Engineered Durum Wheat Germplasm with Multiple Alien Introgressions: Agronomic and Quality Performance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Plot Trials and Growing Seasons
2.3. Measurement of Agronomic Traits
2.4. Grain and Semolina Quality Tests
2.5. Statistical Analysis
3. Results
3.1. Environmental Conditions
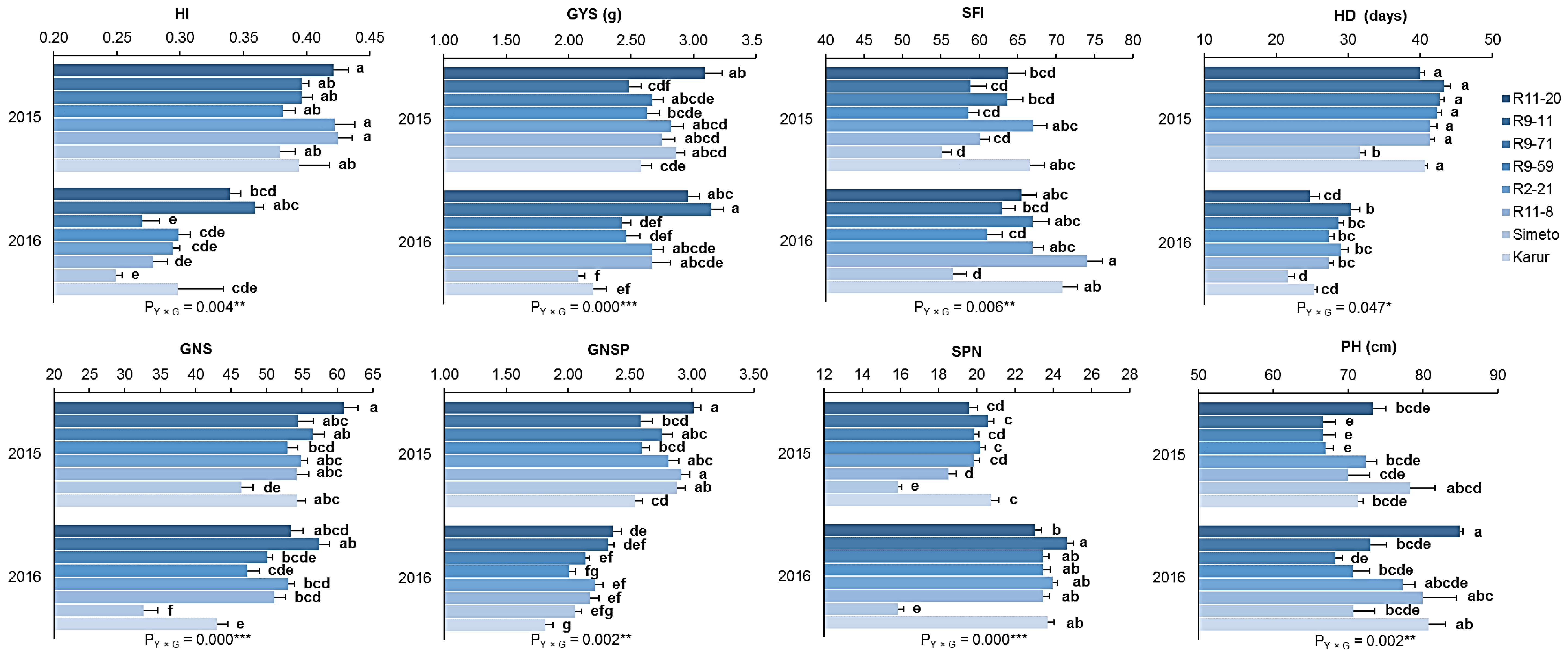
3.2. Yield Assessment
3.2.1. Small-Scale Plots
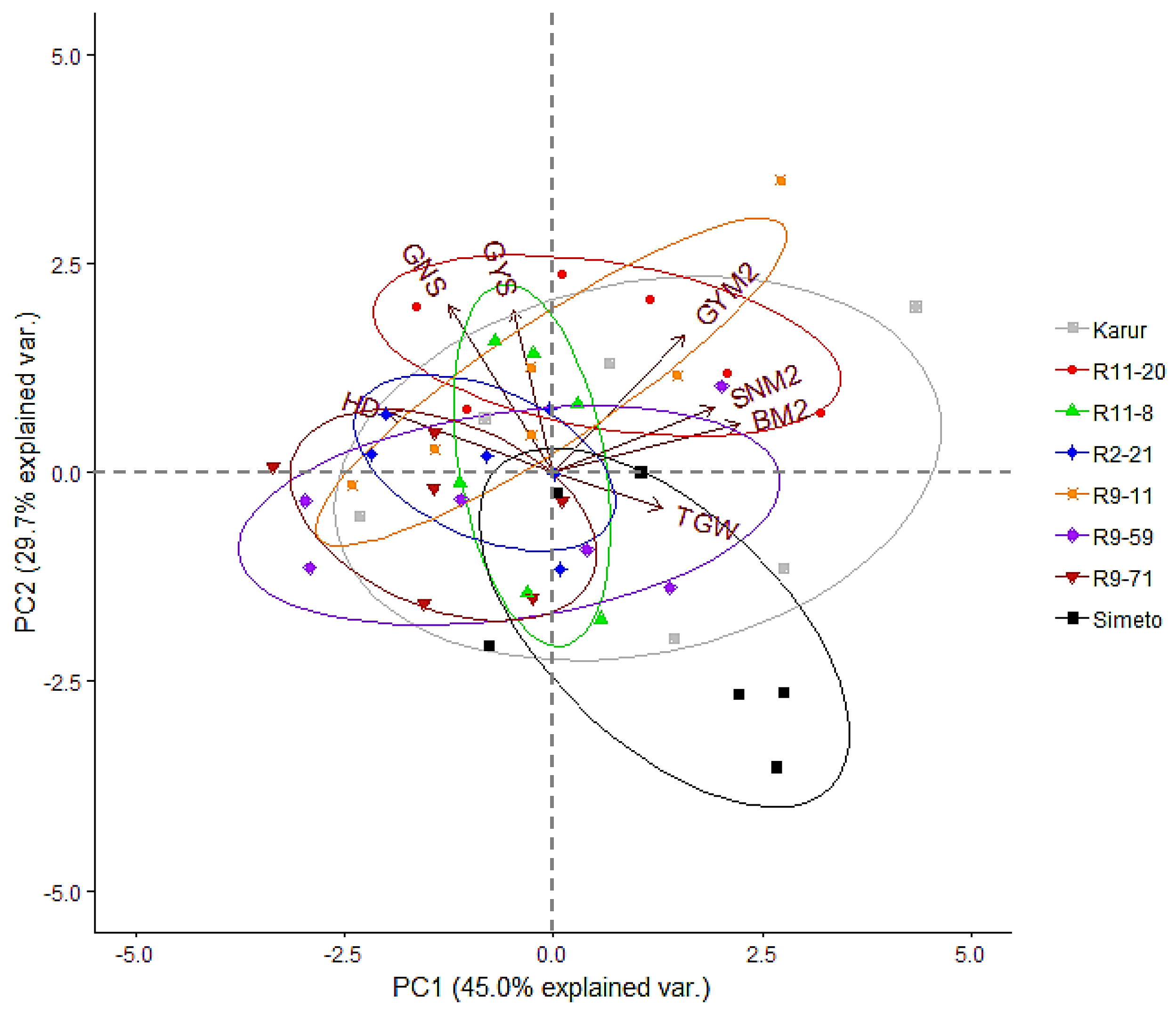
3.2.2. Relationship between Main Yield Components
3.2.3. Large-Scale Plots and Grain Quality of the Most Promising Recombinants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Feldman, M. Historical Aspects and Significance of the Discovery of Wild Wheats: Origin of Wheat, Evolution, Gene Pools. In Proceedings of the Stadler Genetics Symposium, Columbia, MO, USA, 1977; Volume 9, pp. 121–145. [Google Scholar]
- Royo, C.; Soriano, J.M.; Alvaro, F. Wheat: A crop in the bottom of the Mediterranean diet pyramid. In Mediterranean Identities-Environment, Society, Culture; Fuerst-Bjelis, B., Ed.; Intechopen: London, UK, 2017; pp. 381–399. [Google Scholar]
- Ranieri, R. Geography of the durum wheat crop. Pastaria Int. 2015, 6, 24–36. [Google Scholar]
- Bassi, F.M.; Sanchez-Garcia, M. Adaptation and stability analysis of ICARDA durum wheat elites across 18 countries. Crop Sci. 2017, 57, 2419–2430. [Google Scholar] [CrossRef]
