Abstract
Despite the essential roles of soil dissolved organic matter (DOM) and soil microbes in agro-ecosystems, we still have a limited understanding of the extent by which they are impacted by agronomic strategies in ecological and conventional farming. Using three-dimensional fluorescence excitation–emission matrices (3D-EEM) and high-throughput microbial sequencing, the characteristics of soil DOM and microbiota under realistic field conditions were estimated in the farming soils with long-term ecological (EM) and conventional management (CM). Specifically, the role of hedgerows in the ecologically managed land (EMH) was assessed. The total fluorescent intensity of soil DOM in the EMH system was significantly higher than the values in CM and EM systems. Additionally, the five normalized excitation–emission area volumes from regional integration analysis increased in the order CM < EM < EMH. In comparison with CM and EM soils, the hedgerow significantly increased the evenness of the bacterial communities in the EMH system, whereas no differences were found for the alpha-diversity of eukaryotic communities. The composition of soil microbiota was significantly distinct among the three farming systems, with a hedgerow-specific effect on bacterial community and a management-specific effect on eukarya. The predicted functional profiles indicated that the hedgerow showed a higher contribution to the dissimilarity of bacterial functions. Furthermore, the distinction of the soil microbiota was modulated by the soil DOM composition and significantly positive correlations between the microbiota involved in nutrient cycling and soil DOM were observed. The findings in this work strengthen our understanding of the different responses of bacterial and eukaryotic communities under the long-term ecological management and highlight the beneficial roles of hedgerows in increasing organic matter and modulating community assembly.
1. Introduction
Conventional intensive farming systems, largely depending on synthetic fertilizers, pesticides, and herbicides, have contributed greatly to rising crop productivity [1]. However, there is increasing concern for the sustainability of an intensive cultivation system as the intensification leads to ecosystem degradation, soil contamination, diversity loss, and increased greenhouse gas emissions [2,3]. To mitigate these negative impacts and improve sustainable production, ecological or organic farming management based on organic amendments (e.g., crop residue, farmyard compost, and slurry) for plant nutrition and protection has been proposed, which is considered to be low-input and environmentally friendly [4,5]. Thus, understanding how ecological management impacts the agricultural soil is an important subject towards a more sustainable development [6,7,8].
As the key component of terrestrial ecosystems, soil organic matter contributed greatly to soil structure, soil fertility, crop production, and furthers the sustainability of agricultural systems [9,10,11,12]. Although there was evidence that the contents of soil organic matters are increased in agricultural land managed ecologically compared to those from conventional farming, other studies have not observed such differences, which may probably be due to that the overall pool of organic matters in agricultural land is large [13,14,15]. Furthermore, because of the heterogeneity in the structure and function, the responses of soil organic matter to management strategies may be not consistent [16,17,18]. In comparison with total soil organic matter, dissolved organic matter is more sensitive to the disturbance since it is the most reactive and labile fraction, which may give a better indication about soil physicochemical properties and biological activities [11,19,20,21]. However, despite intensive studies examining the quantified changes of total soil organic matters in conventional and organic farming systems, whether and how the management strategies affect the soil dissolved organic matters, particularly the detailed components, remains elusive.
In the terrestrial ecosystem, soil microorganisms play critical roles in driving all biogeochemical cycles on a global scale [22,23,24,25]. It has been reported that agricultural intensification could alter microbial abundance, diversity, and function, which consequently restricts the sustainable productivity of agricultural land [6,26,27,28,29]. Given their importance in agricultural ecosystems, it is vital to understand and manage the soil microbiome through various agricultural strategies to promote beneficial and inhibit detrimental microorganisms. Previous studies have documented that long-term organic fertilization increased soil microbial diversity and heterogeneity, and distinct bacterial and fungal community compositions were observed between organic managed conventional systems [30,31,32]. Even though positive impacts of organic farming were previously reported, the effects of agricultural management on the soil microbiome are, however, complex and commonly diverse, likely due to the enormous complexity of microbial life and the great variation of climatic and edaphic factors across the world [33,34]. Thus, retrieving a universally valid conclusion on the consequences of ecological management practices is difficult.
Recently, hedgerows on agricultural land, initially designed for landscape services, were found to reduce the runoff erosion, and hence decrease the losses of nitrogen and phosphorus in soil [35]. Additionally, hedgerows significantly contributed to the above-ground biodiversity as they provided specific habitats for small mammals, birds, and insects [36,37,38]. For example, Spanns et al. [39] reported that trimmed hedgerows significantly increased the diversity of oribatid mite communities in agricultural land. Although the benefits of hedgerows on the above-ground ecosystems were well studied, little is known about how hedgerows affect the below-ground microbiome in the agriculture land.
The purpose of this study is to describe and estimate the responses of soil dissolved organic matter (DOM) and soil microbial diversity to long-term (10 years) continuous ecological (EM) and conventional farming (CM). The effects of hedgerows in ecologically managed land (EMH) were explored in parallel. Excitation emission matrix fluorescence was employed to identify the components of soil DOM, and the soil microbial diversity assessed using a high-throughput DNA sequencing approach of bacterial and eukaryotic ribosomal markers. We hypothesized that (i) ecological management and hedgerows would have positive effects on soil DOM; (ii) the interaction of ecological management and hedgerows would result in differences in the responses of soil microbial diversity, and (iii) similar responses of bacterial and fungal communities to the farming systems would be observed.
2. Materials and Methods
2.1. Study Site and Soil Sampling
The Anyang research station is located in Henan Province, China (114°15′ E, 35°51′ N), which is characterized by a mean annual precipitation of 606 mm and a mean annual temperature of 13.2 °C. The soil in this station mainly belongs to yellow cinnamon soil (ultisol, USDA), and the cropping regime is wheat-maize rotation. In the conventional system (CM), mineral fertilizers and pesticides were performed to provide plant nutrient and protection. The EM and EMH systems exclusively receive organic fertilizers (farmyard manure and slurry), and the plant protection was performed using mechanical and biological strategies. These fields have been adopted ecologically for 10 years. In addition, the EHM system was bordered by natural indigenous shrubs (mainly including Broussoneia papyrifera and Morus australis) as the hedgerows (1.0–1.5 m width). Four fields of each farming system were chosen, and within each field, two plots were identified with GPS points. For each plot, five soil cores (5 × 20 cm; length × width) were collected and bulked together to form one composite sample. A total of 24 soil samples were collected in July 2019. Soil samples were sieved with 2 mm mesh, and stored at 4 °C for soil chemical analysis and −80 °C for genomic DNA extraction. The soil chemical properties were measured using standard methods, and the details are given in Supplementary materials.
