Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Protein Extraction
2.3. Two-Dimensional Gel Electrophoresis
2.4. Gel Analysis
2.5. In-Gel Digestion and MALDI-TOF/TOF-MS Analysis
2.6. Protein Identification and Database Searching
2.7. Statistical Analysis
3. Results
3.1. Morphological Traits as Influenced by Low-P and Optimum-P Treatments
3.2. Number of Differentially Expressed Proteins
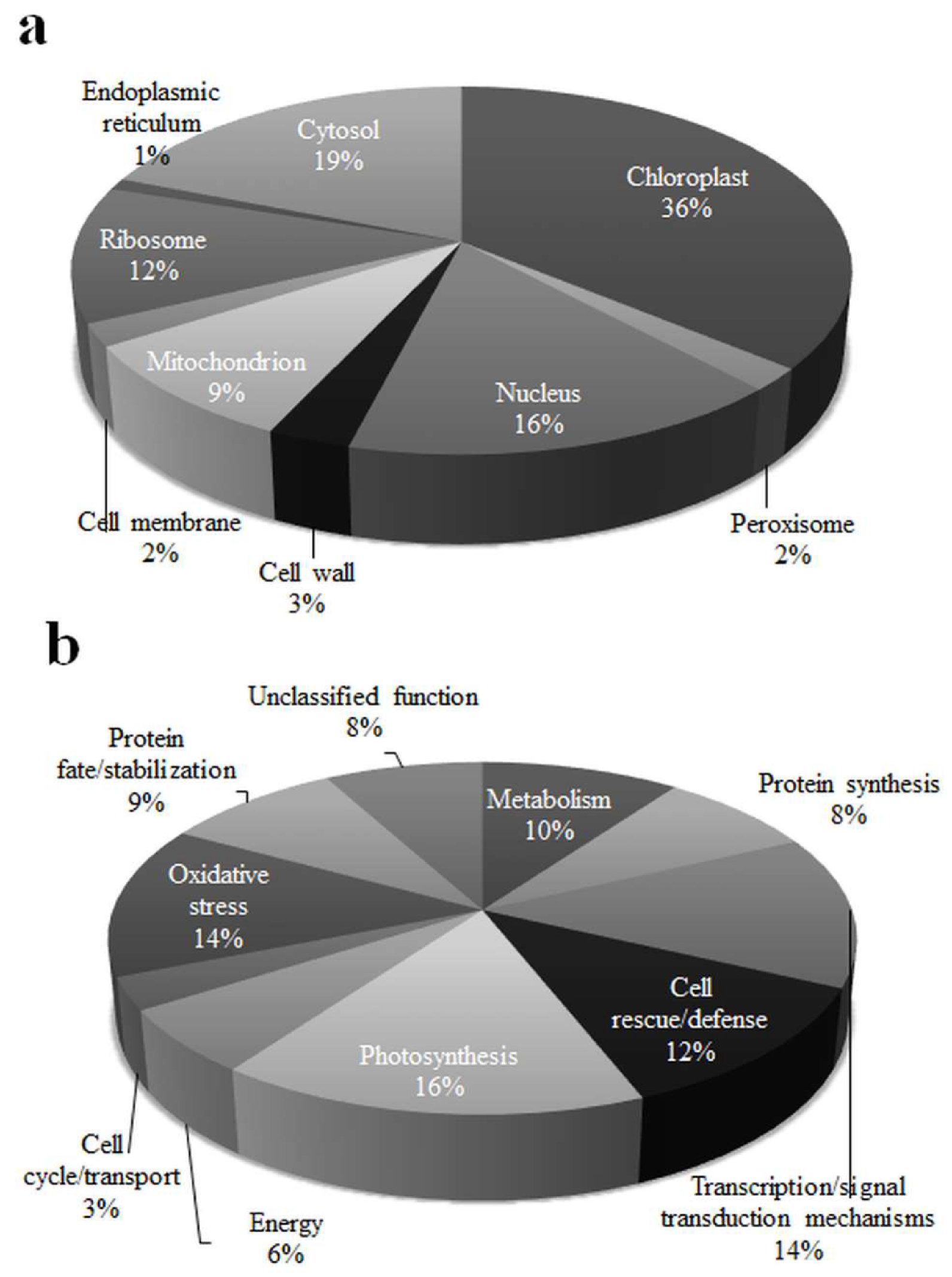
3.3. Spatial and Functional Categorization of Differentially Expressed Proteins
3.4. Differentially Expressed Proteins of Rice Cultivars under Low P Condition
3.4.1. Common Differentially Expressed Proteins of Cv. Nagina and Cv. Panvel under Low P Condition
3.4.2. Differentially Expressed Proteins of Cv. Panvel only under low P Condition
3.4.3. Differentially Expressed Proteins of Cv. Nagina only under Low P Condition
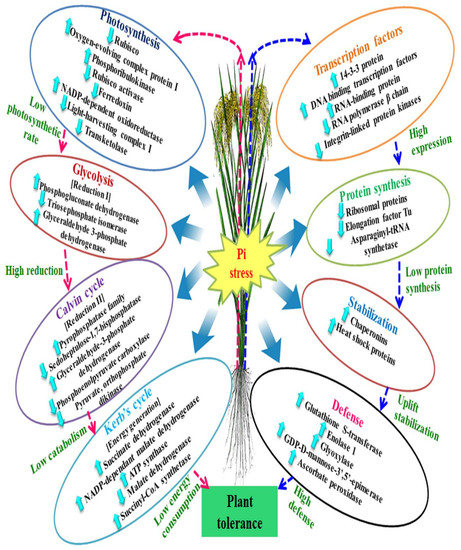
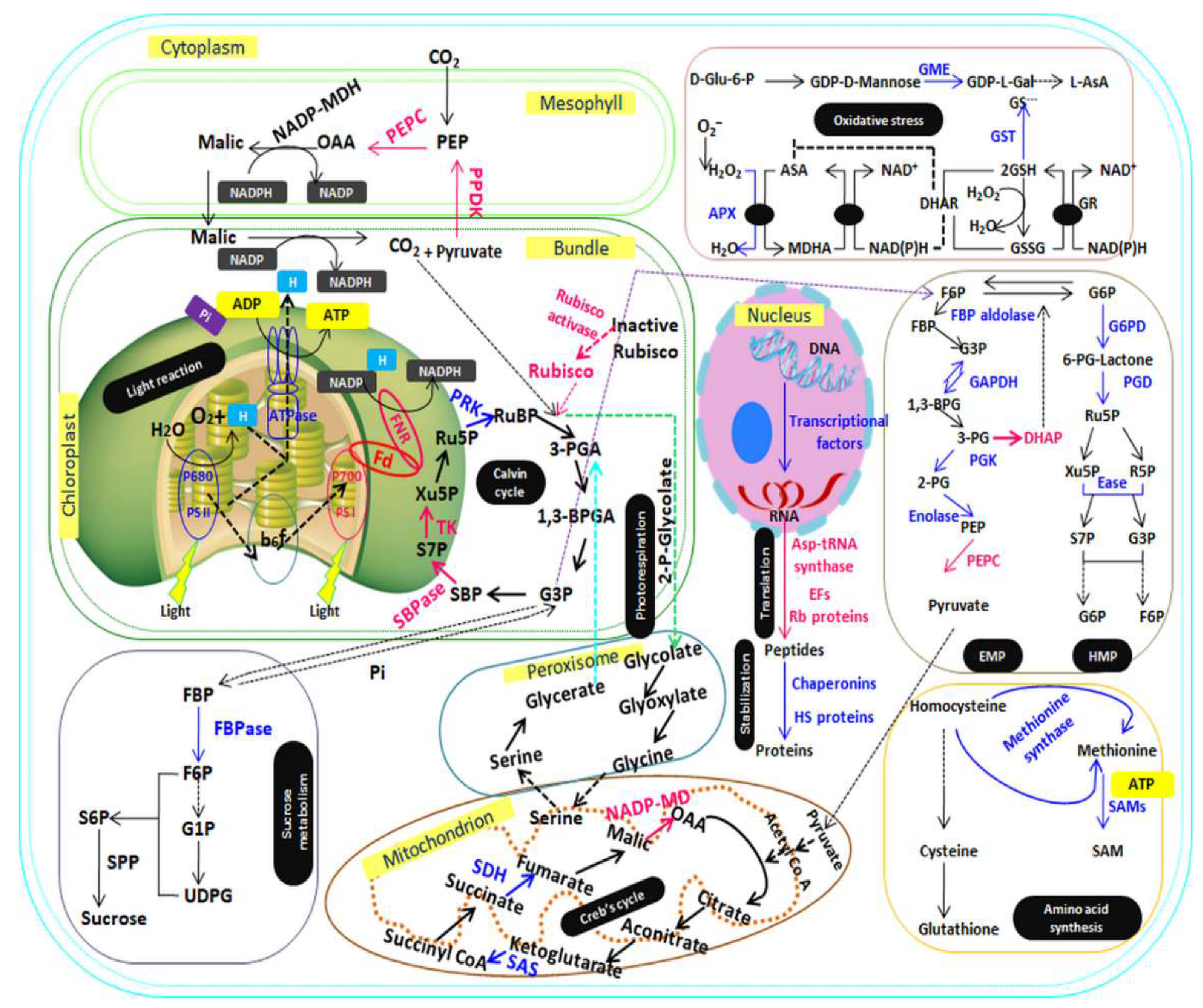
4. Discussion
4.1. Expression Pattern of the Proteins of Energy Metabolism under P-deprivation
4.2. Expression Pattern of the Proteins Involved in Transcription and Translation
4.3. Photosynthesis and CO2 Regulation under P-deficiency
4.4. Expression Proteins of Antioxidant Defense System under P-deficiency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PUE | Phosphate utilization efficiency |
| 2DE | Two-dimensional gel electrophoresis |
| DEP | Differentially expressed proteins |
| MALDI-TOF | Matrix-assisted laser desorption/ionization-time of flight |
References
- Fischer, R.A.; Edmeades, G.O. Breeding and Cereal Yield Progress. Crop. Sci. 2010, 50, S-85–S-98. [Google Scholar] [CrossRef]
- Lynch, J.P. Root Phenes for Enhanced Soil Exploration and Phosphorus Acquisition: Tools for Future Crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Doberman, A.; Fairhurst, T. Rice: Nutrient Disorders and Nutrient Management; IRRI & PPI & PPIC: Makati, Philippines; Singapore, 2000. [Google Scholar]
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Van Kauwenbergh, S.J. World Phosphate Rock Reserves and Resources; IFDC: Muscle Shoals, AL, USA, 2010. [Google Scholar]
- Dawe, D. The potential role of biological nitrogen fixation in meeting future demand for rice and fertilizer. In The Quest for Nitrogen Fixation in Rice; Ladha, J.K., Reddy, P.M., Eds.; International Rice Research Institute: Los Banos, Philippines, 2000; pp. 93–118. [Google Scholar]
- Van Der Velde, M.; Folberth, C.; Balkovič, J.; Ciais, P.; Fritz, S.; Janssens, I.A.; Obersteiner, M.; See, L.; Skalský, R.; Xiong, W.; et al. African crop yield reductions due to increasingly unbalanced Nitrogen and Phosphorus consumption. Glob. Chang. Boil. 2014, 20, 1278–1288. [Google Scholar] [CrossRef]
- Chiera, J.; Thomas, J.; Rufty, T.W. Leaf initiation and development in soybean under phosphorus stress. J. Exp. Bot. 2002, 53, 473–481. [Google Scholar] [CrossRef]
- Brooks, A. Effects of Phosphorus Nutrition on Ribulose-1, 5-Bisphosphate Carboxylase Activation, Photosynthetic Quantum Yield and Amounts of Some Calvin-Cycle Metabolites in Spinach Leaves. Funct. Plant Boil. 1986, 13, 221. [Google Scholar] [CrossRef]