- Sall, A.T.; Chiari, T.; Legesse, W.; Seid-Ahmed, K.; Ortiz, R.; van Ginkel, M.; Bassi, F.M. Durum wheat (Triticum durum Desf.): Origin, cultivation and potential expansion in Sub-Saharan Africa. Agronomy 2019, 9, 263. [Google Scholar] [CrossRef]
- Kearney, J. Food consumption trends and drivers. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2793–2807. [Google Scholar] [CrossRef] [PubMed]
- Reardon, T.; Tschirley, D.; Dolislager, M.; Snyder, J.; Hu, C.; White, S. Urbanization, Diet Change, and Transformation of Food Supply Chains in Asia. 2014. Available online: http://www.fao.org/fileadmin/templates/ags/docs/MUFN/DOCUMENTS/MUS_Reardon_2014.pdf (accessed on 22 October 2019).
- Reardon, T.; Echeverria, R.; Berdegué, J.; Minten, B.; Liverpool-Tasie, S.; Tschirley, D.; Zilberman, D. Rapid transformation of food systems in developing regions: Highlighting the role of agricultural research & innovations. Agric. Syst. 2019, 172, 47–59. [Google Scholar]
- Yüksel, A.N.; Oner, M.D.; Bayram, M. Rediscovery of couscous in the world. Glob. J. Med. Res.–Nutr. Food Sci. 2018, 18, 25–30. [Google Scholar]
- Dahl, C. Global Durum Outlook. Available online: http://www.italmopa.com/wp-content/uploads/2017/05/144_all_1.pdf (accessed on 27 May 2019).
- International Grains Council. Available online: http://www.igc.int/en/default.aspx (accessed on 27 May 2019).
