Genome-Wide Detection of SNP Markers Associated with Four Physiological Traits in Groundnut (Arachis hypogaea L.) Mini Core Collection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Phenotyping
2.2. DNA Extraction and Genotyping
2.3. Data Analysis
Linkage Disequilibrium and Marker-Trait Association
3. Results
3.1. Phenotypic Evaluation
3.2. Marker Data
3.3. Linkage Disequilibrium
3.4. Marker-Trait Association
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Crops. Food and Agriculture Organization of the United Nations. 2017. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 3 February 2018).
- Padi, F.K. Genotype × Environment Interaction for Yield and Reaction to Leaf Spot Infections in Groundnut in Semiarid West Africa. Euphytica 2008, 164, 143–161. [Google Scholar] [CrossRef]
- Suther, M.S.; Patel, D.M. Yield and Nutrient Absorption by Groundnut and Iron Availability in Soil as Influenced by Lime and Soil Water. J. Indian Soc. Soil Sci. 1992, 40, 594–596. [Google Scholar]
- Singh, A.L.; Chaudhari, V. Macronutrient Requirement of Groundnut: Effects on Growth and Yield Components. Indian J. Plant Physiol. 2006, 11, 40–409. [Google Scholar]
- Price, A.H.; Horton, P.; Jones, H.G.; Griffiths, H.; Cairns, J.E. Linking Drought-Resistance Mechanisms to Drought Avoidance in Upland Rice Using a QTL Approach: Progress and New Opportunities to Integrate Stomatal and Mesophyll Responses. J. Exp. Bot. 2002, 53, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Monyo, E.; Ozias-Akins, P.; Liang, X.; Guimaraes, P.; Nigam, S.N.; Upadhyaya, H.D.; Janila, P.; Zhang, X.; Guo, B.; et al. Advances in Arachis Genomics for Peanut Improvement. Biotechnol. Adv. 2012, 30, 639–651. [Google Scholar] [CrossRef]
- Nageswara Rao, R.C.; Talwar, H.S.; Wright, G.C. Rapid Assessment of Specific Leaf Area and Leaf Nitrogen in Peanut (Arachis hypogaea L.) Using a Chlorophyll Meter. J. Agron. Crop. Sci. 2001, 186, 175–182. [Google Scholar] [CrossRef]
- Luis, J.M.; Ozias-Akins, P.; Holbrook, C.C.; Kemerait, R.C., Jr.; Snider, J.L.; Liakos, V. Phenotyping Peanut Genotypes for Drought Tolerance. Peanut Sci. 2016, 43, 36–48. [Google Scholar] [CrossRef]
- Hussain, M.; Malik, M.A.; Farooq, M.; Ashraf, M.Y.; Cheema, M.A. Improving drought tolerance by exogenous application of glycinebetaine and salicylic acid in sunflower. J. Agron. Crop. Sci. 2008, 194, 193–199. [Google Scholar] [CrossRef]