- Tang, X.R.; Yu, T.Q. Effects and mechanisms of P and K nutrients on yield and protein content of fodder rice. Agric. Sci. China 2002, 1, 432–437. [Google Scholar]
- Qiu, H.; Liu, C.; Yu, T.; Mei, X.; Wang, G.; Wang, J.; Cai, Y. Identification of QTL for acid phosphatase activity in root and rhizosphere soil of maize under low phosphorus stress. Euphytica 2014, 197, 133–143. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, J.; Han, Z.; Yang, J.; Ge, C.; Wu, Q. Revealing new insights into different phosphorus-starving responses between two maize (Zea mays) inbred lines by transcriptomic and proteomic studies. Sci. Rep. 2017, 7, 44294. [Google Scholar] [CrossRef]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite Profiling of Low-P Tolerant and Low-P Sensitive Maize Genotypes under Phosphorus Starvation and Restoration Conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liang, W.; Sturrock, C.J.; Pandey, B.K.; Giri, J.; Mairhofer, S.; Wang, D.; Müller, L.; Tan, H.; York, L.M.; et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 2018, 9, 2346. [Google Scholar] [CrossRef]
- Alexova, R.; Millar, A.H. Proteomics of phosphate use and deprivation in plants. Proteomics 2013, 13, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Wang, Y.; Lee, C.H.; Mun, B.G.; Kim, P.J.; Lee, S.Y.; Kim, Y.C.; Kang, K.Y.; Rakwal, R.; Agrawal, G.K.; et al. A comparative proteomics survey of proteins responsive to phosphorous starvation in roots of hydroponically-grown rice seedlings. J. Korean Soc. Appl. Biol. 2011, 54, 667–677. [Google Scholar] [CrossRef]
- Torabi, S.; Wissuwa, M.; Heidari, M.; Naghavi, M.R.; Gilany, K.; Hajirezaei, M.-R.; Omidi, M.; Yazdi-Samadi, B.; Ismail, A.M.; Salekdeh, G.H. A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 2009, 9, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S. Routine Procedure for Growing Rice Plants in Culture Solution. In Laboratory Manual for Physiological Studies of Rice; Yoshida, S., Forno, D.A., Cock, J.H., Eds.; International Rice Research Institute: Los Banos, Philippines, 1976; pp. 61–66. [Google Scholar]
- Wu, C.; Wang, Q.; Xue, S.; Pan, W.; Lou, L.; Li, D.; Hartley, W. Do aeration conditions affect arsenic and phosphate accumulation and phosphate transporter expression in rice (Oryza sativa L.)? Environ. Sci. Pollut. Res. 2016, 25, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.; Pandey, R.N.; Sahoo, R.N.; Gupta, V.K.; Datta, S.C.; Kumar, D. Monitoring nitrogen, phosphorus and sulphur in hybrid rice (Oryza sativa L.) using hyperspectral remote sensing. Precis. Agric. 2016, 18, 736–761. [Google Scholar] [CrossRef]
- Syers, J.K.; Johnston, A.E.; Curtin, D. Efficiency of soil and fertilizer phosphorus use. FAO Fertil. Plant Nutr. Bull. 2008, 18, 108. [Google Scholar]
- Isaacson, T.; Damasceno, C.M.B.; Saravanan, R.S.; He, Y.; Catalá, C.; Saladié, M.; Rose, J.K.C. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Boil. Chem. 1975, 250, 4007–4021. [Google Scholar]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2006, 35, 5–12. [Google Scholar] [CrossRef]
- Wu, C.H. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006, 34, D187–D191. [Google Scholar] [CrossRef]