- Danieli, P.P.; Primi, R.; Ronchi, B.; Ruggeri, R.; Rossini, F.; del Puglia, S.; Cereti, C.F. The potential role of spineless safflower (Carthamus tinctorius L. var. inermis) as fodder crop in central Italy. Ital. J. Agron. 2011, 6, 19–22. [Google Scholar] [CrossRef]
- Rossini, F.; Provenzano, M.E.; Kuzmanović, L.; Ruggeri, R. Jerusalem Artichoke (Helianthus tuberosus L.): A versatile and sustainable crop for renewable energy production in Europe. Agronomy 2019, 9, 528. [Google Scholar] [CrossRef]
- Ceoloni, C.; Kuzmanović, L.; Forte, P.; Gennaro, A.; Bitti, A. Targeted exploitation of gene pools of alien Triticeae species for sustainable and multi-faceted improvement of the durum wheat crop. Crop Pasture Sci. 2014, 65, 96–111. [Google Scholar] [CrossRef]
- Ceoloni, C.; Kuzmanović, L.; Gennaro, A.; Forte, P.; Giorgi, D.; Grossi, M.R.; Bitti, A. Genomes, chromosomes and genes of perennial triticeae of the genus Thinopyrum: The value of their transfer into wheat for gains in cytogenomic knowledge and ‘precision’ breeding. In Advances in Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 333–358. ISBN 9789400775756. [Google Scholar]
- Ceoloni, C.; Kuzmanović, L.; Forte, P.; Virili, M.E.; Bitti, A. Wheat-perennial Triticeae introgressions: Major achievements and prospects. In Alien Introgression in Wheat-Cytogenetics, Molecular Biology, and Genomics; Molnár-Láng, M., Ceoloni, C., Doležel, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 273–313. [Google Scholar]
- Chaudhary, H.K.; Kaila, V.; Rather, S.A.; Badiyal, A.; Hussain, W.; Jamwal, N.S.; Mahato, A. Wheat. In Alien Gene Transfer in Crop Plants, Volume 2: Achievements and Impacts; Pratap, A., Kumar, J., Eds.; Springer: New York, NY, USA, 2014; pp. 1–26. [Google Scholar]
- Feuillet, C.; Langridge, P.; Waugh, R. Cereal breeding takes a walk on the wild side. Trends Genet. 2008, 24, 24–32. [Google Scholar] [CrossRef]
- Sears, E.R. Chromosome Engineering in Wheat. In Proceedings of the Stadler Genetics Symposium 4, Columbia, MO, USA, 21–22 April 1972; Kimber, G., Rédei, G.R., Eds.; pp. 23–38. [Google Scholar]
- Sears, E.R. Transfer of alien genetic material to wheat. In Wheat Science: Today and Tomorrow; Evans, L.T., Peacock, W.J., Eds.; Cambridge University Press: Cambridge, UK, 1981; pp. 75–89. [Google Scholar]
- Ceoloni, C.; Jauhar, P. Chromosome Engineering of the Durum Wheat Genome. In Genetic Resources, Chromosome Engineering, and Crop Improvement: Cereals; Singh, R.J., Jauhar, P.P., Eds.; CRC Press: Boca Raton, FL, USA, 2006; pp. 27–59. ISBN 9780203489260. [Google Scholar]
- Qi, L.; Friebe, B.; Zhang, P.; Gill, B.S. Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 2007, 15, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Sears, E.R. An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 1977, 19, 585–593. [Google Scholar] [CrossRef]
- Giorgi, B. A homoeologous pairing mutant isolated in Triticum durum cv. Cappelli. Mutat. Breed. Newsl. 1978, 11, 4–5. [Google Scholar]
- Ceoloni, C.; Kuzmanović, L.; Ruggeri, R.; Rossini, F.; Forte, P.; Cuccurullo, A.; Bitti, A. Harnessing genetic diversity of wild gene pools to enhance wheat crop production and sustainability: Challenges and opportunities. Diversity 2017, 9, 55. [Google Scholar] [CrossRef]
- Ceoloni, C.; Biagetti, M.; Ciaffi, M.; Forte, P.; Pasquini, M. Wheat chromosome engineering at the 4x level: The potential of different alien gene transfers into durum wheat. Euphytica 1996, 89, 87–97. [Google Scholar] [CrossRef]
- Klindworth, D.L.; Hareland, G.A.; Elias, E.M.; Xu, S.S. Attempted compensation for linkage drag affecting agronomic characteristics of durum wheat 1AS/1DL translocation lines. Crop Sci. 2013, 53, 422–429. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Gennaro, A.; Benedettelli, S.; Dodd, I.C.; Quarrie, S.A.; Ceoloni, C. Structural-functional dissection and characterization of yield-contributing traits originating from a group 7 chromosome of the wheatgrass species Thinopyrum ponticum after transfer into durum wheat. J. Exp. Bot. 2014, 65, 509–525. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Ruggeri, R.; Virili, M.E.; Rossini, F.; Ceoloni, C. Effects of Thinopyrum ponticum chromosome segments transferred into durum wheat on yield components and related morpho-physiological traits in Mediterranean rain-fed conditions. Field Crops Res. 2016, 186, 86–98. [Google Scholar] [CrossRef]
- Oak, M.D.; Tamhankar, S.A. 1BL/1RS translocation in durum wheat and its effect on end use quality traits. J. Plant Biochem. Biotechnol. 2017, 26, 91–96. [Google Scholar] [CrossRef]
- Zarco-Hernandez, J.A.; Santiveri, F.; Michelena, A.; Javier Peña, R. Durum wheat (Triticum turgidum, L.) carrying the 1BL/1RS chromosomal translocation: Agronomic performance and quality characteristics under Mediterranean conditions. Eur. J. Agron. 2005, 22, 33–43. [Google Scholar] [CrossRef]
- Friebe, B.; Heun, M.; Tuleen, N.; Zeller, F.J.; Gill, B.S. Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci. 1994, 34, 621–625. [Google Scholar] [CrossRef]