- Pandey, M.K.; Upadhyaya, H.D.; Rathore, A.; Vadez, V.; Sheshshayee, M.S.; Sriswathi, M.; Govil, M.; Kumar, A.; Gowda, M.V.; Sharma, S.; et al. Genomewide Association Studies for 50 Agronomic Traits in Peanut Using the “reference Set” Comprising 300 Genotypes from 48 Countries of the Semi-Arid Tropics of the World. PLoS ONE 2014, 9, e105228. [Google Scholar] [CrossRef]
- Janila, P.; Venuprasad, R.; Abhishek, R.U.A.; Reddy, K.R.; Waliyar, F.; Nigam, S.N. Genetic Analysis of Resistance to Late Leaf Spot in Interspecific Groundnuts. Euphytica 2013, 193, 13–25. [Google Scholar]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.V.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P.; et al. Achievements and Prospects of Genomics-Assisted Breeding in Three Legume Crops of the Semi-Arid Tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Clevenger, J.; Pandey, M.K.; Wang, H.; Shasidhar, Y.; Chu, Y.; Fountain, J.C.; Choudhary, D.; Culbreath, A.K.; Liu, X.; et al. High-Density Genetic Map Using Whole-Genome Resequencing for Fine Mapping and Candidate Gene Discovery for Disease Resistance in Peanut. Plant Biotechnol. J. 2018, 16, 1954–1967. [Google Scholar] [CrossRef] [PubMed]
- Willing, E.M.; Hoffmann, M.; Klein, J.D.; Weigel, D.; Dreyer, C. Paired-End RAD-Seq for de Novo Assembly and Marker Design without Available Reference. Bioinformatics 2011, 27, 2187–2193. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity Arrays: A Solid State Technology for Sequence Information Independent Genotyping. Nucleic Acids Res. 2001, 29, E25. [Google Scholar] [CrossRef] [PubMed]
- Valdisser, P.; Pereira, W.J.; Almeida Filho, J.E.; Muller, B.S.F.; Coelho, G.R.C.; de Menezes, I.P.P.; Vianna, J.P.G.; Zucchi, M.I.; Lanna, A.C.; Coelho, A.S.G.; et al. In-Depth Genome Characterization of a Brazilian Common Bean Core Collection Using DArTseq High-Density SNP Genotyping. BMC Genom. 2017, 18, 423. [Google Scholar] [CrossRef]
- Al-Abdallat, A.M.; Karadsheh, A.; Hadadd, N.I.; Akash, M.W.; Ceccarelli, S.; Baum, M.; Hasan, M.; Jighly, A.; Abu Elenein, J.M. Assessment of Genetic Diversity and Yield Performance in Jordanian Barley (Hordeum Vulgare L.) Landraces Grown under Rainfed Conditions. BMC Plant Biol. 2017, 17, 191. [Google Scholar] [CrossRef]