- Guda, C. pTARGET: A web server for predicting protein subcellular localization. Nucleic Acids Res. 2006, 34, W210–W213. [Google Scholar] [CrossRef] [PubMed]
- Chankaew, S.; Monkham, T.; Pinta, W.; Sanitchon, J.; Kaewpradit, W.; Srinives, P. Screening tolerance to phosphorus deficiency and validation of phosphorus pptake 1 (Pup1) gene linked markers in Thai indigenous upland rice germplasm. Agronomy 2019, 9, 81. [Google Scholar] [CrossRef]
- Wissuwa, M.; Ae, N. Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed. 2001, 120, 43–48. [Google Scholar] [CrossRef]
- Vejchasarn, P.; Lynch, J.P.; Brown, K.M. Genetic Variability in Phosphorus Responses of Rice Root Phenotypes. Rice 2016, 9, 29. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, H.; Tao, P.; Chen, H. Comparative Proteomic Analyses Provide New Insights into Low Phosphorus Stress Responses in Maize Leaves. PLoS ONE 2014, 9, e98215. [Google Scholar] [CrossRef]
- Zhang, W.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. An Arabidopsis Purple Acid Phosphatase with Phytase Activity Increases Foliar Ascorbate1 [OA]. Plant Physiol. 2007, 146, 431–440. [Google Scholar] [CrossRef]
- Bar-Even, A. Daring metabolic designs for enhanced plant carbon fixation. Plant Sci. 2018, 273, 71–83. [Google Scholar] [CrossRef] [PubMed]
- De Porcellinis, A.J.; Nørgaard, H.; Brey, L.M.F.; Erstad, S.M.; Jones, P.R.; Heazlewood, J.L.; Sakuragi, Y. Overexpression of bifunctional fructose-1,6-bisphosphatase/sedoheptulose-1,7-bisphosphatase leads to enhanced photosynthesis and global reprogramming of carbon metabolism in Synechococcus sp. PCC 7002. Metab. Eng. 2018, 47, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Y.; Chang, Z.; Wang, S.; Ding, Y.; Ding, C. Transcriptomic analysis of field-grown rice (Oryza sativa L.) reveals responses to shade stress in reproductive stage. Plant Growth Regul. 2018, 84, 583–592. [Google Scholar] [CrossRef]
- Huang, S.; Millar, A.H. Succinate dehydrogenase: The complex roles of a simple enzyme. Curr. Opin. Plant Boil. 2013, 16, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Belt, K.; Van Aken, O.; Murcha, M.; Millar, A.H.; Huang, S. An Assembly Factor Promotes Assembly of Flavinated SDH1 into the Succinate Dehydrogenase Complex. Plant Physiol. 2018, 177, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, R. Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand. Photosynth. Res. 2018, 139, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Troncoso-Ponce, M.; Rivoal, J.; Dorion, S.; Sánchez, R.; Venegas-Calerón, M.; Moreno-Pérez, A.J.; Baud, S.; Garces, R.; Martínez-Force, E. Molecular and biochemical characterization of the sunflower (Helianthus annuus L.) cytosolic and plastidial enolases in relation to seed development. Plant Sci. 2018, 272, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Wu, Z.; Zhou, J.; Du, L.; Guo, L.; Wu, Y.; Wu, P. OsPTF1, a Novel Transcription Factor Involved in Tolerance to Phosphate Starvation in Rice1 [w]. Plant Physiol. 2005, 138, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, Q.; Sun, L.; He, Z. The Rice 14-3-3 Gene Family and its Involvement in Responses to Biotic and Abiotic Stress. DNA Res. 2006, 13, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Ludewig, U.; Hawkesford, M.J. Perspective on Wheat Yield and Quality with Reduced Nitrogen Supply. Trends Plant Sci. 2018, 23, 1029–1037. [Google Scholar] [CrossRef]