- Ayala-Navarrete, L.; Bariana, H.S.; Singh, R.P.; Gibson, J.M.; Mechanicos, A.A.; Larkin, P.J. Trigenomic chromosomes by recombination of Thinopyrum intermedium and Th. ponticum translocations in wheat. Theor. Appl. Genet. 2007, 116, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Navarrete, L.I.; Mechanicos, A.A.; Gibson, J.M.; Singh, D.; Bariana, H.S.; Fletcher, J.; Shorter, S.; Larkin, P.J. The Pontin series of recombinant alien translocations in bread wheat: Single translocations integrating combinations of Bdv2, Lr19 and Sr25 disease-resistance genes from Thinopyrum intermedium and Th. ponticum. Theor. Appl. Genet. 2013, 126, 2467–2475. [Google Scholar] [CrossRef] [PubMed]
- Forte, P.; Virili, M.E.; Kuzmanović, L.; Moscetti, I.; Gennaro, A.; D’Ovidio, R.; Ceoloni, C. A novel assembly of Thinopyrum ponticum genes into the durum wheat genome: Pyramiding Fusarium head blight resistance onto recombinant lines previously engineered for other beneficial traits from the same alien species. Mol. Breed. 2014, 34, 1701–1716. [Google Scholar] [CrossRef]
- Ceoloni, C.; Forte, P.; Kuzmanović, L.; Tundo, S.; Moscetti, I.; De Vita, P.; Virili, M.E.; D’Ovidio, R. Cytogenetic mapping of a major locus for resistance to Fusarium head blight and crown rot of wheat on Thinopyrum elongatum 7EL and its pyramiding with valuable genes from a Th. ponticum homoeologous arm onto bread wheat 7DL. Theor. Appl. Genet 2017, 130, 2005–2024. [Google Scholar] [CrossRef]
- Shen, X.; Ohm, H. Molecular mapping of Thinopyrum-derived Fusarium head blight resistance in common wheat. Mol. Breed. 2007, 20, 131–140. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, X.; Hao, Y.; Cai, J.; Ohm, H.W.; Kong, L. A genetic map of Lophopyrum ponticum chromosome 7E, harboring resistance genes to Fusarium head blight and leaf rust. Theor. Appl. Genet. 2011, 122, 263–270. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Mandalà, G.; Tundo, S.; Ciorba, R.; Frangella, M.; Ruggeri, R.; Rossini, F.; Gevi, F.; Rinalducci, S.; Ceoloni, C. Equipping durum wheat—Thinopyrum ponticum recombinant lines with a Thinopyrum elongatum major QTL for resistance to Fusarium diseases through a cytogenetic strategy. Front. Plant Sci. 2019, 10, 1324. [Google Scholar] [CrossRef]
- Singh, R.P.; Huerta-Espino, J.; Rajaram, S.; Crossa, J. Agronomic effects from chromosome translocations 7DL.7Ag and 1BL.1RS in spring wheat. Crop Sci. 1998, 38, 27–33. [Google Scholar] [CrossRef]
- Villareal, R.L.; Bañuelos, O.; Mujeeb-Kazi, A.; Rajaram, S. Agronomic performance of chromosomes 1B and T1BL.1RS near-isolines in the spring bread wheat Seri M82. Euphytica 1998, 103, 195–202. [Google Scholar] [CrossRef]
- Friebe, B.; Jiang, J.; Raupp, W.J.; McIntosh, R.A.; Gill, B.S. Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 1996, 91, 59–87. [Google Scholar] [CrossRef]
- Kim, W.; Johnson, J.W.; Baenziger, P.S.; Lukaszewski, A.J.; Gaines, C.S. Agronomic effect of wheat-rye translocation carrying rye chromatin (1R) from different sources. Crop Sci. 2004, 44, 1254–1258. [Google Scholar] [CrossRef]
- Graybosch, R.A.; Peterson, C.J.; Hansen, L.E.; Worrall, D.; Shelton, D.R.; Lukaszewski, A. Comparative flour quality and protein characteristics of 1BL/1RS and 1AL/1RS wheat-rye translocation lines. J. Cereal Sci. 1993, 17, 95–106. [Google Scholar] [CrossRef]
- Ali, N.; Heslop-Harrison, J.; Ahmad, H.; Graybosch, R.A.; Hein, G.L.; Schwarzacher, T. Introgression of chromosome segments from multiple alien species in wheat breeding lines with wheat streak mosaic virus resistance. Heredity (Edinb.) 2016, 117, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, B.; Tong, Y. The contribution of distant hybridization with decaploid Agropyron elongatum to wheat improvement in China. J. Genet. Genom. 2008, 35, 451–456. [Google Scholar] [CrossRef]
- Wang, Z.G.; An, T.G.; Li, J.M.; Molnár-Láng, M.; Ji, J.; Zhong, G.C.; Mu, S.M. Flourescent in situ hybridization analysis of rye chromatin in the background of “Xiaoyan No. 6”. Acta Bot. Sin. 2004, 46, 436–442. [Google Scholar]
- Micali, S.; Forte, P.; Bitti, A.; D’Ovidio, R.; Ceoloni, C. Chromosome Engineering as a Tool for Effectively Introgressing Multiple Useful Genes from Alien Triticeae into Durum Wheat. In Proceedings of the 10th International Wheat Genetics Symposium, Paestum, Italy, 1–6 September 2003; pp. 896–898. [Google Scholar]
- Gennaro, A.; Forte, P.; Carozza, R.; Savo Sardaro, M.L.; Ferri, D.; Bitti, A.; Borrelli, G.M.; D’Egidio, M.G.; Ceoloni, C. Pyramiding different alien chromosome segments in durum wheat: Feasibility and breeding potential. Isr. J. Plant Sci. 2007, 55, 267–276. [Google Scholar] [CrossRef]
- Ceoloni, C.; Forte, P.; Gennaro, A.; Micali, S.; Carozza, R.; Bitti, A. Recent developments in durum wheat chromosome engineering. Cytogenet. Genome Res. 2005, 109, 328–334. [Google Scholar] [CrossRef]
- Kuzmanović, L.; Ruggeri, R.; Able, J.A.; Bassi, F.M.; Maccaferri, M.; Tuberosa, R.; De Vita, P.; Rossini, F.; Ceoloni, C. Yield of chromosomally engineered durum wheat-Thinopyrum ponticum recombinant lines in a range of contrasting rain-fed environments. Field Crops Res. 2018, 228, 147–157. [Google Scholar] [CrossRef]
- Ceoloni, C.; Del Signore, G.; Ercoli, L.; Donini, P. Locating the alien chromatin segment in common wheat -Aegilops longissima mildew resistant transfers. Hereditas 1992, 116, 239–245. [Google Scholar] [CrossRef]
- Biagetti, M.; Vitellozzi, F.; Ceoloni, C. Physical mapping of wheat-Aegilops longissima breakpoints in mildew-resistant recombinant lines using FISH with highly repeated and low-copy DNA probes. Genome 1999, 42, 1013–1019. [Google Scholar] [CrossRef]