2.2. Soil DOM Extraction and Data Analysis
Water extraction from air-dried soil samples was performed to characterize the soil DOM [40]. Briefly, 5 g of air-dried soil and 25 mL of deionized water were transferred into 50 mL centrifuge tubes, and the vials were shaken at 200 rpm for 30 min on a horizontal shaker. After centrifugation at 4000 rpm for 20 min, the supernatant was filtered through 0.45 μm glass fiber filter paper. The extract was stored in the dark at 4 °C, and analyzed within two days. A Varian Cary Eclipse fluorescence spectrophotometer (Agilent, California, USA) was used for excitation emission matrix (EEM) analysis with emission from 280 to 600 nm at 2 nm step and excitation from 250 to 550 nm at 5 nm step. The EEM data was then normalized to Raman unit (R.U.) by the integrated intensities of the Raman signal and the total fluorescent intensity (TFI) was represented as the overall fluorescence of soil DOM.
Three fluorescent indices, including fluorescent index (FI), humic index (HIX), and biological index (BIX) were calculated to describe the characteristics of soil DOM [41,42,43]. Specifically, FI, the ratio of emission intensity at 470 nm to that at 520 nm with excitation at 370 nm, was used to identify the contribution of microbial activity to soil DOM. The HIX was used to evaluate the complexity and aromaticity of DOM, which was obtained from the ratio of fluorescence intensity at 435–480 nm to the sum intensity at 300–345 nm and 435–480 nm. The BIX was the ratio of fluorescence intensity at emission 380 nm to that at 430 nm, with excitation 310 nm, which can be used for evaluating the relative proportion of organic matter derived from microbe in soil. Additionally, a fluorescence regional integration method for EEM spectra was applied to quantify the DOM components [44]. Five regions, with consistent excitation and emission boundaries, were obtained for each EEM spectra, accounting for the normalized excitation–emission area volumes of aromatic protein I (Region I), aromatic protein II (Region II), fulvic acid-like (Region III), soluble microbial by-product-like (Region IV), and humic acid-like (Region V), respectively.
2.3. DNA Isolation, Amplification, and Sequence Processing
The total DNA was extracted from 0.5 g of each soil sample using the DNeasy PowerSoil isolation kit (Qiagen, Shanghai, China) following the manufacturer’s instructions. The Nano drop spectrophotometric analysis was used to evaluate the quality and concentration of the isolated DNA. The universal 16S rRNA gene forward primer 338F (ACTCCTACGGGAGGCAGCA) and reverse primer 806R (GGACTACHVGGGTWTCTAAT) were used for bacterial amplification, while the 18S rRNA gene was amplified by using the forward primer 817F (TTAGCATGGAATAATRRAATAGGA) and reverse primer 1196R (TCT GGACCTGGTGAGTTTCC). The PCR protocol was carried out in an ABI GeneAmp® 9700 amplification (Thermo Fisher Scientific, Massachusetts, USA) as follows: intimal denaturation at 95 °C for 3 min, then 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, final extension at 72 °C for 10 min. The PCR products were further purified using AxyPrep DNA gel extraction kit (Axygen Biosciences, California, USA), and sequenced on the Illumina Miseq platform by Majorbio BioPharm Technology Co. Ltd. (Shanghai, China).
The raw sequences were filtered and analyzed by Quantitative Insights Into Microbial Ecology (QIIME2, version 2020.02) according to the online introductions [45]. Sequences were demultiplexed and barcodes were trimmed using the cutadapt plugin [46]. The sequences were then denoised using dada2 and clustered into amplicon sequence variants (ASVs) [47]. The taxonomic information of each ASV was assigned using the naive Bayes method using the Silva 132 database as training references [47].
2.4. Statistical Analysis
The alpha-diversity indices, including, observed richness, Smith–Wilson evenness, and Shannon diversity were calculated in QIIME, and the different significances were determined using Kruskal–Wallis and pairwise Wilcoxon tests. To further evaluate the effects of management and hedgerow on alpha-diversity, three different non-parametric multivariate statistical tests, including non-parametric multivariate analysis of variance (Adonis), analysis of similarity (ANOSIM), and multi-response permutation procedure (MRPP) were used based on Euclidean distances [7]. The differences in beta-diversity were measured using Bray–Curtis dissimilarity based on the ASV abundances by principal coordinates analysis (PCoA), and the three tests, i.e., Adonis, ANOSIM, and MRPP were also applied for beta-diversity to maximize comparability with analysis of alpha-diversity [48]. The effects of management and hedgerow on soil chemistry and DOM were examined using PCoA, combined with the Adonis test based on Euclidean distances after z-transformation. Redundancy analysis (RDA) and variance partitioning analysis (VPA) were used to estimate the relationship between beta-diversity and the characteristics of soil chemistry and DOM. Differentially abundant ASVs in CM, EM, and EMH systems were analyzed using the “edgeR” library. The cladogram based on linear discriminant analysis effect size (LEfSe) was used to show the species with significant abundances in the three systems [49]. The functional profiles of soil bacteria and eukarya were analyzed using FAPROTAX [50] and FUNGuild [51], respectively. All the statistical analyses were performed using R software 3.6.2.
