn, P.; Thepwong, K.; Tester, R.; Al-Ghazzewi, F.; Piggott, J.; Shen, N.; Chen, Z.; Chen, F.; Yang, J.; Zhang, D.; et al. Tolerance and nutritional therapy of dietary fibre from konjac glucomannan hydrolysates for patients with inflammatory bowel disease (IBD). Bioact. Carbohydr. Diet. Fibre 2013, 2, 93–98. [Google Scholar] [CrossRef]
- Suwannaporn, P.; Tester, R.F.; Al-Ghazzewi, F.H.; Artitdit, P. Effect of short term administration of konjac glucomannan hydrolysates on adult blood lipid parameters and glucose concentrations. Nutr. Food Sci. 2015, 45, 616–624. [Google Scholar] [CrossRef]
- Arvill, A.; Bodin, L. Effect of short-term ingestion of konjac glucomannan on serum cholesterol in healthy men. Am. J. Clin. Nutr. 1995, 61, 585–589. [Google Scholar] [CrossRef]
- Onishi, N.; Kawamoto, S.; Suzuki, H.; Santo, H.; Aki, T.; Shigeta, S.; Hashimoto, K.; Hide, M.; Ono, K. Dietary Pulverized Konjac Glucomannan Suppresses Scratching Behavior and Skin Inflammatory Immune Responses in NC/Nga Mice. Int. Arch. Allergy Immunol. 2007, 144, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Zalewski, B.M.; Chmielewska, A.; Szajewska, H. The effect of glucomannan on body weight in overweight or obese children and adults: A systematic review of randomized controlled trials. Nutrition 2015, 31, 437–442. [Google Scholar] [CrossRef]
- Al-Ghazzewi, F.H.; Khanna, S.; Tester, R.F.; Piggott, J.R. The potential use of hydrolysed konjac glucomannan as a prebiotic. J. Sci. Food Agric. 2007, 87, 1758–1766. [Google Scholar] [CrossRef]
- Connolly, M.L.; Lovegrove, J.A.; Tuohy, K.M. Konjac glucomannan hydrolysate beneficially modulates bacterial composition and activity within the faecal microbiota. J. Funct. Foods 2010, 2, 219–224. [Google Scholar] [CrossRef]
- Guo, C.; Li, L.; Zhou, H.; Lin, Y.S.; Wu, Y.R. Comparative study on physicochemical properties of konjac glucomannan of three kinds of Amorphophallus. Hunan Agric. Sci. 2012, 9, 95–98. [Google Scholar]
- Zhang, Y.; Zhang, X.; Liu, P.; Feng, C. RAPD Analysis of Amorphophallus Germplasm Resources. J. South Agric. Univ. 2001, 23, 418–421. [Google Scholar]
- Zhang, F.J.; Liu, H.L.; Zhang, J.; Liu, Y.; Zhang, S.L. Research progress of Amorphophallus bulbifera resources. South China Agric. 2013, 7, 64–67. [Google Scholar]
- Zhang, D.H.; Wang, Q.P.; He, Z.G. Amorphophallus muelleri—A New Promising Star from Low Yield Crop to Quality High Yield Crop. Resour. Dev. Mark. 2010, 26, 135–138. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Zuo, Y.J.; Chen, Z.J.; Kondo, K.; Funamoto, T.; Wen, J.; Zhou, S.L. DNA Barcoding of PanaxSpecies. Planta Med. 2010, 77, 182–187. [Google Scholar] [CrossRef]
- Zhang, C.-Y.; Wang, F.-Y.; Yan, H.-F.; Hao, G.; Hu, C.-M.; Ge, X.-J. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 2012, 12, 98–108. [Google Scholar] [CrossRef]
- Alves, T.L.S.; Chauveau, O.; Eggers, L.; Souza-Chies, T.T.; Souza-Chies, T.T. Species discrimination inSisyrinchium(Iridaceae): Assessment of DNA barcodes in a taxonomically challenging genus. Mol. Ecol. Resour. 2013, 14, 324–335. [Google Scholar] [CrossRef]
- Theodoridis, S.; Stefanaki, A.; Tezcan, M.; Aki, C.; Kokkini, S.; Vlachonasios, K.E. DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Cesme-Karaburun Peninsula (Turkey). Mol. Ecol. Resour. 2012, 12, 620–633. [Google Scholar] [CrossRef]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef]
- Yan, L.-J.; Liu, J.; Möller, M.; Zhang, L.; Zhang, X.-M.; Li, D.-Z.; Gao, L.-M. DNA barcoding ofRhododendron(Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. Mol. Ecol. Resour. 2015, 15, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Pompanon, F.; Taberlet, P. DNA barcoding for ecologists. Trends Ecol. Evol. 2009, 24, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, R.; Van Der Bank, M.; Bogarín, D.; Warner, J.; Pupulin, F.; Gigot, G.; Maurin, O.; Duthoit, S.; Barraclough, T.G.; Savolainen, V. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. USA 2008, 105, 2923–2928. [Google Scholar] [CrossRef]