- Abu Zaitoun, S.Y.; Jamous, R.M.; Shtaya, M.J.; Mallah, O.B.; Eid, I.S.; Ali-Shtayeh, M.S. Characterizing Palestinian Snake Melon (Cucumis Melo Var. Flexuosus) Germplasm Diversity and Structure Using SNP and DArTseq Markers. BMC Plant Biol. 2018, 18, 246. [Google Scholar]
- Mousavi-Derazmahalleh, M.; Bayer, P.E.; Hane, J.K.; Valliyodan, B.; Nguyen, H.T.; Nelson, M.N.; Erskine, W.; Varshney, R.K.; Papa, R.; Edwards, D. Adapting Legume Crops to Climate Change Using Genomic Approaches. Plant Cell Environ. 2017, 42, 6–9. [Google Scholar] [CrossRef]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-Wide Association Mapping of Yield and Yield Components of Spring Wheat under Contrasting Moisture Regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef]
- Sneller, C.H.; Mather, D.E.; Crepieux, S. Analytical Approaches and Population Types for Finding and Utilizing QTL in Complex Plant Populations. Crop Sci. 2009, 49, 363–380. [Google Scholar] [CrossRef]
- Baird, S.J. Exploring Linkage Disequilibrium. Mol. Ecol. Resour. 2015, 15, 1017–1019. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, H.D.; Yadav, D.; Dronavalli, N.; Gowda, C.L.L.; Singh, S. Mini Core Germplasm Collections for Infusing Genetic Diversity in Plant Breeding Programs. Electron. J. Plant Breed. 2010, 1, 1294–1309. [Google Scholar]
- Waliyar, F.; Kumar, K.V.K.; Diallo, M.; Traore, A.; Mangala, U.N.; Upadhyaya, H.D.; Sudini, H. Resistance to Pre-Harvest Aflatoxin Contamination in ICRISAT’s Groundnut Mini Core Collection. Eur. J. Plant Pathol. 2016, 145, 901–913. [Google Scholar] [CrossRef]
- Holbrook, C.C.; Anderson, W.F.; Pittman, R.N. Selection of a Core Collection from the U.S. Germplasm Collection of Peanut. Crop Sci. 1993, 33, 859–861. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Bramel, P.J.; Ortiz, R.; Singh, S. Developing a Mini Core of Peanut for Utilization of Genetic Resources. Crop Sci. 2002, 42, 2150–2156. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Dwivedi, S.L.; Nadaf, H.L.; Singh, S. Phenotypic Diversity and Identification of Wild Arachis Accessions with Useful Agronomic and Nutritional Traits. Euphytica 2011, 182, 103. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity Arrays Technology: A Generic Genome Profiling Technology on Open Platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Usinglme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Gangurde, S.S.; Kumar, R.; Pandey, A.K.; Burow, M.; Laza, H.E.; Nayak, S.N.; Guo, B.; Liao, B.; Bhat, R.S.; Madhuri, N.; et al. Climate-Smart Groundnuts for Achieving High Productivity and Improved Quality: Current Status, Challenges, and Opportunities. In Genomic Designing of Climate-Smart Oilseed Crops; Kole, C., Ed.; Springer: Cham, Switzerland, 2019; pp. 133–172. [Google Scholar]
- Tattaris, M.; Reynolds, M.P.; Chapman, S.C. A Direct Comparison of Remote Sensing Approaches for High-Throughput Phenotyping in Plant Breeding. Front. Plant Sci. 2016, 7, 1131. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Reynolds, M.P.; Sansaloni, C. Genome-Wide Association Analyses Identify QTL Hotspots for Yield and Component Traits in Durum Wheat Grown under Yield Potential, Drought, and Heat Stress Environments. Front. Plant Sci. 2018, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Nielen, S.; Vidigal, B.S.; Leal-Bertioli, S.C.; Ratnaparkhe, M.; Paterson, A.H.; Garsmeur, O.; D’Hont, A.; Guimaraes, P.M.; Bertioli, D.J. Matita, a New Retroelement from Peanut: Characterization and Evolutionary Context in the Light of the Arachis A-B Genome Divergence. Mol. Genet. Genom. 2012, 287, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Moretzsohn, M.C.; Gouvea, E.G.; Inglis, P.W.; Leal-Bertioli, S.C.; Valls, J.F.; Bertioli, D.J. A Study of the Relationships of Cultivated Peanut (Arachis hypogaea) and Its Most Closely Related Wild Species Using Intron Sequences and Microsatellite Markers. Ann. Bot. 2013, 111, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The Genome Sequences of Arachis duranensis and Arachis ipaensis, the Diploid Ancestors of Cultivated Peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Denčić, S.; Varshney, R.; Börner, A.; Kobiljski, B. Genome-Wide Association Mapping: A Case Study in Bread Wheat (Triticum aestivum L.). Mol. Breed. 2011, 27, 37–58. [Google Scholar] [CrossRef]
- Mwadzingeni, L.; Shimelis, H.; Rees, D.J.; Tsilo, T.J. Genome-Wide Association Analysis of Agronomic Traits in Wheat under Drought-Stressed and Non-Stressed Conditions. PLoS ONE 2017, 12, e0171692. [Google Scholar] [CrossRef]