3. Results and Discussion
3.1. Variation of Soil Chemical Properties and DOM between the Three Farming Systems
The Adonis analysis indicated that management and hedgerow statistically influenced several soil chemical characteristics (Table S2). For instance, significant effects (p < 0.01) of management, hedgerow, and the interaction of ‘management × hedgerow’ were observed for the contents of TN, TP, and available P. However, TK, available K, alkali-hydrolytic N, and ammonium showed no statistical differences between the three farming systems. Additionally, the farming land managed conventionally or ecologically significantly influenced soil pH and the contents of soil organic matter and nitrate. Previous studies have demonstrated that organic or ecological management could influence soil agroecosystem characters, which was greatly due to the sustained application of organic fertilizer [52]. In our study, farmyard manure and slurry were the exclusive fertilizer in EM and EMH systems, confirming that ecological management may potentially benefit the soil fertility. In comparison with the management mode, few studies have focused on the effects of hedgerows in farming land. Xia et al. [53] observed that contour hedgerows could significantly reduce the runoff and soil erosion, which hence decreased the nitrogen and phosphorus losses. Oshunsanya et al. [54] also reported the similar effects of hedgerows on nitrogen and phosphorus losses. Our results, coupled with previous studies, suggested that the hedgerows on farming land can potentially control soil erosion and increase soil nutrients, further supporting various ecosystem services.
The EEM with regional integration was used to evaluate the differences of DOMs in the three farming systems (Figure 1). Management and hedgerow induced a significant increase in the TFI values. For example, the average TFI values in the EMH system were 23,234 R.U., which were significantly higher than those in CM and EM (13,325 and 18,432 R.U., respectively). The FI values ranged from 1.29 to 1.41, suggesting the soil DOM from our study was derived from terrestrial and allochthonous sources [41,42]. Furthermore, ecological management significantly increased the values of both FI and HIX, revealing a higher humification degree in EM and EMH systems. The elevated HIX values in ecologically managed land may be due to the application of organic fertilizers. In fact, previous studies have demonstrated that organic amendments, e.g., compost, manure, and biochars, can increase the humification degree in soil [55]. No statistical effects of management or hedgerow were observed for the BIX values, and the relatively small BIX values (0.52–0.67) indicated low production of organic matter in the three farming systems.
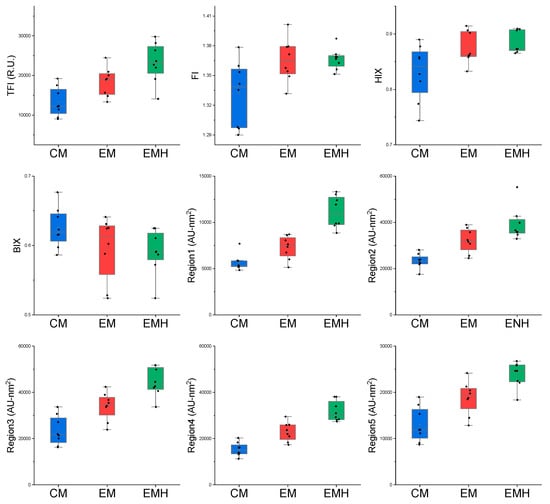
Figure 1.
The values of total Fluorescent Intensity (TFI), fluorescent indices and the area volumes of five regions in conventional management (CM), ecological management (EM), and ecological management with hedgerows (EMH) samples.
In addition to the variation in the values of TFI and fluorescent indices, the excitation–emission area volumes of the five regions were calculated to assess the effects of management and hedgerow on DOM components (Figure 1). It appears that the area volumes of all the five regions increased in the order CM < EM < EMH. For example, the volumes of Regions Ⅰ–Ⅴ in EMH systems were 1.11, 3.68, 4.45, 3.17, and 3.38 × 104 AU-nm2, which were statistically higher than their respective values in CM and EM values. Furthermore, the three multivariate statistical tests indicated management, hedgerow, and their interaction significantly influenced the volumes of Regions I–V (Table S2). According to previous studies (Chen et al., 2013), Regions I, II, and IV can be assigned to be protein-like material, while Regions III and IV were classified as fulvic-like and humic-like components. Therefore, a humification parameter based on the ration between the sum of volumes of Regions III and V and the sum of Regions I, II, and IV was used to characterize the humification of organic substances [56]. Similarly, the humification parameter in EMH was significantly higher than those in EM and CM, indicating that ecological management and hedgerow contributed to the humification process in the farming land.
The relationship between soil chemical properties and DOM characteristics was further assessed based on Pearson’s correlation to explore the potential influences of environmental factors (Figure 2). The TN, TP, and available P were significantly positively correlated to the TFI values and the volumes of Regions I, III, IV, and V, suggesting the elevated contents of N and P may facilitate the DOM production and storage. A negative relationship between pH and DOM was observed. Previous studies have reported that increased soil acidity can greatly reduce DOM leaching [57]. Therefore, the acidification (approximately 0.4 unit) in this study may have decreased DOM leaching and hence led to higher DOC accumulation. Additionally, the management and hedgerow may alter the activities of several certain microbes, potentially influencing the production or consumption of DOM [58,59].
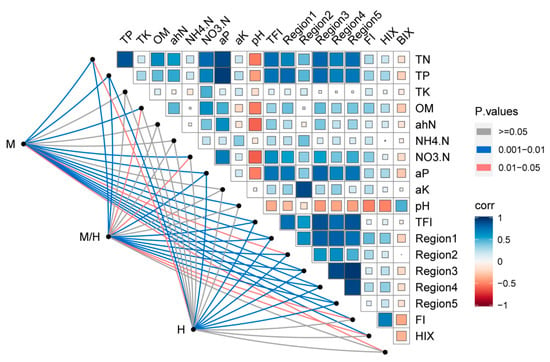
Figure 2.
Spearman’s correlations between environmental factors and soil dissolved organic matter (DOM) properties, and the results of Adonis analysis of management (M), hedgerow (H), and their interaction (M/H). Edge color denotes the statistical significance based on 999 permutations.
3.2. The Farming System Effects on Bacterial and Eukaryotic Communities
After assembling and quality filtering, a total of 1,062,863 (44,285 ± 4455 per sample) bacterial and 1,052,360 (43,848 ± 7495 per sample) eukaryotic high-quality sequences were obtained for the 24 soil samples. Sequence clustering yielded 3935 bacterial and 684 eukaryotic ASVs, respectively. The average Good’s coverage was 0.98 ± 0.03 and 0.99 ± 0.01, respectively, for the bacterial and eukaryotic communities, indicating the sufficient depth of sequencing for all samples.
