Identification and Validation of Quantitative Trait Loci for Grain Number in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Materials and Methods
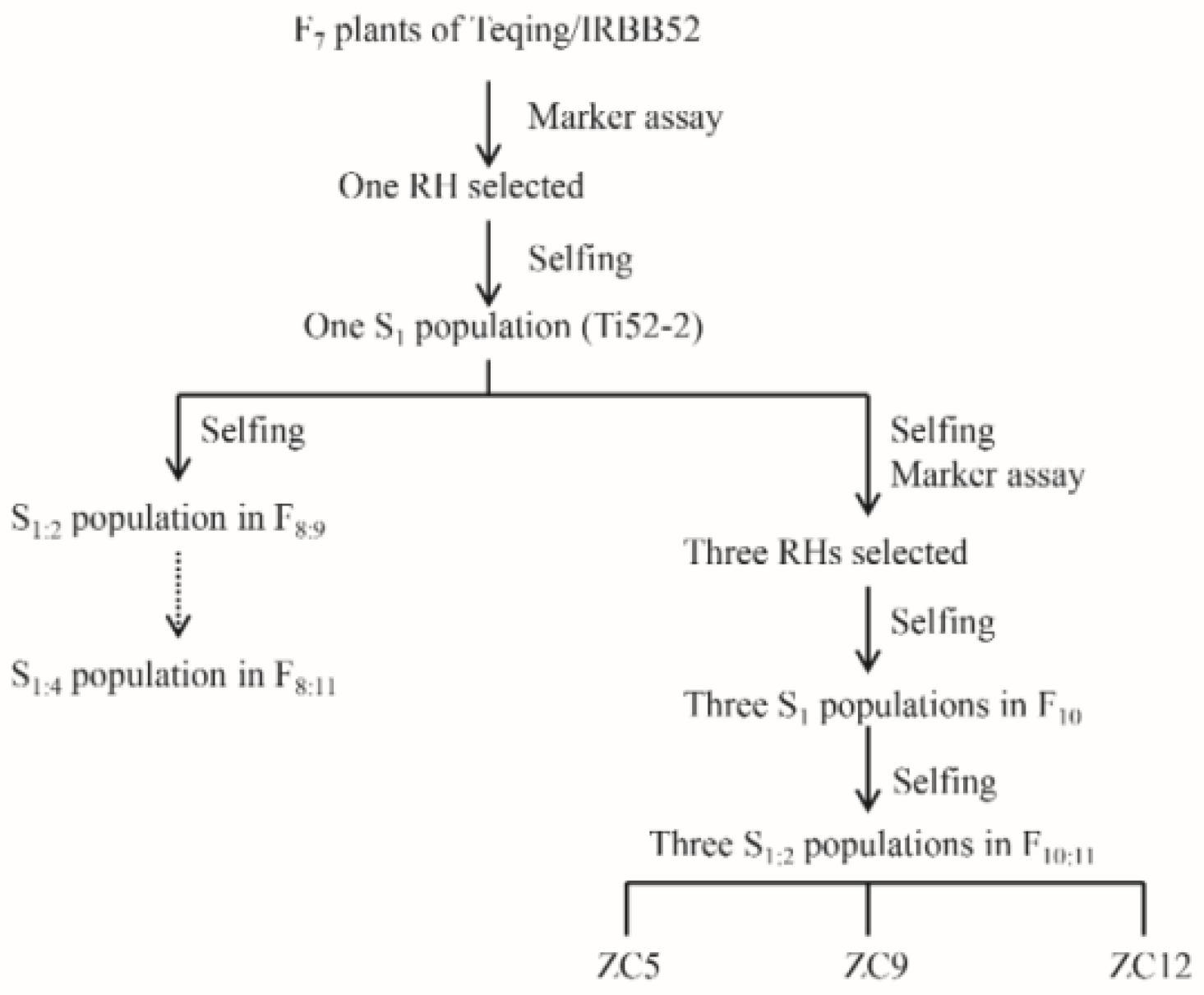
2.1. Plant Materials
2.2. Field Trials and Phenotypic Evaluation
2.3. DNA Marker Analysis
2.4. Data Analysis
3. Results
3.1. Phenotypic Variance
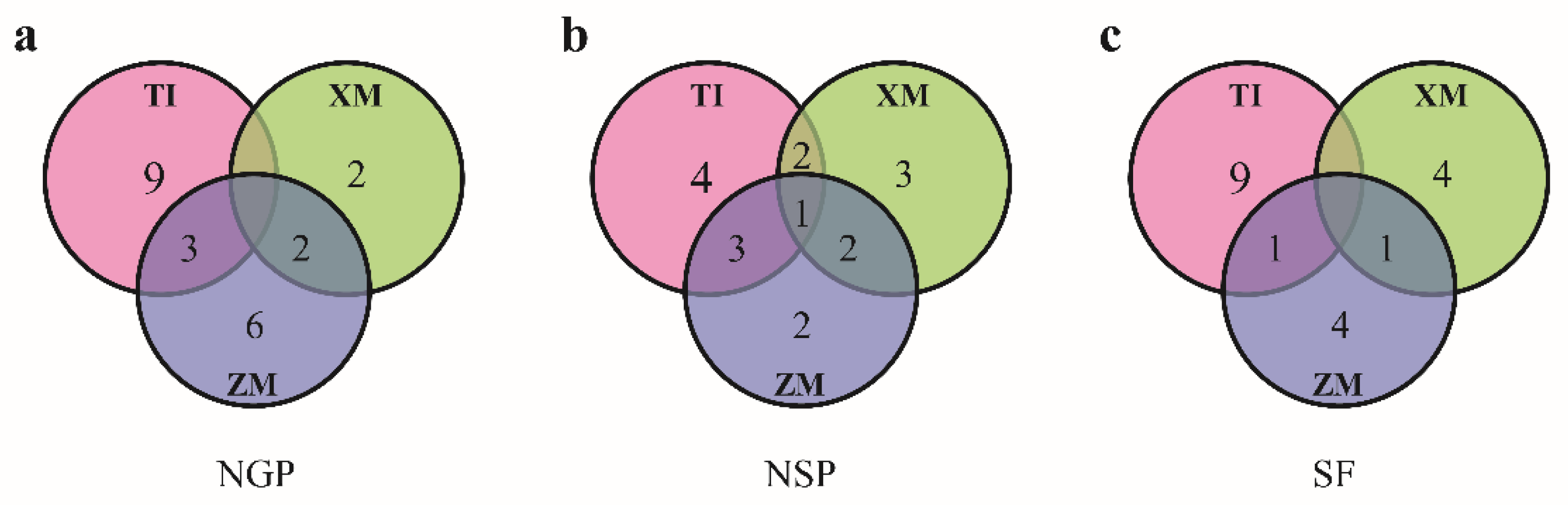
3.2. QTLs Detected in Three RIL Populations
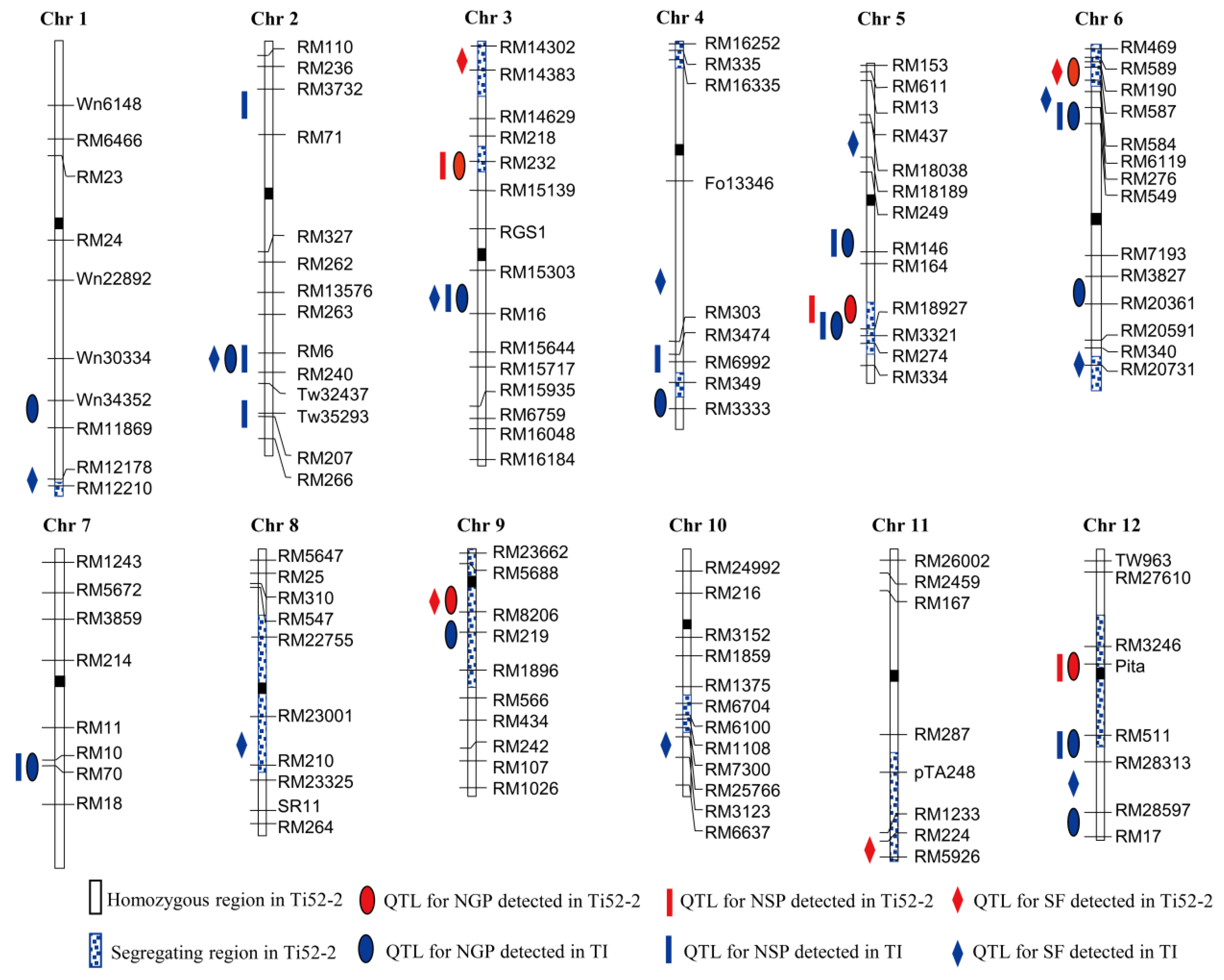
3.3. QTLs Detected in Ti52-2 Population
3.4. Validation of QTLs for NGP in ZC5, ZC9, and ZC12 Populations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, H.; Chen, T.T.; Liu, L.J.; Wang, Z.Q.; Yang, J.C.; Zhang, J.H. Performance in grain yield and physiological traits of rice in the Yangtze River Basin of China during the last 60 yr. J. Integr. Agric. 2013, 12, 57–66. [Google Scholar] [CrossRef]
- Yang, S.H.; Cheng, B.Y.; Shen, W.F.; Liao, X.Y. Progress and strategy of the improvement of indica rice varieties in the Yangtse valley of China. Chin. J. Rice Sci. 2004, 18, 89–93. [Google Scholar]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wu, B.; Xing, Y. Yield-related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 