- Kudlicki, W.; Coffman, A.; Kramer, G.; Hardesty, B. Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing. J. Boil. Chem. 1997, 272, 32206–32210. [Google Scholar] [CrossRef]
- Yakobov, N.; Debard, S.; Fischer, F.; Senger, B.; Becker, H. Cytosolic aminoacyl-tRNA synthetases: Unanticipated relocations for unexpected functions. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2018, 1861, 387–400. [Google Scholar] [CrossRef]
- Hummel, M.; Cordewener, J.H.; de Groot, J.C.; Smeekens, S.; America, A.H.; Hanson, J. Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MS E proteomics. Proteomics 2012, 12, 1024–1038. [Google Scholar] [CrossRef]
- Graifer, D.M.; Karpova, G. Roles of ribosomal proteins in the functioning of translational machinery of eukaryotes. Biochimie 2015, 109, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cardona, T.; Shao, S.; Nixon, P.J. Enhancing photosynthesis in plants: The light reactions. Essays Biochem. 2018, 62, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Dalal, V.K.; Tripathy, B.C. Water-stress induced downsizing of light-harvesting antenna complex protects developing rice seedlings from photo-oxidative damage. Sci. Rep. 2018, 8, 5955. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.-N.; Miyagi, A.; Nishiyama, M.; Yoshida, K.; Hisabori, T.; Kawai-Yamada, M. Ferredoxin/thioredoxin system plays an important role in the chloroplastic NADP status of Arabidopsis. Plant J. 2018, 95, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Medrano, H.; Parry, M.A.J.; Socias, X.; Lawlor, D.W. Long term water stress inactivates Rubisco in subterranean clover. Ann. Appl. Boil. 1997, 131, 491–501. [Google Scholar] [CrossRef]
- Yousuf, P.Y.; Abd Allah, E.F.; Nauman, M.; Asif, A.; Hashem, A.; Alqarawi, A.A.; Ahmad, A. Responsive Proteins in Wheat Cultivars with Contrasting Nitrogen Efficiencies under the Combined Stress of High Temperature and Low Nitrogen. Genes 2017, 8, 356. [Google Scholar] [CrossRef]
- Henkes, S.; Sonnewald, U.; Badur, R.; Flachmann, R.; Stitt, M. A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 2001, 13, 535–551. [Google Scholar] [CrossRef]
- Kim, D.-W.; Rakwal, R.; Agrawal, G.K.; Jung, Y.-H.; Shibato, J.; Jwa, N.-S.; Iwahashi, Y.; Iwahashi, H.; Kim, D.H.; Shim, I.-S.; et al. A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 2005, 26, 4521–4539. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, D.; Tschaplinski, T.J.; Tuskan, G.A. Comparative genomics can provide new insights into the evolutionary mechanisms and gene function in CAM plants. J. Exp. Bot. 2019, 70, 6539–6547. [Google Scholar] [CrossRef]
- Liu, X.; Dai, C.; Zhou, J.; Ren, C.; Li, X.; Zhang, C.; Zhang, J. Phosphoenolpyruvate carboxylase regulation in C4-PEPC -expressing transgenic rice during early responses to drought stress. Physiol. Plant. 2016, 159, 178–200. [Google Scholar] [CrossRef]