For the three alpha-diversity indices estimated in this study, there was no statistically significant difference between the three farming systems for the eukaryotic community (Figure S1). While for the bacterial community, the EMH system showed significantly lower diversity compared to the other two systems, with statistically higher evenness (Figure S2). The three complementary non-parametric multivariate statistical tests further confirmed that management and hedgerow have no significant effect on eukaryotic alpha-diversity (Table S3). All bacterial parameters except richness were significantly influenced by hedgerow, and only bacterial evenness was statistically impacted by the interaction of ‘management × hedgerow’.
Principal coordinate analysis (PCoA) based on Bray–Curtis distances at the ASV level showed that bacterial and eukaryotic communities were significantly distinct between the three farming systems, indicating management and hedgerow were the drivers of microbial beta-diversity (Table 1, Figure 3). The Adonis test showed the bacterial and eukaryotic communities in the farmland managed conventionally and ecologically were on average 10.3% and 18.4% dissimilar, indicating a stronger management effect on eukarya than on bacteria. Additionally, the communities with or without hedgerows were 12.5% and 8.7% dissimilar, respectively, revealing that hedgerow has a stronger effect on bacteria. In fact, the hedgerow effect on the eukaryotic community was not statistically supported by the ANOSIM test (Table 1). The effects of management and hedgerow on dominant bacterial and eukaryotic phyla were further investigated. Even though no significant differences were observed for the relative abundances of the dominant bacterial phyla among the three farming systems, the compositions were statistically distinct (Table S4). Based on the Bray–Curtis distances, the three multivariate statistical tests indicated the management, hedgerow, and their interaction significantly influence the compositions of Actinobacteria, Proteobacteria, Chloroflexi, Gemmatimonadetes, Bacteroidetes, and Planctomycetes. While for eukaryotic phyla, the structure of Ascomycota was significantly impacted. To better illustrate the effects of management and hedgerow on the soil microbiota, we identified bacterial and eukaryotic OTUs that were specifically enriched in the three farming systems. There were 315, 261, and 189 bacterial ASVs significantly enriched in CM, EM, and EMH (Figure 4), respectively. For CM and EM systems, most of the enriched ASVs belonged to Proteobacteria, Acidobacteria, Chloroflexi, and Actinobacteria, while for the EMH system, the enriched ASVs were mostly belonged to Actinobacteria and Chloroflexi. Furthermore, there were 22, 25, and 11 eukaryotic ASVs significantly enriched in CM, EM, and EMH, respectively, which mostly belonged to Ascomycota. Based on LEfSe with a linear discriminant analysis score that exceeded a threshold of 2.0, a total of 58 bacterial and 26 eukaryotic families contributed to the significantly different abundances among the three systems (Figures S3 and S4). Thereinto, 23 bacterial and 10 eukaryotic families were identified as the biomarkers in the EM system, while 11 bacterial and 6 eukaryotic families in the EMH system.

Table 1.
Effects of management and hedgerow on bacterial and eukaryotic beta-diversity as assessed by the three complementary non-parametric multivariate statistical tests.
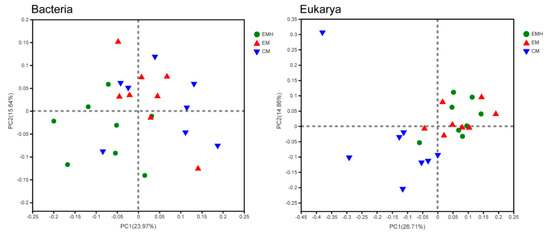
Figure 3.
Principal coordinate analysis (PCoA) based on Bray–Curtis distances showing the differences in bacterial and eukaryotic communities among the three farming systems.
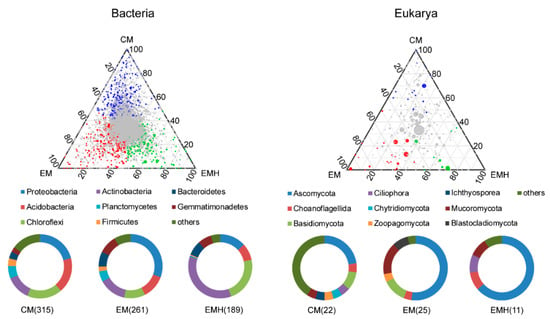
Figure 4.
Ternary plots showing the enriched bacterial and eukaryotic amplicon sequence variants (ASVs) in the three farming systems.
Following the rapid advances in DNA sequencing technologies, intensive studies have examined the responses of microbial communities to agricultural managements, which often observed ambiguous results. For instance, Hartmann et al. [7] and Degrune et al. [34] reported that organic farming showed significantly higher microbial diversity and lower evenness when compared to conventional management. However, Bonanomi et al. [33] observed statistically lower bacterial richness and Shannon diversity in a 20-years organic farming land compared to conventional farming land. Lupatini et al. [32] found that long-terming organic management had a positive effect on both fungal diversity and evenness, but showed no effects on the richness, diversity, and evenness of the protist community. In the current study, only hedgerow showed significant effects on bacterial diversity and evenness, while the management had no effects. Therefore, it is difficult to draw a robust conclusion on the effects of agricultural management on the bulk microbial diversity. One potential reason is that the pedological contexts, agricultural strategies, and sampling designs varied greatly across different studies. For example, previous studies indicated the types of organic input and the crop rotations were the important farming practices affecting soil microbial communities [60]. It has been reported that the response of bacterial evenness was different between short—and long—term organic farming, suggesting that the drawn conclusions strongly depended on the experimental designs [32]. Additionally, although significant differences in the relative abundances of several dominant taxa, including Proteobacteria, Actinobacteria, and Ascomycota, were not observed among the three farming practices, the compositions of these phyla were significantly influenced by management and hedgerow. All these phyla have generally been described as important organic matter decomposers and respond fast under organic carbon-rich conditions [61,62]. The changes in the structures may be strongly linked to the elevated DOM in EM and EMH systems.