- Liu, J.; Möller, M.; Li, D.-Z.; Gao, L.-M.; Zhang, D.-Q. DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol. Ecol. Resour. 2011, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Odrzykoski, I.J.; Matsui, A.; Hasegawa, M.; Akiyama, H.; Jia, Y.; Sabirov, R.; Takahashi, H.; Boufford, D.E.; Murakami, N. Adaptive evolution of rbcL in Conocephalum (Hepaticae, bryophytes). Gene 2009, 441, 169–175. [Google Scholar] [CrossRef]
- Wang, X.-R.; Tsumura, Y.; Yoshimaru, H.; Nagasaka, K.; Szmidt, A.E.; Szmidt, K.N.E. Phylogenetic Relationships of Eurasian Pines (Pinus, Pinaceae) Based on Chloroplast rbcL, matK, rpl20-rps18 Spacer, and trnV Intron Sequences. Am. J. Bot. 1999, 86, 1742. [Google Scholar] [CrossRef]
- Song, J.; Yao, H.; Li, Y.; Li, X.; Lin, Y.; Liu, C.; Han, J.; Xie, C.; Chen, S. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J. Ethnopharmacol. 2009, 124, 434–439. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding rbcL Gene Complements the Non-Coding trnH-psbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Chase, M.W.; Salamin, N.; Wilkinson, M.J.; Dunwell, J.M.; Kesanakurthi, R.P.; Haidar, N.; Savolainen, V. Land plants and DNA barcodes: Short-term and long-term goals. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1889–1895. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.-J.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; CPBOL Group; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; CBOL Plant Working Group; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Gholave, A.R.; Pawar, K.D.; Yadav, S.R.; Bapat, V.A.; Jadhav, J.P. Reconstruction of molecular phylogeny of closely related Amorphophallus species of India using plastid DNA marker and fingerprinting approaches. Physiol. Mol. Biol. Plants 2017, 23, 155–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, Y.; Yin, S.; Yang, H.; Wu, L.; Yan, Y. Genetic diversity and phylogenetic relationships of seven Amorphophallus species in southwestern China revealed by chloroplast DNA sequences. Mitochondrial DNA Part A 2018, 29, 679–686. [Google Scholar] [CrossRef]
- Grob, G.B.J.; Gravendeel, B.; Eurlings, M.C.M. Potential phylogenetic utility of the nuclear FLORICAULA/LEAFY second intron: Comparison with three chloroplast DNA regions in Amorphophallus (Araceae). Mol. Phylogenet. Evol. 2004, 30, 13–23. [Google Scholar] [CrossRef]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Mendes-Rodrigues, C.; Marinho, R.C.; Balao, F.; Arista, M.; Ortiz, P.L.; Carmo-Oliveira, R.; Oliveira, P.E. Reproductive diversity, polyploidy, and geographical parthenogenesis in two Eriotheca (Malvaceae) species from Brazilian Cerrado. Perspect. Plant Ecol. Evol. Syst. 2019, 36, 1–12. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Fay, M.F.; Swensen, S.M.; Chase, M.W. Taxonomic Affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bull. 1997, 52, 111. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, W.; Liu, C.; Zhang, Y.; Chen, Y.; Song, M.; Fan, G.; Liu, X.; Xiang, L.; Zhang, Y. Identification of Hippophae species (Shaji) through DNA barcodes. Chin. Med. 2015, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Del Barrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Shiyang, K.; Vaidya, G.G.; Ng, P.K. DNA Barcoding and Taxonomy in Diptera: A Tale of High Intraspecific Variability and Low Identification Success. Syst. Biol. 2006, 55, 715–728. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.P.; Paulay, G. DNA Barcoding: Error Rates Based on Comprehensive Sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-B.; Wang, Y.P.; Möller, M.; Gao, L.-M.; Wu, D. Applying plant DNA barcodes to identify species of Parnassia (Parnassiaceae). Mol. Ecol. Resour. 2012, 12, 267–275. [Google Scholar] [CrossRef]