- Ceoloni, C.; Ciaffi, M.; Lafiandra, D.; Giorgi, B. Chromosome Engineering as a Means of Transferring 1D Storage Protein Genes from Common to Durum Wheat. In Proceedings of the 8th International Wheat Genetics Symposium, Beijing, China, 20–25 July 1993; pp. 159–163. [Google Scholar]
- Gennaro, A.; Forte, P.; Panichi, D.; Lafiandra, D.; Pagnotta, M.A.; D’Egidio, M.G.; Ceoloni, C. Stacking small segments of the 1D chromosome of bread wheat containing major gluten quality genes into durum wheat: Transfer strategy and breeding prospects. Mol. Breed. 2012, 30, 149–167. [Google Scholar] [CrossRef]
- Ceoloni, C.; Forte, P.; Ciaffi, M.; Nenno, M.; Bitti, A.; De Vita, P.; D’Egidio, M. Chromosomally Engineered Durum Wheat: The Potential of Alien Gene Introgressions Affecting Disease Resistance and Quality. In Proceedings of the Durum Wheat Improvement in the Mediterranean Region: New Challenges, 12–14 April 2000; Royo, C., Nachit, M., Di Fonzo, N., Araus, J.L., Eds.; Options Méditerranéennes, Série A: Séminaires Méditerranéens. pp. 363–371. [Google Scholar]
- D’Ovidio, R.; Masci, S. The low-molecular-weight glutenin subunits of wheat gluten. J. Cereal Sci. 2004, 39, 321–339. [Google Scholar] [CrossRef]
- Quaranta, F.; Belocchi, A.; Fornara, M.; Ripa, C.; D’Egidio, M.G. Le Varietà di Frumento duro in Italia: Risultati della rete Nazionale di Sperimentazione 1999–2012; Consiglio per la Ricerca e la Sperimentazione in Agricoltura, Rome-Italy: Rome, Italy, 2013; ISBN 978-88-97081-31-9. [Google Scholar]
- Rossini, F.; Provenzano, M.E.; Sestili, F.; Ruggeri, R. Synergistic effect of sulfur and nitrogen in the organic and mineral fertilization of durum wheat: Grain yield and quality traits in the Mediterranean environment. Agronomy 2018, 8, 189. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Graziani, M.; Maccaferri, M.; Royo, C.; Salvatorelli, F.; Tuberosa, R. QTL dissection of yield components and morpho-physiological traits in a durum wheat elite population tested in contrasting thermo-pluviometric conditions. Crop Pasture Sci. 2014, 65, 80–95. [Google Scholar] [CrossRef]
- Ravel, C.; Dardevet, M.; Leenhardt, F.; Bordes, J.; Joseph, J.L.; Perretant, M.R.; Exbrayat, F.; Poncet, C.; Balfourier, F.; Chanliaud, E.; et al. Improving the yellow pigment content of bread wheat flour by selecting the three homoeologous copies of Psy1. Mol. Breed. 2013, 31, 87–99. [Google Scholar] [CrossRef]
- Ficco, D.B.M.; Mastrangelo, A.M.; Trono, D.; Borrelli, G.M.; De Vita, P.; Fares, C.; Beleggia, R.; Platani, C.; Papa, R. The colours of durum wheat: A review. Crop Pasture Sci. 2014, 65, 1–15. [Google Scholar] [CrossRef]
- Quaranta, F.; Basili, O.; Belocchi, A.; Bottazzi, P.; Cacciatori, P.; Caprara, F.; Ciccoritti, R.; Fabbrini, L.; Mariotti, R.; Mazzon, V.; et al. Risultati della 42a sperimentazione nazionale 2014–2015. Le varietà di grano duro per le semine 2015. Centro Italia versante tirrenico. Inf. Agr. 2015, 33, 21–24. [Google Scholar]
- Quaranta, F.; Basili, O.; Belocchi, A.; Bottazzi, P.; Cacciatori, P.; Caprara, F.; Arcangeli, A.; Fabbrini, L.; Locatelli, M.; Mariotti, R.; et al. Risultati della 43a sperimentazione nazionale 2015–2016. Le varietà di grano duro per le semine 2016. Centro Italia versante tirrenico. Inf. Agr. 2016, 33, 20–23. [Google Scholar]
- Quaranta, F.; Arcangeli, A.; Basili, O.; Belocchi, A.; Bottazzi, P.; Cacciatori, P.; Fabbrini, L.; Moscaritolo, S.; Mariotti, R.; Mazzon, V.; et al. Speciale Grano Duro. Dettaglio regionale dei risultati: Centro Italia versante tirrenico. Inf. Agr. 2017, 31, 49–51. [Google Scholar]
- Troccoli, A.; Borrelli, G.M.; De Vita, P.; Fares, C.; Di Fonzo, N. Durum wheat quality: A multidisciplinary concept. J. Cereal Sci. 2000, 32, 99–113. [Google Scholar] [CrossRef]
- Yesli, A.; Latati, M.; Tellah, S.; Abdellaoui, Z.; Ounane, G. Physicochemical and rheological properties and bread-making potential of durum flour and semolina. J. Food Agric. Environ. 2017, 15, 14–20. [Google Scholar]
- Ren, T.H.; Chen, F.; Yan, B.J.; Zhang, H.Q.; Ren, Z.L. Genetic diversity of wheat-rye 1BL.1RS translocation lines derived from different wheat and rye sources. Euphytica 2012, 183, 133–146. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Liu, C.; Yang, Z.; Deng, K.; Peng, J.; Zhou, J.; Li, G.; Tang, Z.; Ren, Z. Analysis of DNA methylation variation in wheat genetic background after alien chromatin introduction based on methylation-sensitive amplification polymorphism. Chin. Sci. Bull. 2008, 53, 58–69. [Google Scholar] [CrossRef]
- Dong, Z.Y.; Wang, Y.M.; Zhang, Z.J.; Shen, Y.; Lin, X.Y.; Ou, X.F.; Han, F.P.; Liu, B. Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor. Appl. Genet. 2006, 113, 196–205. [Google Scholar] [CrossRef]
- Sall, A.T.; Cisse, M.; Gueye, H.; Kabbaj, H.; Ndoye, I.; Filali-Maltouf, A.; Belkadi, B.; El-Mourid, M.; Ortiz, R.; Bassi, F.M. Heat tolerance of durum wheat (Triticum durum Desf.) elite germplasm tested along the Senegal river. J. Agric. Sci. 2018, 10, 217–233. [Google Scholar]
- Sall, A.T.; Bassi, F.M.; Cisse, M.; Gueye, H.; Ndoye, I.; Filali-Maltouf, A.; Ortiz, R. Durum wheat breeding: In the heat of the Senegal river. Agriculture 2018, 8, 99. [Google Scholar] [CrossRef]
- Bhattacharya, S. Wheat rust back in Europe. Nature 2017, 542, 145–146. [Google Scholar] [CrossRef]
- RustTracker.org—A Global Wheat Rust Monitoring System. Available online: https://rusttracker.cimmyt.org/?p=7083 (accessed on 10 December 2019).
