Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Pathogen Stress Treatment
2.2. Identifying Functional Genes Adjacent to Differentially Expressed Long Noncoding Transcripts (lncRNAs)
2.3. Real-Time Quantitative PCR Analysis
3. Results
3.1. Identification of Transcription Factor Genes Adjacent to Differentially Expressed Long Non-Coding RNA in Wheat Responding to Pathogen Infection
3.2. Co-Expression of Long Non-Coding RNAs with Adjacent Functional Genes
3.3. Co-Expression of Long Non-Coding RNAs and Allele-Specific Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DE | differentially expressed |
| ncRNA | non-coding RNA |
| lincRNAs | long intergenic non-coding RNA |
| linncRNAs | long intron non-coding RNA |
References
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Romero-Barrios, N.; Jegu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 1–10. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.Y.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef]
- Yuan, J.; Li, J.; Yang, Y.; Tan, C.; Zhu, Y.; Hu, L.; Qi, Y.; Lu, Z.J. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 2018, 93, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2010, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, W.; Hao, J.; Lv, S.; Wang, C.; Tong, W.; Wang, Y.; Wang, Y.; Liu, X.; Ji, W. Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genomics 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed]
- Laurent, S.G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef]
- Beltran, M.; Puig, I.; Pena, C.; Garcia, J.M.; Alvarez, A.B.; Pena, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, J.; Lv, Q.; Qin, J.; Huang, Y.; Yu, M.; Zhong, M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett. 2019, 440–441, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 2018, 9, 3516. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Diloknawarit, P.; Park, B.S.; Chua, N.H. ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol. 2019, 221, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- Bourras, S.; McNally, K.E.; Ben-David, R.; Parlange, F.; Roffler, S.; Praz, C.R.; Oberhaensli, S.; Menardo, F.; Stirnweis, D.; Frenkel, Z.; et al. Multiple Avirulence Loci and Allele-Specific Effector Recognition Control the Pm3 Race-Specific Resistance of Wheat to Powdery Mildew. Plant Cell 2015, 27, 2991–3012. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Han, D.J.; Wang, Q.L.; Chen, X.M.; Zeng, Q.D.; Wu, J.H.; Xue, W.B.; Zhan, G.M.; Huang, L.L.; Kang, Z.S. Emerging Yr26-Virulent Races of Puccinia striiformis f. tritici Are Threatening Wheat Production in the Sichuan Basin, China. Plant Dis. 2015, 99, 754–760. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, M.; Chen, X.; Kang, Z. Role of Alternate Hosts in Epidemiology and Pathogen Variation of Cereal Rusts. Annu. Rev. Phytopathol. 2016, 54, 207–228. [Google Scholar] [CrossRef]
- Xue, F.; Ji, W.; Wang, C.; Zhang, H.; Yang, B. High-density mapping and marker development for the powdery mildew resistance gene PmAS846 derived from wild emmer wheat (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2012, 124, 1549–1560. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 661–674. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. Biochim. Biophys. Acta 2016, 1859, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Sharma, S.; Taneja, M.; Tyagi, S.; Singh, K.; Upadhyay, S.K. Survey of High Throughput RNA-Seq Data Reveals Potential Roles for lncRNAs during Development and Stress Response in Bread Wheat. Front. Plant Sci. 2017, 8, 1019. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol. Prog. 2011, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, S.; Wang, C.; Ji, W. The role of transcription factor in wheat defense against pathogen and its prospect in breeding. J. Plant Biol. Crop Res. 2018, 1, 1005. [Google Scholar]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126. [Google Scholar] [CrossRef]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Juan, L.; Wang, G.; Radovich, M.; Schneider, B.P.; Clare, S.E.; Wang, Y.; Liu, Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med. Genomics 2013, 6, S7. [Google Scholar] [CrossRef] [PubMed]