- Azuma, H.; García-Franco, J.G.; Rico-Gray, V.; Thien, L.B.; Thien, V.R.-G.B. Molecular phylogeny of the Magnoliaceae: The biogeography of tropical and temperate disjunctions. Am. J. Bot. 2001, 88, 2275–2285. [Google Scholar] [CrossRef]
- Hamilton, M.B. Patterns and Relative Rates of Nucleotide and Insertion/Deletion Evolution at Six Chloroplast Intergenic Regions in New World Species of the Lecythidaceae. Mol. Biol. Evol. 2003, 20, 1710–1721. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Fazekas, A.J.; Steeves, R.A.D.; Janovec, J. Testing candidate plant barcode regions in the Myristicaceae. Mol. Ecol. Resour. 2008, 8, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y. The Relationship Between Microsatellite Slippage Mutation Rate and the Number of Repeat Units. Mol. Biol. Evol. 2003, 20, 2123–2131. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shi, L.; Han, J.; Li, G.; Lu, H.; Hou, J.; Zhou, X.; Meng, F.; Downie, S.R. Identification of species in the angiosperm family Apiaceae using DNA barcodes. Mol. Ecol. Resour. 2014, 14, 1231–1238. [Google Scholar] [CrossRef]
- Sui, X.-Y.; Huang, Y.; Tan, Y.; Guo, Y.; Long, C.L. Molecular Authentication of the Ethnomedicinal Plant Sabia parviflora and Its Adulterants by DNA Barcoding Technique. Planta Med. 2011, 77, 492–496. [Google Scholar] [CrossRef]
- Steven, G.N.; Subramanyam, R. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol. Ecol. Resour. 2009, 9 (Suppl. S1), 172–180. [Google Scholar] [CrossRef]
- Dong, L.-N.; Wortley, A.H.; Wang, H.; Li, D.-Z.; Lu, L. Efficiency of DNA barcodes for species delimitation: A case in Pterygiella Oliv. (Orobanchaceae). J. Syst. Evol. 2011, 49, 189–202. [Google Scholar] [CrossRef]
- Yan, H.-F.; Hao, G.; Hu, C.-M.; Ge, X.-J. DNA barcoding in closely related species: A case study of Primula L. sect. Proliferae Pax (Primulaceae) in China. J. Syst. Evol. 2011, 49, 225–236. [Google Scholar] [CrossRef]
- Casiraghi, M.; Labra, M.; Ferri, E.; Galimberti, A.; De Mattia, F. DNA barcoding: A six-question tour to improve users’ awareness about the method. Brief. Bioinform. 2010, 11, 440–453. [Google Scholar] [CrossRef]
- Newton, C.R.; Graham, A.; Heptinstall, L.E.; Powell, S.J.; Summers, C.; Kalsheker, N.; Smith, J.C.; Markham, A.F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989, 17, 2503–2516. [Google Scholar] [CrossRef]
| Regions | Primer Pairs | Sequence 5′–3′ | Cited | Thermocycling Conditions |
|---|---|---|---|---|
| matK | 390f | CGATCTATTCATTCAATATTTC | [36] | 94 °C 4 min; (35 cycles: 94 °C 60 s; 53 °C 30 s; 72 °C 60 s); 72 °C 10 min |
| 1326r | TCTAGCACACGAAAGTCGAAGT | |||
| trnH-psbA | psbA | GTTATGCATGAACGTAATGCTC | [37,38] | 94 °C 4 min; (34 cycles: 94 °C 60 s; 55 °C 60 s; 72 °C 60 s); 72 °C 10 min |
| trnH2 | CGCGCATGGTGGATTCACAATCC | |||
| rbcL | 1F | ATGTCACCACAAACAGAAACT | [39] | 94 °C 4 min; (35 cycles: 94 °C 60 s; 55 °C 30 s; 72 °C 60 s); 72 °C 10 min |
| 724R | TCGCATGTACCTGCAGTAGC | |||
| trnL-trnF | trnL-e | GGTTCAAGTCCCTCTATCCC | [40] | 95 °C 4 min; (35 cycles: 94 °C 45 s; 52 °C 45 s; 72 °C 60 s); 72 °C 10 min |
| trnF-f | ATTTGAACTGGTGACACGAG | |||
| ITS2 | ITS2-F | GCGATACTTGGTGTGAAT | [41] | 94 °C 4 min; (35 cycles: 94 °C 45 s; 56 °C 30 s; 72 °C 45 s); 72 °C 10 min |
| ITS2-R | GACGCTTCTCCAGACTACAAT | |||
| nad1 | F | GATCGGCCATAAATGTACTCC | 70 °C 4 min; 94 °C 4min; (36 cycles: 94 °C 20 s; 55 °C 20 s; 72 °C 90 s); 72 °C 5 min | |