- Ceoloni, C.; Del Signore, G.; Pasquini, M.; Testa, A. Transfer of Mildew Resistance from Triticum longissimaum into Wheat by Induced Homoeologous Recombination. In Proceedings of the Seventh International Wheat Genetics Symposium, Cambridge, UK, 13–19 July 1988; Miller, T., Koebner, R., Eds.; pp. 221–226. [Google Scholar]
- Kwiatek, M.; Belter, J.; Majka, M.; Wiśniewska, H. Allocation of the S-genome chromosomes of Aegilops variabilis Eig. carrying powdery mildew resistance in triticale (× Triticosecale Wittmack). Protoplasma 2016, 253, 329–343. [Google Scholar] [CrossRef]
- Basandrai, A.K.; Basandrai, D. Powdery mildew of wheat and its management. In Management of Wheat and Barley Diseases; Singh, D.P., Ed.; Apple Academic Press Inc.: Oakville, ON, Canada; Waretown, NJ, USA, 2018; Chapter 5; ISBN 978-1-77188-546-1. [Google Scholar]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Bebber, D.; Ramotowski, M.; Gurr, S. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 2013, 3, 985–988. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Jin, Y.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Govindan, V. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 2011, 49, 465–481. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Jin, Y.; Lagudah, E.S.; Ayliffe, M.A.; Bhavani, S.; Rouse, M.N.; Pretorius, Z.A.; Szabo, L.J.; Huerta-Espino, J.; et al. Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology 2015, 105, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Sissons, M. Role of durum wheat composition on the quality of pasta and bread. Food 2008, 2, 75–90. [Google Scholar]
- Zhang, W.; Dubcovsky, J. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor. Appl. Genet. 2008, 116, 635–645. [Google Scholar] [CrossRef] [PubMed]
- ARVALIS - Institut du Végétal. Choisir & Décider Céréales à Paille – Variétés et Interventions D’automne 2018 Synthèse Nationale. Available online: www.arvalis-infos.fr (accessed on 2 July 2019).
- Gennaro, A.; Borrelli, G.M.; D’Egidio, M.G.; De Vita, P.; Ravaglia, S.; Ceoloni, C. A Chromosomally Engineered Durum Wheat-Thinopyrum ponticum Recombinant Line with Novel and Promising Attributes for Varietal Development. In Proceedings of the 10th International Wheat Genetics Symposium, Paestum, Italy, 1–6 September 2003; Pogna, N.E., Romanò, M., Pogna, E.A., Galterio, G., Eds.; pp. 881–883. [Google Scholar]
- Rogers, W.J.; Rickatson, J.M.; Sayers, E.J.; Law, C.N. Dosage effects of chromosomes of homoeologous groups 1 and 6 upon bread-making quality in hexaploid wheat. Theor. Appl. Genet. 1990, 80, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Pogna, N.E.; Mazza, M.; Redaelli, R.; Ng, P.K.W. Gluten Quality and Storage Protein Composition of Durum Wheat Lines Containing the Gli-D1/Glu-D3 Loci. In Proceedings of the 6th International Gluten Workshop, Melbourne, Australia, 1–6 September 1996; Wrigley, C.W., Ed.; pp. 18–22. [Google Scholar]
- Molnár-Láng, M.; Ceoloni, C.; Doležel, J. (Eds.) Alien Introgression in Wheat—Cytogenetics, Molecular Biology, and Genomics; Springer: Cham, Switzerland, 2015; pp. 1–385. ISBN 978-3-319-23493-9. [Google Scholar]
- Prohens, J.; Gramazio, P.; Plazas, M.; Dempewolf, H.; Kilian, B.; Díez, M.J.; Fita, A.; Herraiz, F.J.; Rodríguez-Burruezo, A.; Soler, S.; et al. Introgressiomics: A new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 2017, 213, 158. [Google Scholar] [CrossRef]
| Recombinant Chromosome | Donor Species | Alien Segment Size (% arm) | Alien Genes | Recombinant Line | |||||
|---|---|---|---|---|---|---|---|---|---|
| R11-20 | R9-11 | R9-71 | R9-59 | R2-21 | R11-8 | ||||
| 7AS·7AL-7el1L | Th. ponticum | 23 | Lr19+Sr25+Yp | + | + | + | + | + | + |