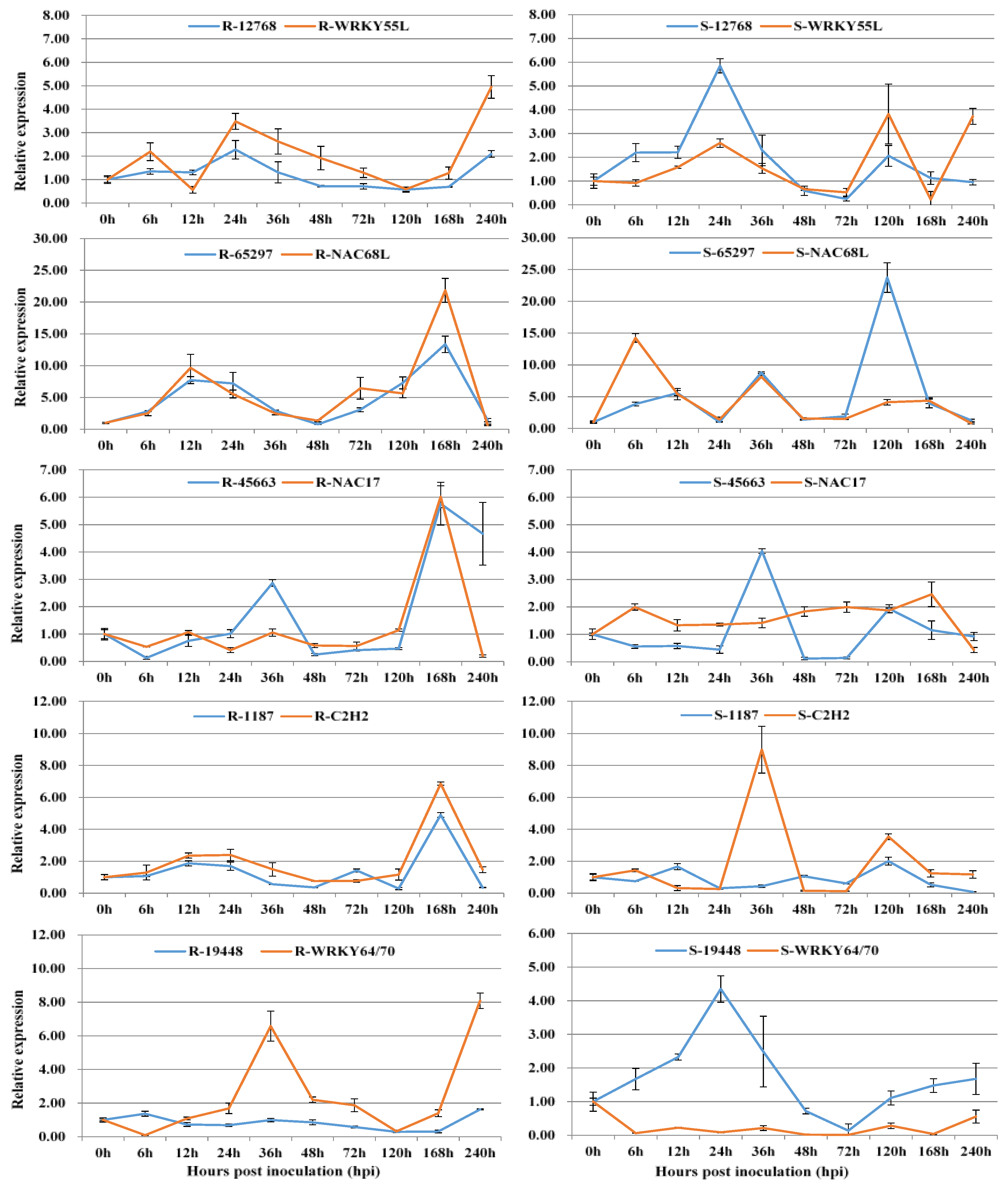
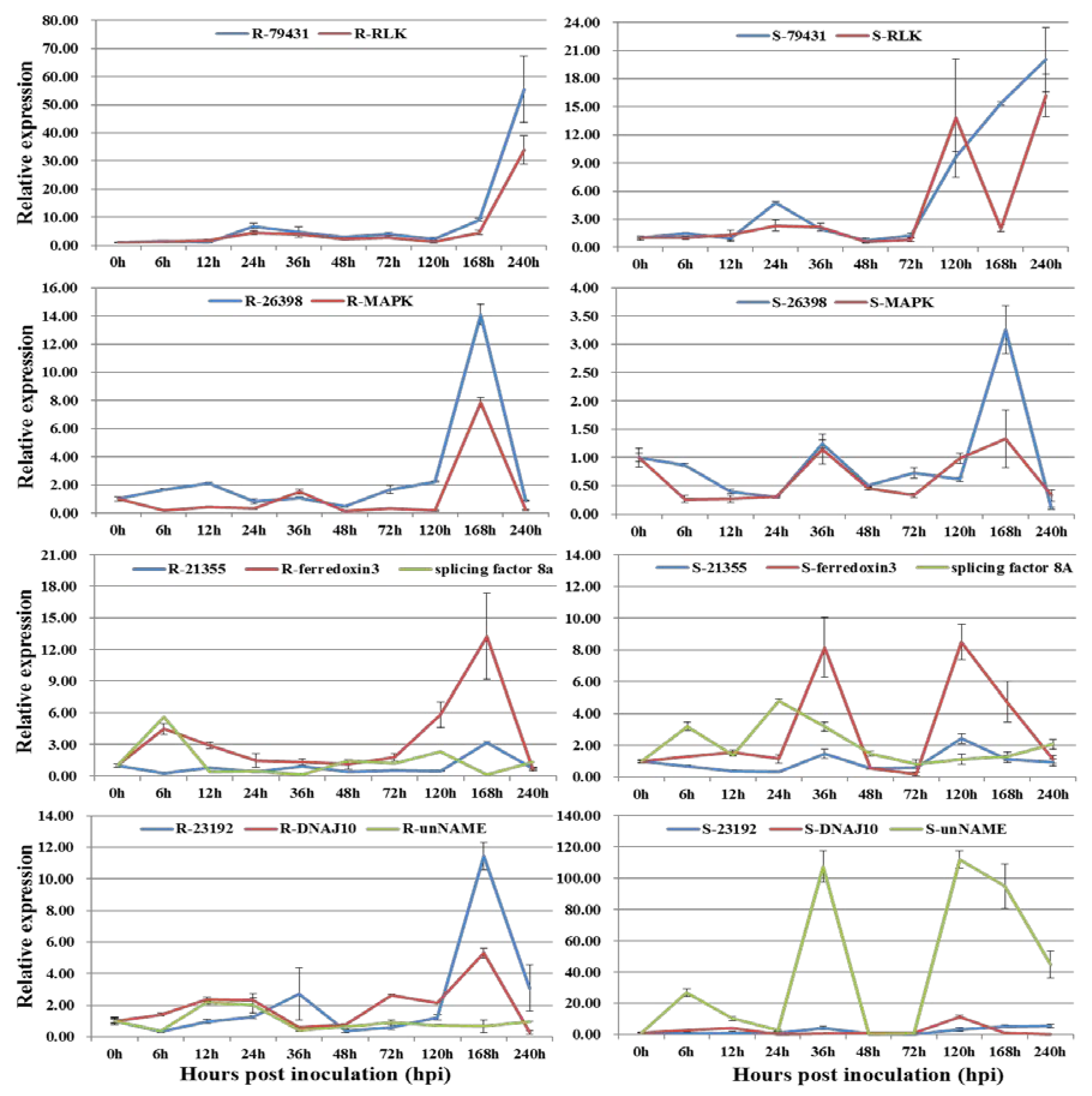
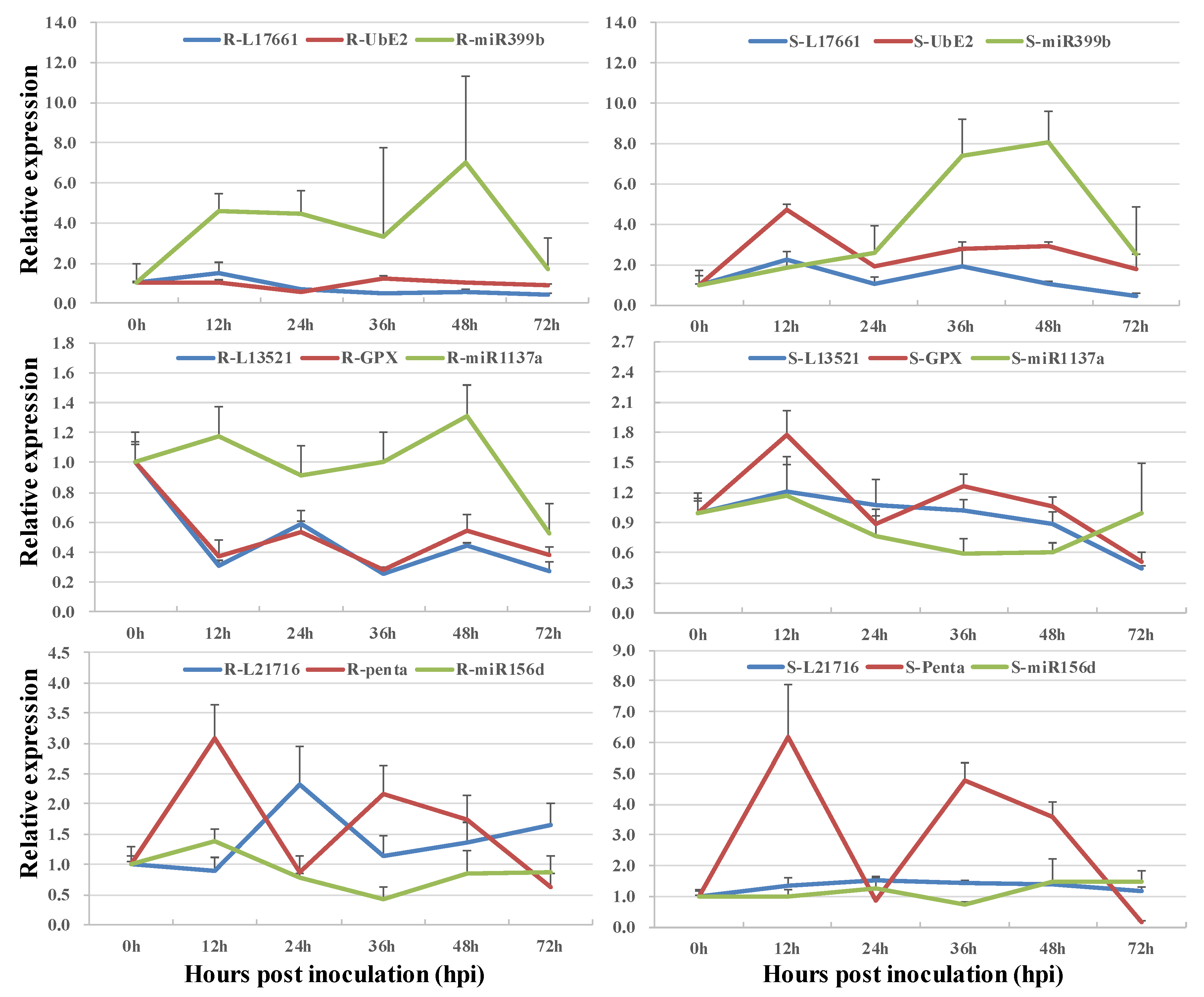
| LncRNA ID | Adjacent Functional Gene | TF Type | LncRNA Type |
|---|---|---|---|
| T4_Unigene_BMK.9130 | Ta_TraesCS1A01G200500.1 | B3 | LincRNA |
| T16_Unigene_BMK.1187 | Ta_TraesCS1B01G146800.1 | C2H2 | LincRNA |
| T10_Unigene_BMK.12768 | Ta_TraesCS1B01G243100.1 | WRKY55L | LincRNA |
| T10_Unigene_BMK.12768 | Ta_TraesCS1B01G243200.1 | AP2/ERF-ERF | LincRNA |
| T4_Unigene_BMK.17456 | Ta_TraesCS1B01G273100.1 * | CSD | LuncRNA |
| T4_Unigene_BMK.17456 | Ta_TraesCS1D01G262500.1 | CSD | LincRNA |
| T19_Unigene_BMK.34110 | Ta_TraesCS2A01G319700.1 | GNAT | LincRNA |
| T13_Unigene_BMK.49502 | Ta_TraesCS3A01G421400.1 * | bHLH | LpncRNA |
| T16_Unigene_BMK.67438 | Ta_TraesCS3A01G432900.1 | MADS-M-type | LinncRNA |
| T4_Unigene_BMK.30836 | Ta_TraesCS3D01G136600.1 | NF-X1 | LincRNA |
| T10_Unigene_BMK.65297 | Ta_TraesCS3D01G333100.1 * | NAC68L/4L | LinncRNA |
| T16_Unigene_BMK.92879 | Ta_TraesCS3D01G365300.1 | B3 | LincRNA |
| T4_Unigene_BMK.9309 | Ta_TraesCS4A01G211100.1 | MYB | LinncRNA |
| T13_Unigene_BMK.19448 | Ta_TraesCS4D01G172200.1 | WRKY64/70 | LincRNA |
| T13_Unigene_BMK.40522 | Ta_TraesCS4D01G265400.1 | GNAT | LincRNA |
| T19_Unigene_BMK.49358 | Ta_TraesCS5A01G312000.1 * | AP2/ERF-ERF | LpncRNA |
| T4_Unigene_BMK.45663 | Ta_TraesCS5D01G279100.2 * | NAC17L | LinncRNA |
| T4_Unigene_BMK.47960 | Ta_TraesCS6A01G085800.1 | BES1 | LincRNA |
| T16_Unigene_BMK.71332 | Ta_TraesCS6B01G219200.1 | mTERF | LincRNA |
| T13_Unigene_BMK.34604 | Ta_TraesCS6B01G237700.1 | AP2/ERF-ERF | LincRNA |
| T19_Unigene_BMK.51118 | Ta_TraesCS6D01G121100.1 | AP2/ERF-ERF | LpncRNA |
| T19_Unigene_BMK.54493 | Ta_TraesCS6D01G217800.1 | AP2/ERF-ERF | LincRNA |