3.3. The Farming System Effects on Predictive Functional Profiling
In addition to the composition analysis of the microbial community, the functional profiles based on bacterial and eukaryotic gene data were predicted using FAPROTAX and FUNGuild. The pattern of predicted functional profiles under different farming systems was visualized using PCoA based on the Bray–Curtis dissimilarity (Figure 5). A close clustering of EM and CM samples on bacterial functions was observed, which was separated from the EMH samples. The primary principal coordinate explained 74.4% of the total variance. The three complementary non-parametric multivariate statistical tests further indicated that hedgerow significantly (p = 0.001) influenced the bacterial functions (Table S5). Furthermore, the most prominent predicted functions obtained via FAPROTAX analysis were chemoheterotrophy (37.7%) and aerobic chemoheterotrophy (35.3%), and the relative abundances of these two categories were higher in EMH and EM soil than in the CM soil (Figure S5). The abundances of the category nitrification, aerobic nitrite oxidation, and aerobic ammonia oxidation were also relatively higher in EMH and EM soil samples. It has been well documented that chemoheterotrophic bacteria need to obtain carbon and energy from the oxidation of soil organic compounds [63]. The higher abundances of chemoheterotrophy and aerobic chemoheterotrophy in EM and EMH soils potentially suggested that the contents of available organic matters were higher in these two farming systems, which can be explained by the higher TFI values and area volumes of Regions I–IV. Nitrification, i.e., the conversion of ammonium via nitrite to nitrate, coupled with aerobic nitrite oxidation ammonia oxidation, were relatively higher in EM and EMH systems, likely indicating that ecological management could facilitate the process of nitrogen fixation. For FUNGuild analysis, the CM samples were separated with EM and EMH along the primary coordinate, which explained 47.58% of the total variance. The multivariate statistical tests indicated that the profiles of the eukaryotic trophic mode were statistically (p = 0.001) impacted by management and the interaction of management and hedgerow. The dominant trophic modes, including pathotrophs and saprotrophs, also exhibited high variability in relative abundance among the three farming systems (Figure S6). Through the functional prediction, we can conclude the agricultural strategies influence not only the composition of soil microbiome but also the microbial functions in soil. However, it should be noted that the classification of the microbial ASVs into functional groups was performed based on the previous literatures, which may show several limitations. For instance, only small parts of the microbial ASVs, 1569 bacterial and 99 eukaryotic ASVs in this study, were classified into the functional groups. Additionally, previous studies also indicated the function from cultured members of one certain taxon may be falsely generalized to all the members [50,61].
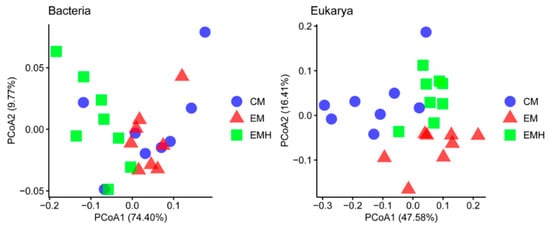
Figure 5.
Principal coordinate analysis (PCoA) based on Bray–Curtis distances showing the differences in predicted functional profiles among the three farming systems.
3.4. Relationship between Soil Microbial Communities, Soil Properties, and DOM Composition
The redundancy analysis (RDA) and VPA were used to estimate the contributions of soil environmental properties and DOM composition to the soil microbial communities (Figure 6). For the EEM results, only the area volumes of Regions I–V were used. While for soil environmental properties, TK, available K, alkali-hydrolytic N, and ammonium were not included, due to that management or hedgerow has no effects on these parameters. The soil DOM compositions and combined environmental parameters explained 35.60% and 32.22% of bacterial community variation, respectively. For the eukaryotic community, the DOM and environmental parameters explained 23.92% and 19.06%, respectively. The results strongly suggested DOM was responsible for shaping the bacterial and eukaryotic community structures.
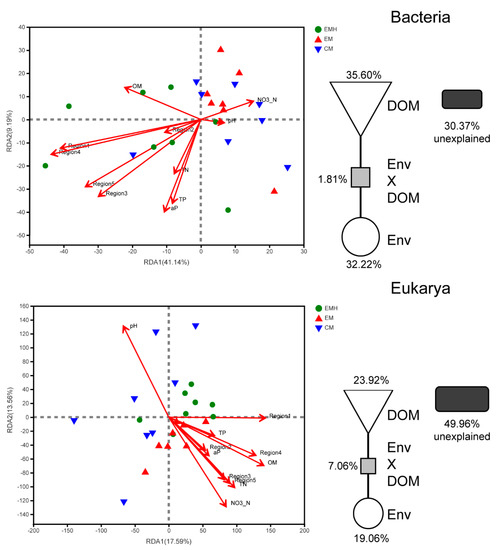
Figure 6.
The redundancy analysis (RDA) and variance partitioning analysis (VPA) analysis showing the relationship between environmental factors, soil dissolved organic matter (DOM), and soil microbial communities.
To further investigate the interconnections between soil microbial communities and abiotic factors, the coefficient analysis based on the Spearman correlation between family-level relative abundances and the environmental variables was carried out (Figure 7). In general, more positive correlations were observed between bacterial families and environmental variables, whereas more negative associations were found for eukaryotic taxa. The family Beijerinckiaceae, Pseudonocardiaceae, Azospirillaceae, and Streptomycetaceae were significantly (p < 0.05) positively correlated with the area volumes of Regions I–V, potentially suggesting that these taxa may play an important role in the nitrogen and carbon cycles. In fact, members of these bacteria are known to be involved in the degradation and conversion of organic compounds, and have been widely observed in composts or compost-amended soils. For instance, Silva et al. [64] reported that Pseudonocardiaceae and Streptomycetaceae were significantly abundant in the mature compost, which contributed to the high contents of humic-like acids. Additionally, Azospirillaceae has been reported as free-living nitrogen-fixing bacteria that is associated with promoting plant growth [65], indicating a high content of soil DOM may be beneficial for crop growth. Furthermore, the FAPROTAX analysis also indicated ASVs belonging to these families contributed to the functions of chemoheterotrophy and aerobic chemoheterotrophy. For eukarya, Sympoventuriaceae, Cucurbitariaceae, and Stachybotryaceae were statistically (p < 0.05) negatively correlated with the volume of DOM. Previous studies have indicated members of these eukarya can be saprobic or parasitic [66]. The negative associations may suggest that DOM induced in EM and EMH systems potentially controls the fungal pathogens. Furthermore, strong positive correlations between DOM and the family Rhizinaceae, Nectriaceae, and Arachnomycetaceae (p < 0.05) were observed, which may be attributed to the fact that these taxa are complex carbon-decomposing fungi and the elevated DOM contributes to their abundances [66]. In summary, strong correlations between specific microbial taxa and DOM were observed, suggesting that ecological management and hedgerow can promote the presence of beneficial microbiome and hence facilitate the nutrient transformation and pathogen control.