- Zhang, Y.; Giuliani, R.; Zhang, Y.; Zhang, Y.; Araújo, W.L.; Wang, B.; Liu, P.; Sun, Q.; Cousins, A.; Edwards, G.; et al. Characterization of maize leaf pyruvate orthophosphate dikinase using high throughput sequencing. J. Integr. Plant Boil. 2018, 60, 670–690. [Google Scholar] [CrossRef]
- Liu, F.; Huang, N.; Wang, L.; Ling, H.; Sun, T.; Ahmad, W.; Muhammad, K.; Guo, J.; Xu, L.; Gao, S.; et al. A Novel L-ascorbate Peroxidase 6 Gene, ScAPX6, Plays an Important Role in the Regulation of Response to Biotic and Abiotic Stresses in Sugarcane. Front. Plant Sci. 2018, 8, 2262. [Google Scholar] [CrossRef]
- Tao, J.; Wu, H.; Li, Z.; Huang, C.; Xu, X. Molecular Evolution of GDP-D-Mannose Epimerase (GME), a Key Gene in Plant Ascorbic Acid Biosynthesis. Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.; Coates, A.R.M.; Henderson, B. Chaperonin 60 unfolds its secrets of cellular communication. Cell Stress Chaperones 2002, 7, 317–329. [Google Scholar] [CrossRef]
- Alavilli, H.; Lee, H.; Park, M.; Yun, D.-J.; Lee, B.-H. Enhanced multiple stress tolerance in Arabidopsis by overexpression of the polar moss peptidyl prolyl isomerase FKBP12 gene. Plant Cell Rep. 2017, 37, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Kumar, D.; Jain, M.; Chaudhary, P.; Deswal, R.; Sarin, N.B. Ectopic overexpression of a salt stress-induced pathogenesis-related class 10 protein (PR10) gene from peanut (Arachis hypogaea L.) affords broad spectrum abiotic stress tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. (PCTOC) 2011, 109, 19–31. [Google Scholar] [CrossRef]


| Physiological Traits | Cv. Nagina 22 | Cv. Panvel | Statistical Analysis (p < 0.05) | ||||
|---|---|---|---|---|---|---|---|
| Optimum-P | Low-P | Optimum-P | Low-P | C | T | C × T | |
| Root length (cm) | 14.2 ± 2.55 | 18.5 ± 2.78 ** | 12.8 ± 1.98 | 15.3 ± 1.67 * | 0.031 | 0.012 | 0.033 |
| Shoot length (cm) | 32.5 ± 3.66 | 24.3 ± 3.11 * | 36.4 ± 4.23 | 31.6 ± 0.34 * | 0.033 | 0.021 | 0.035 |
| Plant dry weight (g plant−1) | 2.04 ± 0.47 | 1.24 ± 0.35 * | 2.59 ± 0.61 | 2.14 ± 0.37 * | 0.007 | 0.015 | 0.026 |
| Leaf P concentration (mg g−1 DW) | 1.63 ± 0.24 | 1.04 ± 0.21 *** | 2.05 ± 0.32 | 1.57 ± 0.22 ** | 0.013 | 0.003 | 0.007 |
| Root P concentration (mg g−1 DW) | 1.78 ± 0.23 | 1.22 ± 0.20 ** | 2.14 ± 0.33 | 1.72 ± 0.25 * | 0.021 | 0.005 | 0.011 |
| Phosphorus use efficiency (%) | 12.96 ± 1.24 | 8.61 ± 1.03 ** | 15.97 ± 2.01 | 12.54 ± 1.73 * | 0.002 | 0.000 | 0.006 |
| Distribution of DEPs | Cv. Panvel | Cv. Nagina 22 | Cv. Panvel + Cv. Nagina 22 | Total DEPs |
|---|---|---|---|---|
| Up regulated | 21 | 7 | 14 | 42 |
| Down regulated | 4 | 10 | 7 | 21 |
| S. N. | Accession No. | Name of Protein | Exp. MW (kDa) | Exp. Pi | M.S. | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative spot intensity (Optimum P:Low P) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nagina 22 | Panvel | ||||||||||
| 1 | ABL74560 | Fructose-bisphosphate aldolase | 45.6 | 6.45 | 82 | 12 | Cytosol | Energy | Upregulated | 1.00:3.13 | 1.00:2.91 |
| 2 | XP_015646992 | Succinate dehydrogenase flavoprotein subunit | 65.8 | 6.12 | 134 | 15 | Mitochondrion | Energy | Upregulated | 1.00:3.22 | 1.00:3.50 |