| 1AL·1AS−1DS | T. aestivum | 17 | Gli-D1/Glu-D3 | + | + | + | + | + | ‒ |
| 3BL·3BS-3S’S | Ae. longissima | <20 | Pm13 | + | + | + | ‒ | ‒ | ‒ |
| Season | 2014–15 | 2015–16 | 2016–17 |
|---|---|---|---|
| Sowing date | 15 Jan 2015 | 17 Nov 2015 | 6 Dec 2016 |
| Harvest date | 30 Jun 2015 | 30 Jun 2016 | 28 Jun 2017 |
| Crop cycle length (days) | 166 | 226 | 204 |
| Sowing density (seed m-2) | 350 | 350 | 350 |
| Plot size (m2) | 2.25 | 2.25 | 10.5 |
| Total rainfall (mm) | 238 | 383 | 176 |
| Mean temperature at heading (°C) | 18.6 | 13.2 | 12.1 |
| Sowing to heading | |||
| Rainfall (mm) | 215 | 257 | 122 |
| Tmin (°C) | 4.8 | 4.2 | 3.4 |
| Tmean (°C) | 9.6 | 9.3 | 8.7 |
| Tmax (°C) | 15.1 | 14.9 | 14.6 |
| Heading to harvest | |||
| Rainfall (mm) | 22 | 126 | 54 |
| Tmin (°C) | 12.9 | 12.2 | 12.5 |
| Tmean (°C) | 19.7 | 17.9 | 19.7 |
| Tmax (°C) | 26.8 | 24.3 | 27.0 |
| Factor | G | Y | G x Y | R(Y) | Error | ||||
|---|---|---|---|---|---|---|---|---|---|
| df | 7 | 1 | 7 | 4 | 28 | ||||
| GYM2 | 12704.4 | * | 8991.0 | 7932.3 | 1341.9 | 3871.5 | |||
| BM2 | 53432.8 | * | 1197633.8 | ** | 31304.2 | 17905.1 | 16021.7 | ||
| HI | 0.003 | *** | 0.128 | *** | 0.002 | ** | 0.002 | * | 0.000 |
| GNM2 | 5963918.1 | ** | 470143.0 | 2046131.3 | 242809.3 | 1357093.7 | |||
| SNM2 | 1993.7 | * | 3700.1 | * | 550.7 | 252.9 | 776.3 | ||
| PH | 98.3 | *** | 297.8 | *** | 56.1 | ** | 31.0 | 12.2 | |
| HD | 59.4 | *** | 2227.7 | *** | 5.0 | * | 0.792 | 2.1 | |
| TGW | 120.9 | *** | 103.2 | *** | 9.4 | 8.7 | 5.4 | ||
| df | 7 | 1 | 7 | 4 | 364 | ||||
| GYS | 2.1 | *** | 2.5 | ** | 1.9 | *** | 0.3 | 0.2 | |
| GNS | 1470.5 | *** | 3260.0 | *** | 341.2 | *** | 106.2 | 60.2 | |
| GNSP | 1.1 | *** | 37.6 | *** | 0.3 | ** | 0.1 | 0.1 | |
| SFI | 926.3 | *** | 1457.7 | *** | 220.5 | ** | 433.5 | *** | 76.5 |
| SL | 59.9 | *** | 0.004 | 0.504 | 1.381 | * | 0.465 | ||
| SPN | 222.5 | *** | 1047.7 | *** | 26.1 | *** | 5.7 | 2.629 | |
| CHAFF | 0.3 | *** | 1.7 | *** | 0.1 | 0.2 | *** | 0.035 |
| Genotype | R11-20 | R9-11 | R9-71 | R9-59 | R2-21 | R11-8 | Simeto | Karur | h2 | K-S Test | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7el1/1D/3Sl a) | + + + | + + + | + + + | + + ‒ | + + ‒ | + ‒ ‒ | ‒ ‒ ‒ | ‒ ‒ ‒ | (p-value) | |||||||||
| GYM2 (g) | 354.9 | a | 339.8 | ab | 233.2 | b | 261.2 | ab | 261.7 | ab | 276.6 | ab | 249.0 | ab | 327.9 | ab | 0.38 | 0.196 |
| BM2 (g) | 954.2 | a | 907.8 | ab | 714.8 | b | 793.0 | ab | 752.0 | ab | 793.1 | ab | 830.1 | ab | 969.6 | a | 0.41 | 0.058 |
| HI | 0.38 | a | 0.38 | a | 0.33 | bc | 0.34 | abc | 0.36 | ab | 0.35 | abc | 0.31 | c | 0.35 | abc | 0.33 | 0.084 |
| GNM2 | 6691.9 | a | 6663.0 | a | 4827.8 | ab | 5145.1 | ab | 5269.2 | ab | 5372.6 | ab | 3992.8 | b | 6645.2 | a | 0.66 | 0.188 |
| SNM2 | 203.5 | ab | 190.8 | ab | 161.5 | b | 172.3 | ab | 177.8 | ab | 189.8 | ab | 186.7 | ab | 219.9 | a | 0.72 | 0.226 |
| PH (cm) | 79.1 | a | 69.8 | bcd | 67.5 | d | 68.8 | cd | 74.8 | abc | 75.0 | abc | 74.5 | abc | 76.0 | ab | 0.43 | 0.052 |
| HD (days) | 32.3 | c | 36.8 | a | 35.7 | ab | 34.8 | abc | 35.2 | ab | 34.3 | abc | 26.7 | d | 33.0 | bc | 0.92 | 0.369 |
| TGW (g) | 53.0 | b | 50.7 | bc | 48.3 | c | 50.6 | bc | 49.7 | bc | 51.6 | b | 62.4 | a | 49.0 | bc | 0.92 | 0.442 |
| GYS (g) | 3.02 | a | 2.81 | ab | 2.55 | bcd | 2.55 | bcd | 2.75 | abc | 2.71 | bc | 2.47 | cd | 2.39 | d | 0.06 | 0.001 |
| GNS | 57.1 | a | 55.9 | a | 53.3 | abc | 50.1 | bc | 54.0 | ab | 52.7 | abc | 39.6 | d | 48.7 | c | 0.77 | 0.000 |
| GNSP | 2.69 | a | 2.45 | bc | 2.45 | bc | 2.30 | cd | 2.52 | ab | 2.55 | ab | 2.47 | bc | 2.18 | d | 0.69 | 0.053 |