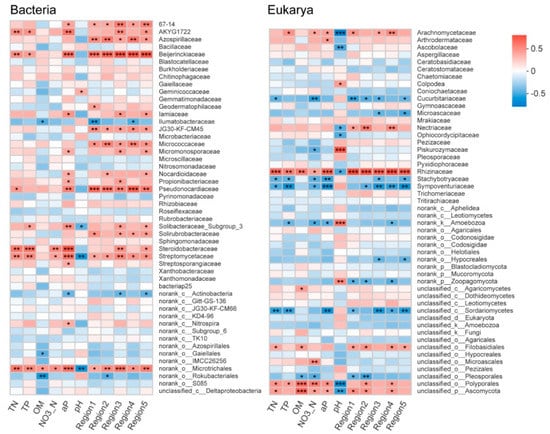
Figure 7.
Spearman’s correlations between environmental factors, soil DOM, and microbial taxa.
It must be noted that only the fluorescence characteristics of DOM obtained by EEM was considered to investigate the effects of ecological management on soil in this study. Other technologies, such as Fourier-Transform Ion-Cyclotron-Resonance Mass Spectrometry (FT-ICR-MS), which can analyze the molecular composition of DOM, may can be used for further studies to help us to predict the carbon cycling under ecological management. Additionally, climate variables (e.g., precipitation and temperature) may also affect the soil microenvironmental conditions and microbial community structure and activity. The effects of ecological management under different climate conditions should be further studied.
4. Conclusions
Taking the advantage of EEM fluorescence and molecular biology informatics, the characteristics of soil DOM and microbial communities under different agricultural strategies were explored. Our results indicated that long-term ecological management and hedgerow significantly increased the content of soil DOM compositions and the degree of humification. Compared to the conventional management, the organic farming system increased the evenness of bacterial community, whereas the alpha-diversity of eukaryotic community was similar among the three farming systems. Both management and hedgerow contributed to the distinct bacterial composition in the three farming systems, however, no significant effects of hedgerow on the eukaryotic community were observed. Higher abundances of functions related to nutrient cycles were observed in EM and EMH systems. In addition, specific microbial taxa, which were involved in organic decomposition and nutrient transformation, were found to be strongly correlated with the soil DOM compositions, suggesting that ecological management and hedgerow may be beneficial for improving nutrient availability and crop growth.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/10/9/1316/s1.
Author Contributions
R.S.: study design, data collection; C.S.: data analysis, data interpretation; Y.S.: iterature search; Q.W.: figures; H.L.: study design; J.W.: data analysis, study design, writing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polaasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.A.; DeFries, R.; Asner, G.P.; Barford, C.; Bonan, G.; Carpenter, S.R.; Chapin, F.S.; Coe, M.T.; Daily, G.C.; Gibbs, H.K.; et al. Global Consequences of Land Use. Science 2005, 309, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a Cultivated Planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G.A. An Underground Revolution: Biodiversity and Soil Ecological Engineering for Agricultural Sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Gomiero, T.; Pimentel, D.; Paoletti, M.G. Environmental Impact of Different Agricultural Management Practices: Conventional vs. Organic Agriculture. Crit. Rev. Plant Sci. 2011, 30, 95–124. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural Intensification Reduces Microbial Network Complexity and the Abundance of Keystone Taxa in Roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct Soil Microbial Diversity under Long-Term Organic and Conventional Farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.W.I.; Torn, M.S.; Abiven, S.; Dittmar, T.; Guggenberger, G.; Janssens, I.A.; Kleber, M.; Kögel-Knabner, I.; Lehmann, J.; Manning, D.A.C.; et al. Persistence of Soil Organic Matter as an Ecosystem Property. Nature 2011, 478, 49–56. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Z.; Ye, Q.; Liang, Y.; Liu, M.; Dang, Z.; Wang, Y.; Liu, C. Chemodiversity of Soil Dissolved Organic Matter. Environ. Sci. Technol. 2020, 54, 6174–6184. [Google Scholar] [CrossRef]
- Kellerman, A.M.; Kothawala, D.N.; Dittmar, T.; Tranvik, L.J. Persistence of Dissolved Organic Matter in Lakes Related to Its Molecular Characteristics. Nat. Geosci. 2015, 8, 454–457. [Google Scholar] [CrossRef]
- Lehmann, J.; Kleber, M. The Contentious Nature of Soil Organic Matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C.M. Agricultural Management and Plant Selection Interactively Affect Rhizosphere Microbial Community Structure and Nitrogen Cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef]
- Fließbach, A.; Oberholzer, H.R.; Gunst, L.; Mäder, P. Soil Organic Matter and Biological Soil Quality Indicators after 21 Years of Organic and Conventional Farming. Agric. Ecosyst. Environ. 2007, 118, 273–284. [Google Scholar] [CrossRef]
- Gattinger, A.; Muller, A.; Haeni, M.; Skinner, C.; Fliessbach, A.; Buchmann, N.; Mäder, P.; Stolze, M.; Smith, P.; Scialabba, N.E.H.; et al. Enhanced Top Soil Carbon Stocks under Organic Farming. Proc. Natl. Acad. Sci. USA 2012, 109, 18226. [Google Scholar] [CrossRef] [PubMed]
- Lal, R. Soil Carbon Management and Climate Change. Carbon Manag. 2013, 4, 439–462. [Google Scholar] [CrossRef]
- Cotrufo, M.F.; Soong, J.L.; Horton, A.J.; Campbell, E.E.; Haddix, M.L.; Wall, D.H.; Parton, W.J. Formation of Soil Organic Matter via Biochemical and Physical Pathways of Litter Mass Loss. Nat. Geosci. 2015, 8, 776–779. [Google Scholar] [CrossRef]