- Nigam, S.N.; Upadhyaya, H.D.; Chandra, S.; Nageswara Rao, R.C.; Wright, G.C.; Reddy, A.G.S. Gene Effects for Specific Leaf Area and Harvest Index in Three Crosses of Groundnut (Arachis hypogaea). Ann. Appl. Biol. 2001, 139, 301–306. [Google Scholar] [CrossRef]
- Qin, H.; Feng, S.; Chen, C.; Guo, Y.; Knapp, S.; Culbreath, A.; He, G.; Wang, M.L.; Zhang, X.; Holbrook, C.C.; et al. An Integrated Genetic Linkage Map of Cultivated Peanut (Arachis hypogaea L.) Constructed from Two RIL Populations. Theor. Appl. Genet. 2012, 124, 653–664. [Google Scholar] [CrossRef]
- Farre, A.; Benito, I.L.; Cistue, L.; de Jong, J.H.; Romagosa, I.; Jansen, J. Linkage Map Construction Involving a Reciprocal Translocation. Theor. Appl. Genet. 2011, 122, 1029–1037. [Google Scholar] [CrossRef]
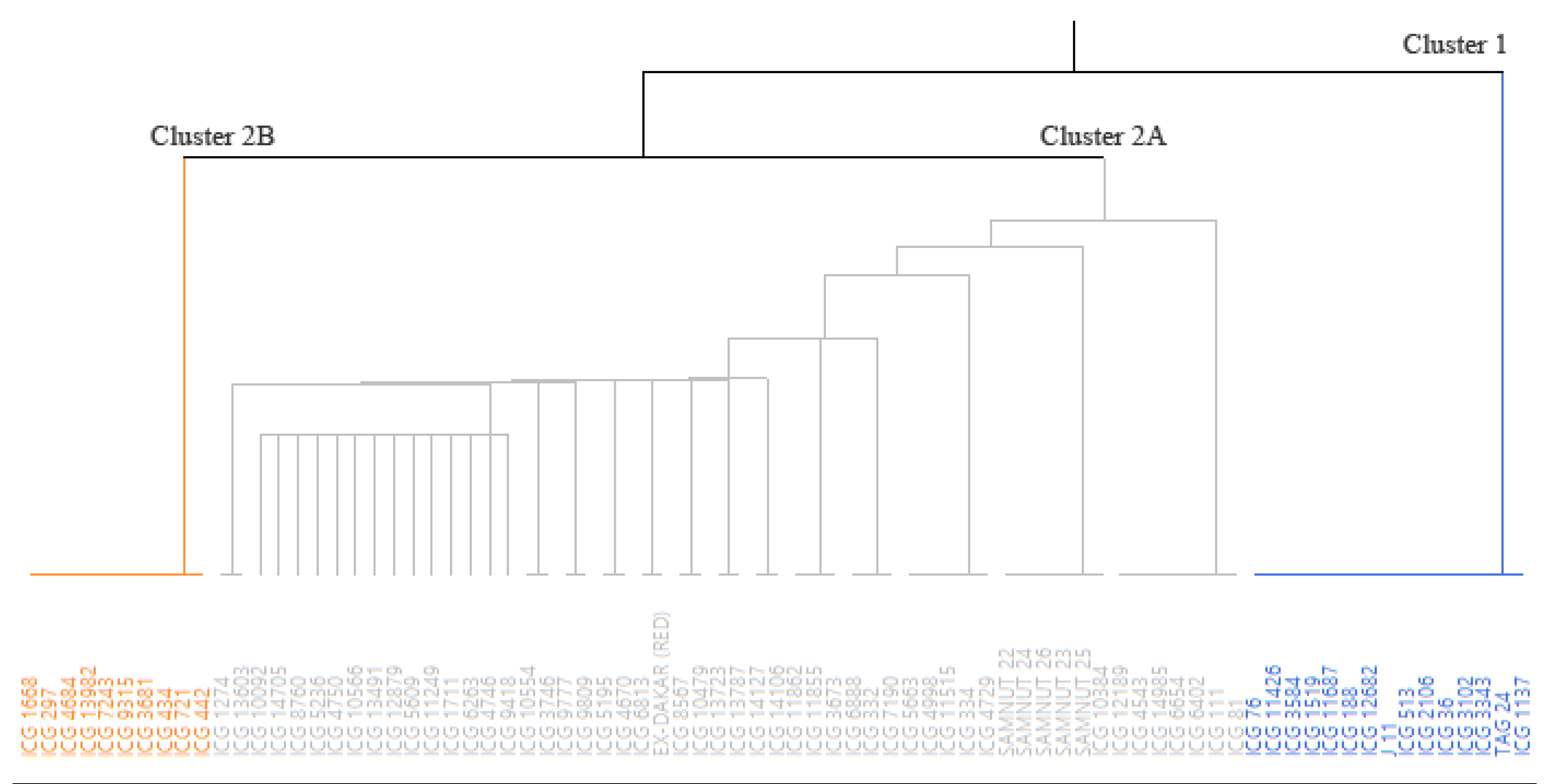
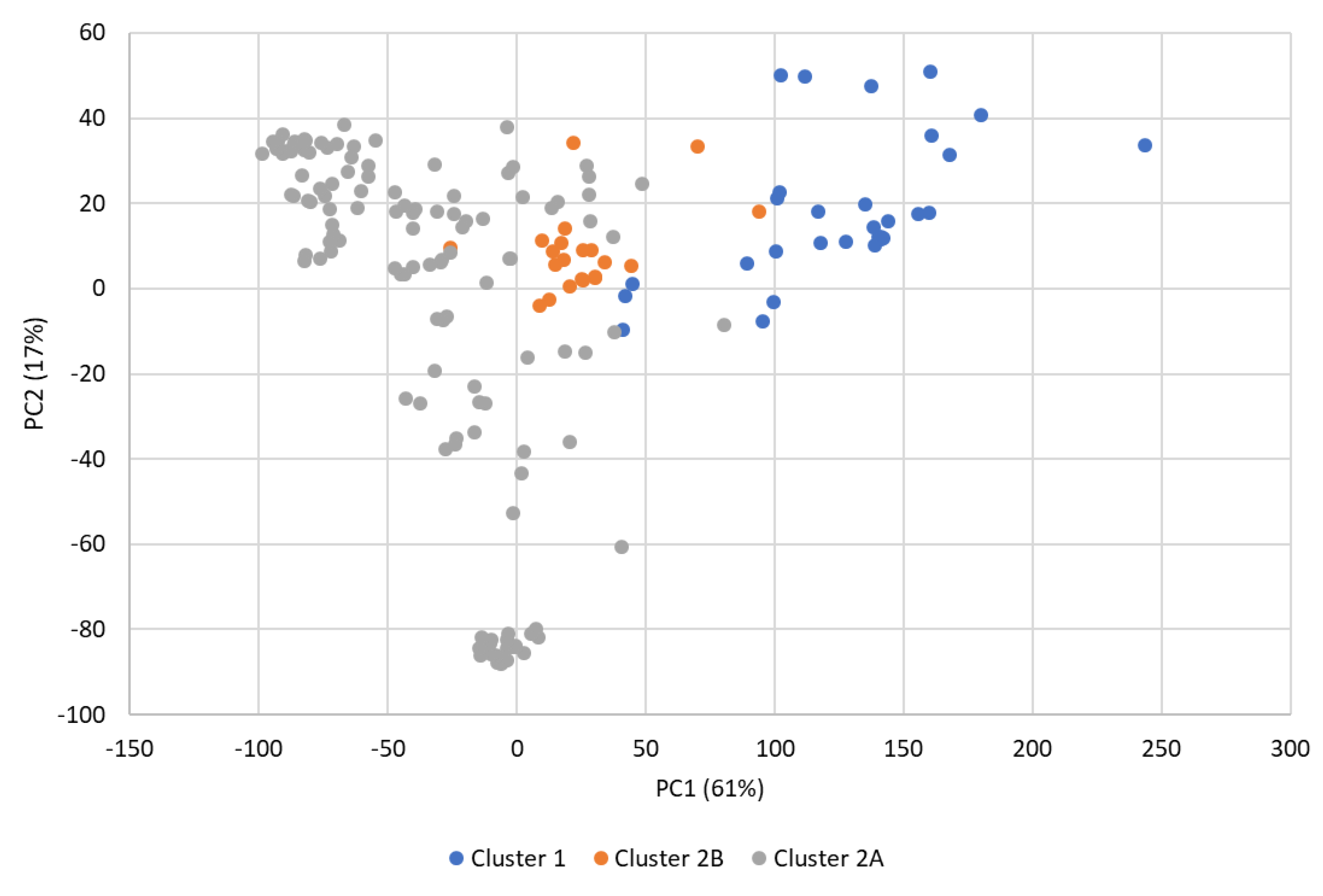
| Effect | df | Canopy Temperature | Leaf Area Index | SPAD Chlorophyll Meter Reading | NDVI |
|---|---|---|---|---|---|
| Rep (Environment) | 2 | 679.62 | 7.28 | 18.73 | 0.47 |
| Environment | 1 | 4820.38 ** | 54.69 ** | 7149.59 ** | 13.81 ** |
| Lines | 124 | 7.82 ** | 0.66 | 49.14 ** | 0.43 ** |
| Environment × Lines | 124 | 7.69 | 0.49 | 26.09 ** | 0.001 |
| Error | 248 | 7.25 | 0.84 | 15.42 | 0.02 |
| Heritability | 0.22 | 0.03 | 0.55 | 0.25 |
| SPAD Chlorophyll Meter Reading | Leaf Area Index | Canopy Temperature | Normalized Difference Vegetative Index (NDVI) | |
|---|---|---|---|---|
| SPAD (Soil Plant Analysis Development) chlorophyll reading | ||||
| Leaf area index | −0.03 | |||
| 0.72 | ||||
| Canopy temperature | −0.16 | −0.19 | ||
| 0.08 | 0.03 | |||
| NDVI | 0.08 | 0.12 | −0.25 | |
| 0.37 | 0.19 | 0.01 |
| Trait | Marker | Chromosome | Position (Mbp) | p-Value | Marker R2 | Allelic Effect | |
|---|---|---|---|---|---|---|---|
| Leaf area index | M1 | 100028731 | A03 | 112.30 | 0.0007 | 0.073 | −1.33 |
| M1 | 100028731 | B03 | 114.70 | 0.00084 | 0.068 | −1.31 | |
| M2 | 100055743 | A06 | 29.59 | 0.00089 | 0.069 | 1.96 | |
| M2 | 100055743 | B07 | 39.39 | 0.00083 | 0.068 | 1.97 | |
| Canopy temperature | M3 | 100002202 | A02 | 14.18 | 0.00099 | 0.098 | −1.74 |
| M3 | 100002202 | B02 | 17.44 | 0.00089 | 0.097 | −1.95 | |
| M4 | 100006533 | A03 | 12.02 | 2.06 × 10−5 | 0.166 | −4.27 | |