| 3 | BAA00147 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 54.1 | 6.31 | 348 | 15 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.51 | 1.00: −3.23 |
| 4 | AAS93256 | Glutathione S-transferase | 34.2 | 5.49 | 94 | 11 | Cytoplasm | Oxidative stress | Upregulated | 1.00:2.40 | 1.00:2.91 |
| 5 | XP_015643741 | Enolase 1 | 57.8 | 5.59 | 107 | 11 | Chloroplast | Cell rescue/defense | Upregulated | 1.00:3.00 | 1.00:3.02 |
| 6 | BAT16624 | NADP-dependant malate dehydrogenase | 49.3 | 6.14 | 114 | 13 | Mitochondrion, chloroplast, cytosol | Metabolism | Downregulated | 1.00: −3.32 | 1.00: −3.10 |
| 7 | 2002393A | Photosystem II oxygen-evolving complex protein 1 | 37.2 | 6.87 | 159 | 13 | Chloroplast | Photosynthesis | Upregulated | 1.00:2.54 | 1.00:3.00 |
| 8 | BAV53208 | Small ribosomal protein 4 | 53.2 | 6.93 | 89 | 9 | Ribosome, mitochondrion | Protein synthesis | Downregulated | 1.00: −3.43 | 1.00: −3.11 |
| 9 | AEP20544 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | 53.3 | 6.19 | 96 | 11 | Chloroplast | Photosynthesis | Downregulated | 1.00: −4.35 | 1.00: −3.72 |
| 10 | AAP12937 | Transposon protein, putative, CACTA, En/Spmsub-class | 56.2 | 6.8 | 86 | 16 | Nucleus | - | Upregulated | 1.00:2.97 | 1.00:3.68 |
| 11 | Q948T6 | Glyoxylase I 7 | 33.4 | 6.3 | 86 | 14 | Peroxisome | Oxidative stress | Upregulated | 1.00:4.20 | 1.00:5.20 |
| 12 | ABR25753 | Chaperonin GroEL | 65.3 | 4.91 | 87 | 10 | Cytoplasm | Protein fate/stabilization | Upregulated | 1.00:3.53 | 1.00:5.11 |
| 13 | AAM74563 | Elongation factor Tu | 52.1 | 4.98 | 123 | 16 | Cytosol, plastid, mitochondrion | Protein synthesis | Downregulated | 1.00: −2.33 | 1.00: −2.72 |
| 14 | NP_973937 | DNA binding transcription factors | 27.8 | 5.7 | 96 | 8 | Nucleus | Transcription/STMs | Upregulated | 1.00:3.22 | 1.00:4.61 |
| 15 | Q2QLY4 | Methionine synthase | 55.1 | 6.5 | 112 | 10 | Cytosol | Metabolism | Upregulated | 1.00:4.21 | 1.00:4.52 |
| 16 | CAD41255 | OSJNBa0067K08.7 | 33.4 | 5.33 | 141 | 8 | - | - | Downregulated | 1.00: −3.84 | 1.00: −3.51 |
| 17 | D7LVK3 | ABA-responsive element binding protein 3 | 32.8 | 5.93 | 123 | 11 | Nucleus | Transcription/STMs | Upregulated | 1.00:4.42 | 1.00:4.61 |
| 18 | P69667 | Ribosomal protein L23 | 10.13 | 7.02 | 81 | 8 | Mitochondrion | Protein synthesis | Downregulated | 1.00: −2.11 | 1.00: −2.02 |
| 19 | Q6NPS8 | NADPH-dependent FMN reductase | 32.1 | 6.55 | 82 | 8 | Nucleus, cytoplasm | Cell cycle/transport | Upregulated | 1.00:3.62 | 1.00:3.64 |
| 20 | AAX85991 | Protein disulfide isomerase | 57.1 | 5.3 | 152 | 6 | Endoplasmic reticulum | Protein fate/stabilization | Upregulated | 1.00:2.71 | 1.00:3.22 |
| 21 | ANG44638 | Maturase K | 20.9 | 6.36 | 87 | 7 | Nucleus | Cell rescue/defense | Upregulated | 1.00:4.84 | 1.00:4.80 |
| S. N. | Accession No. | Name of Protein | Exp. Mw (kDa) | Exp. Pi | M. S. | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative Spot Intensity (Optimum P: Low P) |
|---|---|---|---|---|---|---|---|---|---|---|
| 22 | ACA50522 | 14¨C3¨C3 protein | 34.2 | 4.93 | 112 | 9 | Nucleus | Transcription/STMs | Upregulated | 1.00:3.60 |
| 23 | BAD07865 | Phosphoribulokinase | 46.1 | 5.45 | 91 | 14 | Chloroplast, cytosol | Photosynthesis | Upregulated | 1.00:3.22 |