- Doetterl, S.; Stevens, A.; Six, J.; Merckx, R.; Van Oost, K.; Casanova Pinto, M.; Casanova-Katny, A.; Muñoz, C.; Boudin, M.; Zagal Venegas, E.; et al. Soil Carbon Storage Controlled by Interactions between Geochemistry and Climate. Nat. Geosci. 2015, 8, 780–783. [Google Scholar] [CrossRef]
- Ghimire, R.; Lamichhane, S.; Acharya, B.S.; Bista, P.; Sainju, U.M. Tillage, Crop Residue, and Nutrient Management Effects on Soil Organic Carbon in Rice-Based Cropping Systems: A Review. J. Integr. Agric. 2017, 16, 1–15. [Google Scholar] [CrossRef]
- Borisover, M.; Lordian, A.; Levy, G.J. Water-Extractable Soil Organic Matter Characterization by Chromophoric Indicators: Effects of Soil Type and Irrigation Water Quality. Geoderma 2012, 179, 28–37. [Google Scholar] [CrossRef]
- Chantigny, M.H.; Harrison-Kirk, T.; Curtin, D.; Beare, M. Temperature and Duration of Extraction Affect the Biochemical Composition of Soil Water-Extractable Organic Matter. Soil Biol. Biochem. 2014, 75, 161–166. [Google Scholar] [CrossRef]
- Gao, J.; Liang, C.; Shen, G.; Lv, J.; Wu, H. Spectral Characteristics of Dissolved Organic Matter in Various Agricultural Soils throughout China. Chemosphere 2017, 176, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel Soil Bacteria Possess Diverse Genes for Secondary Metabolite Biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of Biodiversity in the Earth Mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Huang, Y.; Feng, S.Z.; Ge, Y.H.; Zhang, W.; He, X.; Wang, K. Soil organic carbon mineralization with fresh organic substrate and inorganic carbon additions in a red soil is controlled by fungal diversity along a pH gradient. Geoderma 2018, 321, 79–89. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6. [Google Scholar] [CrossRef]
- de Vries, F.T.; Thébault, E.; Liiri, M.; Birkhofer, K.; Tsiafouli, M.A.; Bjørnlund, L.; Jørgensen, H.B.; Brady, M.V.; Christensen, S.; de Ruiter, P.C.; et al. Soil Food Web Properties Explain Ecosystem Services across European Land Use Systems. Proc. Natl. Acad. Sci. USA 2013, 110, 14296–14301. [Google Scholar] [CrossRef]
- Ding, L.J.; Su, J.Q.; Sun, G.X.; Wu, J.S.; Wei, W.X. Increased Microbial Functional Diversity under Long-Term Organic and Integrated Fertilization in a Paddy Soil. Appl. Microbiol. Biotechnol. 2018, 102, 1969–1982. [Google Scholar] [CrossRef]
- Postma-Blaauw, M.B.; de Goede, R.G.M.; Bloem, J.; Faber, J.H.; Brussaard, L. Soil Biota Community Structure and Abundance under Agricultural Intensification and Extensification. Ecology 2010, 91, 460–473. [Google Scholar] [CrossRef]
- Coller, E.; Cestaro, A.; Zanzotti, R.; Bertoldi, D.; Pindo, M.; Larger, S.; Albanese, D.; Mescalchin, E.; Donati, C. Microbiome of Vineyard Soils Is Shaped by Geography and Management. Microbiome 2019, 7, 140. [Google Scholar] [CrossRef]
- Lupatini, M.; Korthals, G.W.; de Hollander, M.; Janssens, T.K.S.; Kuramae, E.E. Soil Microbiome Is More Heterogeneous in Organic than in Conventional Farming System. Front. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Lupatini, M.; Korthals, G.W.; Roesch, L.F.W.; Kuramae, E.E. Long-Term Farming Systems Modulate Multi-Trophic Responses. Sci. Total Environ. 2019, 646, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; De Filippis, F.; Cesarano, G.; La Storia, A.; Ercolini, D.; Scala, F. Organic Farming Induces Changes in Soil Microbiota That Affect Agro-Ecosystem Functions. Soil Biol. Biochem. 2016, 103, 327–336. [Google Scholar] [CrossRef]
- Degrune, F.; Boeraeve, F.; Dufrêne, M.; Cornélis, J.T.; Frey, B.; Hartmann, M. The Pedological Context Modulates the Response of Soil Microbial Communities to Agroecological Management. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef]
- Holden, J.; Grayson, R.P.; Berdeni, D.; Bird, S.; Chapman, P.J.; Edmondson, J.L.; Firbank, L.G.; Helgason, T.; Hodson, M.E.; Hunt, S.F.P.; et al. The Role of Hedgerows in Soil Functioning within Agricultural Landscapes. Agric. Ecosyst. Environ. 2019, 273, 1–12. [Google Scholar] [CrossRef]
- Marshall, E.J.P.; West, T.M.; Kleijn, D. Impacts of an Agri-Environment Field Margin Prescription on the Flora and Fauna of Arable Farmland in Different Landscapes. Agric. Ecosyst. Environ. 2006, 113, 36–44. [Google Scholar] [CrossRef]
- Morandin, L.A.; Kremen, C. Hedgerow Restoration Promotes Pollinator Populations and Exports Native Bees to Adjacent Fields. Ecol. Appl. 2013, 23, 829–839. [Google Scholar] [CrossRef]
- Morandin, L.A.; Long, R.F.; Kremen, C. Pest Control and Pollination Cost–Benefit Analysis of Hedgerow Restoration in a Simplified Agricultural Landscape. J. Econ. Entomol. 2016, 109, 1020–1027. [Google Scholar] [CrossRef]
- Spaans, F.; Caruso, T.; Hammer, E.C.; Montgomery, I. Trees in Trimmed Hedgerows but Not Tree Health Increase Diversity of Oribatid Mite Communities in Intensively Managed Agricultural Land. Soil Biol. Biochem. 2019, 138, 107568. [Google Scholar] [CrossRef]
- Chai, L.; Huang, M.; Fan, H.; Wang, J.; Jiang, D.; Zhang, M.; Huang, Y. Urbanization Altered Regional Soil Organic Matter Quantity and Quality: Insight from Excitation Emission Matrix (EEM) and Parallel Factor Analysis (PARAFAC). Chemosphere 2019, 220, 249–258. [Google Scholar] [CrossRef]