| T16_Unigene_BMK.22544 | Ta_TraesCS7A01G326400.1 | TUB | LinncRNA |
| T16_Unigene_BMK.22544 | Ta_TraesCS7B01G227000.1 * | TUB | LinncRNA |
| T16_Unigene_BMK.22544 | Ta_TraesCS7D01G323100.1 | TUB | LinncRNA |
| T16_Unigene_BMK.23889 | Ta_TraesCS7D01G166500.1 | MYB | LincRNA |
| T13_Unigene_BMK.30347 | Ta_TraesCS7D01G269300.1 | bZIP | LincRNA |
| LncRNA ID | Adjacent Functional Gene | Definition | LncRNA Type |
|---|---|---|---|
| T16.92969 | TraesCS5B01G026400 | uncharacterized protein LOC109734965 | lpncRNA |
| T13.33010 | TraesCS5B01G036800 * | chloroplast stem-loop binding protein of 41 kDa | linncRNA |
| T16.69540 | TraesCS5B01G076100 * | Putative lipid-transfer protein DIR1 | luncRNA |
| T13.22353 | TraesCS5B01G098000 | peptidyl-prolyl cis-trans isomerase G | linncRNA |
| T13.42814 | TraesCS5B01G117000 | uncharacterized protein LOC109768056 isoform X1 | linncRNA |
| T4.63565 | TraesCS5B01G121400 | 2-oxoisovalerate dehydrogenase | linncRNA |
| T19.46503 | TraesCS5B01G134500 | uncharacterized protein LOC109774113 | lincRNA |
| T19.46503 | TraesCS5B01G134600 * | uncharacterized protein LOC109774111 isoform X1 | lincRNA |
| T16.29097 | TraesCS5B01G177300 | mediator complex subunit 25 | linncRNA |
| T16.6266 | TraesCS5B01G208100 * | cysteine endopeptidase EP gamma | linncRNA |
| T7.1464 | TraesCS5B01G232600 * | 1-aminocyclopropane-1-carboxylate oxidase 1-like | lpncRNA |
| T16.68333 | TraesCS5B01G300100 | uncharacterized protein LOC109733149 isoform X2 | lincRNA |
| T16.68333 | TraesCS5B01G300200 * | CDGSH iron-sulfur domain-containing protein NEET | lincRNA |
| T10.61842 | TraesCS5B01G302500 | GDP-mannose 3,5-epimerase 2 | linncRNA |
| T1.39963 | TraesCS5B01G404600 * | subtilisin-like protease SBT1.7 | linncRNA |
| T1.37489 | TraesCS5B01G453800 * | Lr10 disease-resistance locus receptor-like protein kinase 1.5 | linncRNA |
| T10.43083 | TraesCS5B01G478100 * | uncharacterized protein LOC109760008 | linncRNA |