| R | CCCCATATATTCCCGGAGC |
| Barcode | Number of Samples | Aligned Length, bp | Number of Variable Sites, (%) | Number of Parsimony Informative Sites, (%) | Intraspecific Distances (Average), % | Interspecific Distances (Average), % |
|---|---|---|---|---|---|---|
| ITS2 | 25 | 501 | 35, (6.99) | 35, (6.99) | 0–0.58, (0.08) a | 0–6.23, (4.06) b |
| matK | 25 | 830 | 13, (1.68) | 13, (1.68) | 0–0.36, (0.08) ab | 0–1.58, (0.81) c |
| rbcL | 25 | 681 | 10, (1.47) | 8, (1.17) | 0–0.29, (0.02) b | 0–1.47, (0.73) d |
| nad1 | 25 | 1255 | 9, (0.72) | 9, (0.72) | 0, (0) d | 0–0.72, (0.43) e |
| trnH-psbA | 24 | 660 | 169, (25.61) | 169, (25.61) | 0–0.15, (0.03) b | 0–22.74, (14.93) a |
| trnL-trnF | 25 | 410 | 7, (1.71) | 7, (1.71) | 0, (0) d | 0–1.73, (0.92) c |
| Barcode Region | N Groups | N Sequences | Best Match, N (%) | Best Close Match, N (%) | No Match (%) | Threshold, % | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | |||||
| ITS2 | 4 | 25 | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0.57 |
| matK | 4 | 25 | 24 (96.0%) | 0 (0.0%) | 1 (4.0%) | 24 (96.0%) | 0 (0.0%) | 1 (4.0%) | 0 (0.0%) | 0.35 |
| rbcL | 4 | 25 | 0 (0.0%) | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 0.58 |
| nad1 | 4 | 25 | 0 (0.0%) | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 0.00 |
| trnH-psbA | 4 | 24 | 24 (100.0%) | 0 (0.0%) | 0 (0.0%) | 24 (100.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2.68 |
| trnL-trnF | 4 | 25 | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 25 (100.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0.00 |
| ITS2 | matK | rbcL | nad1 | trnH-psbA | trnL-trnF | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Paired | Max | Min | Max | Min | Max | Min | Max | Min | Max | Min | Max | Min |
| A.k-A.a | 0 | 0.39% | 0.12% | 0.36% | 0.29% | 0 | 0 | 0 | 0.15% | 21.01% | 0 | 0.24% |
| A.k-A.m | 0.58% | 4.24% | 0.36% | 1.09% | 0 | 1.18% | 0 | 0.71% | 0 | 21.94% | 0 | 1.48% |
| A.k-A.b | 0 | 6.00% | 0.24% | 1.21% | 0 | 1.18% | 0 | 0.71% | 0 | 22.25% | 0 | 0.98% |
| A.k-A.y | 0 | 6.00% | 0.24% | 1.21% | 0 | 1.18% | 0 | 0.71% | 0 | 22.25% | 0 | 0.98% |
| A.a-A.m | 0.58% | 4.67% | 0.36% | 0.97% | 0.29% | 1.18% | 0 | 0.71% | 0.15% | 13.77% | 0 | 1.73% |
| A.a-A.b | 0 | 6.23% | 0.24% | 1.09% | 0.29% | 1.18% | 0 | 0.71% | 0.15% | 14.06% | 0 | 1.23% |
| A.a-A.y | 0 | 6.23% | 0.24% | 1.09% | 0.29% | 1.18% | 0 | 0.71% | 0.15% | 14.06% | 0 | 1.23% |
| A.m-A.b | 0.58% | 3.03% | 0.36% | 0.12% | 0 | 0 | 0 | 0 | 0.12% | 11.03% | 0 | 0.49% |
| A.m-A.y | 0.58% | 3.03% | 0.36% | 0.12% | 0 | 0 | 0 | 0 | 0 | 11.03% | 0 | 0.49% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; She, X.; Liu, E.; Harijati, N.; Cheng, T.; Hu, Z.; Jin, S.; Diao, Y. Screening of the Candidate DNA Barcodes for Three Important Amorphophallus Species Identification. Agronomy 2020, 10, 1366. https://doi.org/10.3390/agronomy10091366
Zhao C, She X, Liu E, Harijati N, Cheng T, Hu Z, Jin S, Diao Y. Screening of the Candidate DNA Barcodes for Three Important Amorphophallus Species Identification. Agronomy. 2020; 10(9):1366. https://doi.org/10.3390/agronomy10091366
Chicago/Turabian StyleZhao, Chufeng, Xuan She, Erxi Liu, Nunung Harijati, Teng Cheng, Zhongli Hu, Surong Jin, and Ying Diao. 2020. "Screening of the Candidate DNA Barcodes for Three Important Amorphophallus Species Identification" Agronomy 10, no. 9: 1366. https://doi.org/10.3390/agronomy10091366
APA StyleZhao, C., She, X., Liu, E., Harijati, N., Cheng, T., Hu, Z., Jin, S., & Diao, Y. (2020). Screening of the Candidate DNA Barcodes for Three Important Amorphophallus Species Identification. Agronomy, 10(9), 1366. https://doi.org/10.3390/agronomy10091366