| M4 | 100006533 | B03 | 14.68 | 2.19 × 10−5 | 0.163 | −4.22 | |
| M5 | 100057474 | A05 | 4.01 | 0.00017 | 0.128 | −2.86 | |
| M5 | 100057474 | B05 | 3.92 | 0.00018 | 0.125 | −2.87 | |
| M6 | 100007799 | A05 | 4.75 | 0.00078 | 0.102 | −2.33 | |
| M6 | 100007799 | B05 | 4.84 | 0.00046 | 0.109 | −2.56 | |
| M7 | 100008002 | A08 | 7.71 | 0.00097 | 0.098 | −2.29 | |
| M7 | 100008002 | B07 | 113.51 | 0.00057 | 0.105 | −2.52 | |
| M8 | 100007218 | A10 | 78.51 | 0.00069 | 0.104 | −1.51 | |
| M8 | 100007218 | B10 | 103.61 | 0.00062 | 0.104 | −1.49 | |
| M9 | 100002387 | B01 | 135.56 | 0.00097 | 0.096 | −4.75 | |
| SPAD chlorophyll meter reading | M10 | 100000269 | B05 | 17.00 | 0.00099 | 0.208 | −11.65 |
| NDVI | M11 | 100118763 | A04 | 67.54 | 0.00069 | 0.070 | 0.16 |
| M11 | 100118763 | B02 | 106.74 | 0.001 | 0.066 | 0.16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaibu, A.S.; Sneller, C.; Motagi, B.N.; Chepkoech, J.; Chepngetich, M.; Miko, Z.L.; Isa, A.M.; Ajeigbe, H.A.; Mohammed, S.G. Genome-Wide Detection of SNP Markers Associated with Four Physiological Traits in Groundnut (Arachis hypogaea L.) Mini Core Collection. Agronomy 2020, 10, 192. https://doi.org/10.3390/agronomy10020192
Shaibu AS, Sneller C, Motagi BN, Chepkoech J, Chepngetich M, Miko ZL, Isa AM, Ajeigbe HA, Mohammed SG. Genome-Wide Detection of SNP Markers Associated with Four Physiological Traits in Groundnut (Arachis hypogaea L.) Mini Core Collection. Agronomy. 2020; 10(2):192. https://doi.org/10.3390/agronomy10020192
Chicago/Turabian StyleShaibu, Abdulwahab S., Clay Sneller, Babu N. Motagi, Jackline Chepkoech, Mercy Chepngetich, Zainab L. Miko, Adamu M. Isa, Hakeem A. Ajeigbe, and Sanusi G. Mohammed. 2020. "Genome-Wide Detection of SNP Markers Associated with Four Physiological Traits in Groundnut (Arachis hypogaea L.) Mini Core Collection" Agronomy 10, no. 2: 192. https://doi.org/10.3390/agronomy10020192
APA StyleShaibu, A. S., Sneller, C., Motagi, B. N., Chepkoech, J., Chepngetich, M., Miko, Z. L., Isa, A. M., Ajeigbe, H. A., & Mohammed, S. G. (2020). Genome-Wide Detection of SNP Markers Associated with Four Physiological Traits in Groundnut (Arachis hypogaea L.) Mini Core Collection. Agronomy, 10(2), 192. https://doi.org/10.3390/agronomy10020192