| SFI | 64.6 | abc | 60.9 | bcd | 65.3 | ab | 59.8 | cd | 67.0 | a | 67.0 | a | 55.9 | d | 68.7 | a | 0.76 | 0.101 |
| SL (cm) | 8.7 | abc | 9.2 | a | 8.6 | bc | 9.0 | ab | 9.2 | a | 8.0 | d | 5.8 | e | 8.3 | cd | 0.99 | 0.219 |
| SPN | 21.3 | bc | 22.7 | a | 21.7 | abc | 21.8 | abc | 21.9 | abc | 21.0 | c | 15.9 | d | 22.2 | ab | 0.88 | 0.233 |
| CHAFF (g) | 0.90 | ab | 0.94 | a | 0.82 | bcd | 0.84 | ab | 0.82 | bc | 0.81 | bcd | 0.70 | d | 0.71 | cd | 0.79 | 0.000 |
| Trait | Genotype | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R11−20 | R9−11 | R2_21 | Kanakis | Achille | Ramirez | Karur | Dylan | Simeto | ||||||||||
| 7el1/1D/3Sl a) | + + + | + + + | + + ‒ | ‒ ‒ ‒ | ‒ ‒ ‒ | ‒ ‒ ‒ | ‒ ‒ ‒ | ‒ ‒ ‒ | ‒ ‒ ‒ | |||||||||
| HD (days) | 29 | ab | 31 | a | 31 | a | 25 | de | 28 | bc | 26 | cd | 30 | ab | 23 | f | 23 | ef |
| PH (cm) | 72.7 | ab | 60.7 | c | 72.7 | ab | 77.7 | a | 75.0 | ab | 74.7 | ab | 72.0 | ab | 69.3 | abc | 66.3 | bc |
| GY (t ha−1) | 4.63 | ab | 3.49 | c | 3.67 | bc | 5.22 | a | 4.36 | abc | 4.17 | bc | 3.97 | bc | 3.95 | bc | 3.93 | bc |
| SNM2 | 263.3 | 284.4 | 254.4 | 262.2 | 230.0 | 254.4 | 252.2 | 247.8 | 240.0 | ns | ||||||||
| GNM2 | 9775.0 | a | 7570.0 | abc | 7609.1 | abc | 9426.0 | ab | 7893.5 | abc | 8374.5 | abc | 9059.6 | ab | 7327.4 | bc | 6223.9 | c |
| TGW (g) | 47.5 | bcd | 46.1 | cd | 48.3 | bcd | 55.2 | abc | 55.3 | ab | 49.8 | bcd | 43.8 | d | 54.1 | bc | 63.5 | a |
| TW (kg hL−1) | 81.3 | cd | 80.5 | d | 82.0 | c | 85.4 | a | 86.2 | a | 86.1 | a | 82.1 | c | 83.7 | b | 83.8 | b |
| Trait | Recombinant Line | ANCOVA p value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| R11−20 | R9−11 | R2−21 | Karur | ||||||
| 7el1/1D/3Sl a) | + + + | + + + | + + ‒ | ‒ ‒ ‒ | |||||
| Grain protein content (%) | 15.4 | 16.4 | 15.0 | 16.1 | 0.072 | ||||
| Grain moisture (%) | 11.1 | 11.2 | 11.5 | 11.4 | 0.151 | ||||
| Yellow index (b*) | 31.6 | 30.1 | 31.7 | 29.1 | 0.202 | ||||
| Brown index (100 - L*) | 10.4 | b | 10.0 | b | 11.6 | a | 10.2 | b | 0.002** |
| Semolina yield (%) | 64.2 | 64.1 | 64.3 | 64.5 | 0.932 | ||||
| Ash content (%) | 0.82 | d | 0.85 | c | 0.87 | b | 0.91 | a | 0.000*** |
| Gluten index (%) | 98.5 | a | 95.1 | a | 98.2 | a | 81.8 | b | 0.001** |
| Water absorption (14%) | 60.8 | 61.7 | 59.7 | 62.1 | 0.526 | ||||
| Peak time (min) | 5.0 | a | 5.1 | a | 5.3 | a | 4.2 | b | 0.002** |
| Stability (min) | 18.0 | b | 16.8 | b | 20.7 | a | 10.5 | c | 0.001** |
| Eicc (FU) | 30.0 | c | 37.0 | b | 23.0 | d | 45.0 | a | 0.000*** |
| FQN | 170.0 | b | 150.0 | c | 200.0 | a | 100.0 | d | 0.000*** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuzmanović, L.; Rossini, F.; Ruggeri, R.; Pagnotta, M.A.; Ceoloni, C. Engineered Durum Wheat Germplasm with Multiple Alien Introgressions: Agronomic and Quality Performance. Agronomy 2020, 10, 486. https://doi.org/10.3390/agronomy10040486
Kuzmanović L, Rossini F, Ruggeri R, Pagnotta MA, Ceoloni C. Engineered Durum Wheat Germplasm with Multiple Alien Introgressions: Agronomic and Quality Performance. Agronomy. 2020; 10(4):486. https://doi.org/10.3390/agronomy10040486
Chicago/Turabian StyleKuzmanović, Ljiljana, Francesco Rossini, Roberto Ruggeri, Mario A. Pagnotta, and Carla Ceoloni. 2020. "Engineered Durum Wheat Germplasm with Multiple Alien Introgressions: Agronomic and Quality Performance" Agronomy 10, no. 4: 486. https://doi.org/10.3390/agronomy10040486
APA StyleKuzmanović, L., Rossini, F., Ruggeri, R., Pagnotta, M. A., & Ceoloni, C. (2020). Engineered Durum Wheat Germplasm with Multiple Alien Introgressions: Agronomic and Quality Performance. Agronomy, 10(4), 486. https://doi.org/10.3390/agronomy10040486