| 24 | Q0DYB1 | Inorganic pyrophosphatase family protein | 33.2 | 6.1 | 110 | 12 | Cytoplasm | Metabolism | Upregulated | 1.00:4.63 |
| 25 | BAD67774 | Phosphogluconate dehydrogenase | 54.2 | 5.89 | 104 | 14 | Cytoplasm | Metabolism | Upregulated | 1.00:2.92 |
| 26 | BAF92702 | Chaperonin 60 β precursor | 55.1 | 5.4 | 128 | 8 | Mitochondrion | Protein fate/stabilization | Upregulated | 1.00:2.51 |
| 27 | AAB33001 | Sedoheptulose-1,7-bisphosphatase | 41.9 | 5.81 | 143 | 14 | Chloroplast | Metabolism | Downregulated | 1.00: −2.11 |
| 28 | 3E5R_A | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic precursor | 55.2 | 6.47 | 94 | 13 | Chloroplast | Energy | Upregulated | 1.00:3.52 |
| 29 | ABA18619 | 59-epimerase | 43.5 | 5.89 | 104 | 8 | Cytosol | Oxidative stress | Upregulated | 1.00:3.23 |
| 30 | BAG24017 | RNA-binding protein | 49.7 | 5.30 | 106 | 9 | Cytosol, nucleus | Transcription/STMs | Upregulated | 1.00:4.50 |
| 31 | M1NZ56 | Pyrroline-5-carboxylate synthetase | 28.1 | 6.7 | 114 | 13 | Cytoplasm | Cell rescue/defense | Upregulated | 1.00:3.11 |
| 32 | ACJ54888 | Heat shock protein 40 | 28.7 | 5.8 | 96 | 8 | Nucleus, mitochondrion, ER | Protein fate/stabilization | Upregulated | 1.00:3.50 |
| 33 | AAF85973 | PR-10b protein | 17.2 | 6.7 | 23 | 9 | Nucleus | - | Upregulated | 1.00:2.55 |
| 34 | ABA39947 | UDP-glucose epimerase | 42.5 | 5.78 | 102 | 8 | Cytosol | Oxidative stress | Upregulated | 1.00:1.63 |
| 35 | BAS78758 | Os02g0493300 | 56.4 | 6.15 | 87 | 8 | - | - | Upregulated | 1.00:1.80 |
| 36 | AJB98433 | 6-Phosphogluconolactonase | 34.2 | 5.46 | 87 | 10 | Cytosol | Metabolism | Upregulated | 1.00:3.41 |
| 37 | Q40693 | Heat shock protein 70 | 27.5 | 5.3 | 92 | 9 | Nucleus, mitochondrion | Protein fate/stabilization | Upregulated | 1.00:1.50 |
| 38 | BAA11351 | Ribosomal protein S19 | 12.6 | 6.8 | 58 | 13 | Ribosome, mitochondrion | Protein synthesis | Downregulated | 1.00: −3.22 |
| 39 | ABI74568 | Phosphoglycerate kinase | 46.2 | 5.48 | 167 | 14 | Cytosol | Energy | Upregulated | 1.00:3.53 |
| 40 | ABA92415 | NADP-dependent oxidoreductase P1 | 40.3 | 6.31 | 119 | 13 | Chloroplast | Photosynthesis | Upregulated | 1.00:3.28 |
| 41 | AAQ23061 | Heat shock responsive transcription factor | 37.9 | 5.1 | 54 | 9 | Nucleus, cytoplasm | Protein fate/stabilization | Upregulated | 1.00:3.97 |
| 42 | BAC00625 | Malate dehydrogenase | 37.7 | 5.41 | 86 | 9 | Mitochondrion | Metabolism | Downregulated | 1.00: −3.24 |
| 43 | AAK92626 | ATPase | 74.8 | 6.48 | 157 | 11 | Plasma membrane | Energy | Upregulated | 1.00:3.32 |
| 44 | BAD17324 | Flavonol synthase | 35.9 | 5.43 | 104 | 10 | Cytoplasm, nucleus | Oxidative stress | Upregulated | 1.00:4.61 |
| 45 | Q6AVA8 | Pyruvate orthophosphate dikinase | 73.3 | 5.15 | 128 | 22 | Chloroplast, cytoplasm | Metabolism | Downregulated | 1.00: ─3.00 |
| 46 | Q6K9N6 | Succinyl-CoA synthetase beta subunit | 44.2 | 5.64 | 63 | 10 | Mitochondrion | Metabolism | Upregulated | 1.00:3.62 |
| S. N. | Accession No. | Name of Protein | Exp. Mw (kDa) | Exp. Pi | M. S. | No. of Matched Peptides | Location | Process | Mode of Regulation | Relative Spot Intensity (Optimum P: Low P) |