- Huguet, A.; Vacher, L.; Relexans, S.; Saubusse, S.; Froidefond, J.M.; Parlanti, E. Properties of Fluorescent Dissolved Organic Matter in the Gironde Estuary. Org. Geochem. 2009, 40, 706–719. [Google Scholar] [CrossRef]
- McKnight, D.M.; Boyer, E.W.; Westerhoff, P.K.; Doran, P.T.; Kulbe, T.; Andersen, D.T. Spectrofluorometric Characterization of Dissolved Organic Matter for Indication of Precursor Organic Material and Aromaticity. Limnol. Oceanogr. 2001, 46, 38–48. [Google Scholar] [CrossRef]
- Zsolnay, A.; Baigar, E.; Jimenez, M.; Steinweg, B.; Saccomandi, F. Differentiating with Fluorescence Spectroscopy the Sources of Dissolved Organic Matter in Soils Subjected to Drying. Chemosphere 1999, 38, 45–50. [Google Scholar] [CrossRef]
- Chen, W.; Westerhoff, P.; Leenheer, J.A.; Booksh, K. Fluorescence Excitation−Emission Matrix Regional Integration to Quantify Spectra for Dissolved Organic Matter. Environ. Sci. Technol. 2003, 37, 5701–5710. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Matson, P.A.; Parton, W.J.; Power, A.G.; Swift, M.J. Agricultural Intensification and Ecosystem Properties. Science 1997, 277, 504–509. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Guo, X.; Feng, J.; Shi, Z.; Zhou, X.; Yuan, M.; Tao, X.; Hale, L.; Yuan, T.; Wang, J.; Qin, Y.; et al. Climate Warming Leads to Divergent Succession of Grassland Microbial Communities. Nat. Clim. Chang. 2018, 8, 813–818. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Williams, A.; Börjesson, G.; Hedlund, K. The Effects of 55 Years of Different Inorganic Fertiliser Regimes on Soil Properties and Microbial Community Composition. Soil Biol. Biochem. 2013, 67, 41–46. [Google Scholar] [CrossRef]
- Xia, L.; Hoermann, G.; Ma, L.; Yang, L. Reducing Nitrogen and Phosphorus Losses from Arable Slope Land with Contour Hedgerows and Perennial Alfalfa Mulching in Three Gorges Area, China. CATENA 2013, 110, 86–94. [Google Scholar] [CrossRef]
- Oshunsanya, S.O.; Li, Y.; Yu, H. Vetiver Grass Hedgerows Significantly Reduce Nitrogen and Phosphorus Losses from Fertilized Sloping Lands. Sci. Total Environ. 2019, 661, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Barthod, J.; Rumpel, C.; Dignac, M.F. Composting with Additives to Improve Organic Amendments. A Review. Agron. Sustain. Dev. 2018, 38, 17. [Google Scholar] [CrossRef]
- He, X.S.; Xi, B.D.; Li, X.; Pan, H.W.; An, D.; Bai, S.G.; Li, D.; Cui, D.Y. Fluorescence Excitation–Emission Matrix Spectra Coupled with Parallel Factor and Regional Integration Analysis to Characterize Organic Matter Humification. Chemosphere 2013, 93, 2208–2215. [Google Scholar] [CrossRef]
- Hagedorn, F.; Kammer, A.; Schmidt, M.W.I.; Goodale, C.L. Nitrogen Addition Alters Mineralization Dynamics of 13C-Depleted Leaf and Twig Litter and Reduces Leaching of Older DOC from Mineral Soil. Glob. Chang. Biol. 2012, 18, 1412–1427. [Google Scholar] [CrossRef]
- Wang, J.J.; Bowden, R.D.; Lajtha, K.; Washko, S.E.; Wurzbacher, S.J.; Simpson, M.J. Long-Term Nitrogen Addition Suppresses Microbial Degradation, Enhances Soil Carbon Storage, and Alters the Molecular Composition of Soil Organic Matter. Biogeochemistry 2019, 142, 299–313. [Google Scholar] [CrossRef]
- Wang, J.J.; Liu, Y.; Bowden, R.D.; Lajtha, K.; Simpson, A.J.; Huang, W.L.; Simpson, M.J. Long-Term Nitrogen Addition Alters the Composition of Soil-Derived Dissolved Organic Matter. ACS Earth Space Chem. 2020, 4, 189–201. [Google Scholar] [CrossRef]
- Lori, M.; Symnaczik, S.; Mäder, P.; Deyn, G.D.; Gattinger, A. Organic Farming Enhances Soil Microbial Abundance and Activity—A Meta-Analysis and Meta-Regression. PLoS ONE 2017, 12, e0180442. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative Metagenomic, Phylogenetic and Physiological Analyses of Soil Microbial Communities across Nitrogen Gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Bahkali, A.H.; Guo, L.D.; Hyde, K.D. A Molecular, Morphological and Ecological Re-Appraisal of Venturiales―A New Order of Dothideomycetes. Fungal Divers. 2011, 51, 249–277. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.E.F.; Lopes, A.R.; Cunha-Queda, A.C.; Nunes, O.C. Comparison of the Bacterial Composition of Two Commercial Composts with Different Physicochemical, Stability and Maturity Properties. Waste Manag. 2016, 50, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Sridevi, G.; Minocha, R.; Turlapati, S.A.; Goldfarb, K.C.; Brodie, E.L.; Tisa, L.S.; Minocha, S.C. Soil Bacterial Communities of a Calcium-Supplemented and a Reference Watershed at the Hubbard Brook Experimental Forest (HBEF), New Hampshire, USA. FEMS Microbiol. Ecol. 2012, 79, 728–740. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, B.X.; Ren, H.; Zhang, J. Composition and Functional Diversity of Microbial Community across a Mangrove-Inhabited Mudflat as Revealed by 16S RDNA Gene Sequences. Sci. Total Environ. 2018, 633, 518–528. [Google Scholar] [CrossRef]
- McGee, K.M.; Eaton, W.D.; Shokralla, S.; Hajibabaei, M. Determinants of Soil Bacterial and Fungal Community Composition toward Carbon-Use Efficiency across Primary and Secondary Forests in a Costa Rican Conservation Area. Microb. Ecol. 2019, 77, 148–167. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).