| T13.86345 | TraesCS5B01G488300 | protein synthesis inhibitor II-like | linncRNA |
| T13.38179 | TraesCS5B01G535800 | pirin-like protein isoform X1 | lpncRNA |
| T13.49097 | TraesCS5B01G547000 | cinnamoyl-CoA reductase 1-like | luncRNA |
| T16.26398 | TraesCS5B01G565100 | MAP kinase kinase | linncRNA |
| T16.83333 | TraesCS1B01G069300 | uncharacterized protein LOC109765977 | linncRNA |
| T16.13852 | TraesCS1B01G110200 | Alanyl-tRNA synthetase | lincRNA |
| T16.13852 | TraesCS1B01G110300 | hypothetical protein BRADI_2g36145v3 | lincRNA |
| T16.32365 | TraesCS1B01G113500 | glutathione S-transferase 4-like | luncRNA |
| T16.1187 | TraesCS1B01G146700 | uncharacterized protein LOC109755951 | lincRNA |
| T19.42425 | TraesCS1B01G163000 | uncharacterized protein LOC100846051 isoform | lincRNA |
| T19.42425 | TraesCS1B01G163100 | uncharacterized protein LOC109772407 | lincRNA |
| T16.3724 | TraesCS1B01G174600 * | uncharacterized protein LOC109772407 | linncRNA |
| T16.16515 | TraesCS1B01G200700 | endonuclease MutS2 isoform X1 | lincRNA |
| T16.16515 | TraesCS1B01G200800 | polyprotein/retrotransposon protein, unclassified | LincRNA |
| T16.89858 | TraesCS1B01G276800 * | putative proteinase inhibitor-related protein | luncRNA |
| T7.4304 | TraesCS1B01G276900 * | wali6/Al-inducible genes | luncRNA |
| T13.23369 | TraesCS1B01G289100 | tankyrase-1-like isoform X4 | linncRNA |
| T13.24210 | TraesCS1B01G289600 * | guanylyl cyclase | linncRNA |
| T13.23192 | TraesCS1B01G384100 | chaperone protein dnaJ 10-like | lincRNA |
| T13.23192 | TraesCS1B01G384200 | unnamed protein product | lincRNA |
| T10.79431 | TraesCS1B01G394900 | Lr10 disease-resistance locus receptor-like protein kinase 1.2 | linncRNA |
| T13.51457 | TraesCS1B01G410300 * | tropinone reductase homolog At5g06060-like isoform X3 | linncRNA |
| T16.7005 | TraesCS1B01G410300 * | tropinone reductase homolog At5g06060-like isoform X3 | luncRNA |
| T16.21355 | TraesCS1B01G416400 | Pre-mRNA-processing-splicing factor 8A | lincRNA |