|---|---|---|---|---|---|---|---|---|---|---|
| 47 | P93431 | Rubisco activase chloroplast precursor | 52.21 | 4.93 | 148 | 6 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.26 |
| 48 | XP_015641702 | Ferredoxin-nitrite reductase, chloroplastic | 71.3 | 6.15 | 94 | 11 | Chloroplast | Photosynthesis | Downregulated | 1.00: −4.43 |
| 49 | BAP76084 | Phosphoenolpyruvate carboxylase | 25.9 | 6.87 | 98 | 7 | Chloroplast, mitochondrion | Metabolism | Downregulated | 1.00: −3.26 |
| 50 | Q01859 | F1-ATP synthase, beta subunit | 66.4 | 5.39 | 165 | 18 | Mitochondrion | Energy | Upregulated | 1.00:4.70 |
| 51 | A3C4S4 | GDP-D-mannose-3′,5′-epimerase | 54.3 | 6.63 | 102 | 11 | Cytosol | Oxidative stress | Upregulated | 1.00:3.32 |
| 52 | ABA96472 | FKBP-type peptidyl-prolyl cis-trans isomerase | 27.2 | 6.84 | 86 | 8 | Nucleus | Cell rescue/defense | Upregulated | 1.00:3.83 |
| 53 | AAS46111 | RNA polymerase β chain | 81.1 | 6.93 | 215 | 10 | Chloroplast | Transcription/STMs | Downregulated | 1.00: −3.90 |
| 54 | AAB65699 | Ferredoxin | 44.5 | 6.35 | 147 | 16 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.44 |
| 55 | A2Y7D9 | Chloroplast light-harvesting complex I protein LHCA3 | 31.1 | 6.46 | 85 | 7 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.37 |
| 56 | P48494 | Triosephosphate isomerase | 21.2 | 5.5 | 151 | 13 | Cytoplasm | Metabolism | Downregulated | 1.00: −3.70 |
| 57 | - | Unnamed protein product | 46.3 | 5.49 | 97 | 14 | - | - | Upregulated | 1.00:3.80 |
| 58 | XP_015643207 | Transketolase chloroplastic | 17.8 | 6.12 | 84 | 16 | Chloroplast | Photosynthesis | Downregulated | 1.00: −3.52 |
| 59 | ABL74559 | Glyceraldehyde 3-phosphate dehydrogenase | 53.8 | 6.72 | 84 | 8 | Cytoplasm | Energy | Upregulated | 1.00:3.45 |
| 60 | AET04420 | Integrin-linked protein kinase family protein | 34.2 | 6.11 | 104 | 12 | Ribosome | Transcription/STMs | Downregulated | 1.00: −3.11 |
| 61 | XP_015621402 | Asparaginyl-tRNA synthetase, cytoplasmic 3 | 65.1 | 6.13 | 173 | 23 | Cytoplasm | Protein synthesis | Downregulated | 1.00: −3.24 |
| 62 | AAP13093 | Ascorbate peroxidase cytoplasmic | 31.4 | 5.75 | 92 | 12 | Cytoplasm | Oxidative stress | Upregulated | 1.00:4.42 |
| 63 | AAB63591 | Chaperonin 10 Kd subunit | 25.8 | 5.66 | 82 | 8 | Cytoplasm | Protein stabilization | Upregulated | 1.00:3.54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tantray, A.Y.; Ali, H.M.; Ahmad, A. Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency. Agronomy 2020, 10, 1028. https://doi.org/10.3390/agronomy10071028
Tantray AY, Ali HM, Ahmad A. Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency. Agronomy. 2020; 10(7):1028. https://doi.org/10.3390/agronomy10071028
Chicago/Turabian StyleTantray, Aadil Yousuf, Hayssam M. Ali, and Altaf Ahmad. 2020. "Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency" Agronomy 10, no. 7: 1028. https://doi.org/10.3390/agronomy10071028
APA StyleTantray, A. Y., Ali, H. M., & Ahmad, A. (2020). Analysis of Proteomic Profile of Contrasting Phosphorus Responsive Rice Cultivars Grown under Phosphorus Deficiency. Agronomy, 10(7), 1028. https://doi.org/10.3390/agronomy10071028