| T16.21355 | TraesCS1B01G416500 | ferredoxin-3, chloroplastic-like isoform X2 | lincRNA |
| T13.26624 | TraesCS1B01G433300 * | Chlorophyll a-b binding protein WCAB precursor | linncRNA |
| T16.538 | TraesCS1B01G433300 * | Chlorophyll a-b binding protein WCAB precursor | linncRNA |
| miRNA | lncRNA Mimic | lncRNA | Functional Gene |
|---|---|---|---|
| tae-miR1137a | T16.13521 | TraesCS6A02G246400.1 | |
| ath-miR414 | T13.49993 | TraesCS7B02G044200.1 | |
| ath-miR5658 | T13.33064 | TraesCS1D02G123200.1 | |
| osa-miR1439 | T19.34869 | 1B: 392147777-392149747 | |
| hvu-miR5049f | T1.48244 | TraesCS1B02G377700.1 | |
| bdi-miR394 | T1.37489 | TraesCS3D02G428200.1 | |
| tae-miR167a | T10.71969 | 7A:267192134-267192333 | |
| ath-miR390a-3p | T10.3513 | TraesCS6D02G306000.1 | |
| ata-miR160a-3p | T19.51118 | TraesCS1B02G080500.1 | |
| ata-miR395c-5p | T13.34604 | 1D:247796039-247796286 | |
| ath-miR399b | T13.17661 | TraesCS1B02G415800.1 | |
| ata-miR156d-3p | T13.21716 | TraesCS2D02G400500.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Wang, G.; Wang, S.; Nie, X.; Wang, C.; Wang, Y.; Zhang, H.; Ji, W. Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection. Agronomy 2020, 10, 896. https://doi.org/10.3390/agronomy10060896
Hu W, Wang G, Wang S, Nie X, Wang C, Wang Y, Zhang H, Ji W. Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection. Agronomy. 2020; 10(6):896. https://doi.org/10.3390/agronomy10060896
Chicago/Turabian StyleHu, Weiguo, Guanghao Wang, Siwen Wang, Xiaojun Nie, Changyou Wang, Yajuan Wang, Hong Zhang, and Wanquan Ji. 2020. "Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection" Agronomy 10, no. 6: 896. https://doi.org/10.3390/agronomy10060896
APA StyleHu, W., Wang, G., Wang, S., Nie, X., Wang, C., Wang, Y., Zhang, H., & Ji, W. (2020). Co-Regulation of Long Non-Coding RNAs with Allele-Specific Genes in Wheat Responding to Powdery Mildew Infection. Agronomy, 10(6), 896. https://doi.org/10.3390/agronomy10060896