2012, 54, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Li, J.X.; Yu, S.B.; Xu, C.G.; Tan, Y.F.; Gao, Y.J.; Li, X.H.; Zhang, Q. Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid. Theor. Appl. Genet. 2000, 101, 248–254. [Google Scholar] [CrossRef]
- Yu, S.B.; Li, J.X.; Xu, C.G.; Tan, Y.F.; Gao, Y.J.; Li, X.H.; Zhang, Q.; Saghai Maroof, M.A. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 1997, 94, 9226–9231. [Google Scholar] [CrossRef]
- Hittalmani, S.; Shashidhar, H.E.; Bagali, P.G.; Huang, N.; Sidhu, J.S.; Singh, V.P.; Khush, G.S. Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 2002, 125, 207–214. [Google Scholar] [CrossRef]
- Lu, C.; Shen, L.; Tan, Z.; Xu, Y.; He, P.; Chen, Y.; Zhu, L. Comparative mapping of QTLs for agronomic traits of rice across environments by using a doubled-haploid population. Theor. Appl. Genet. 1997, 94, 145–150. [Google Scholar] [CrossRef]
- Xiao, J.; Li, J.; Yuan, L.; Tanksley, S.D. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef]
- Zhuang, J.Y.; Fan, Y.Y.; Rao, Z.M.; Wu, J.L.; Xia, Y.W.; Zheng, K.L. Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice. Theor. Appl. Genet. 2002, 105, 1137–1145. [Google Scholar] [CrossRef]
- Liu, T.; Mao, D.; Zhang, S.; Xu, C.; Xing, Y. Fine mapping SPP1, a QTL controlling the number of spikelets per panicle, to a BAC clone in rice (Oryza sativa). Theor. Appl. Genet. 2009, 118, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, L.; Liu, T.; Xu, C.; Xing, Y. Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds. Theor. Appl. Genet. 2009, 118, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Zhu, Z.; Zhang, B.; Tan, L.; Fu, Y.; Wang, X.; Sun, C.Q. Fine mapping of a quantitative trait locus for grain number per panicle from wild rice (Oryza rufipogon Griff.). Theor. Appl. Genet. 2006, 113, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.X.; Zhu, M.Z.; Shi, M.; Gao, J.P.; Lin, H.X. Fine mapping and candidate gene analysis of spd6, responsible for small panicle and dwarfness in wild rice (Oryza rufipogon Griff.). Theor. Appl. Genet. 2009, 119, 827–836. [Google Scholar] [CrossRef]
- Hu, Z.; Cao, L.; Sun, X.; Zhu, Y.; Zhang, T.; Jiang, L.; Liu, Y.; Dong, S.; Sun, D.; Yang, J.; et al. Fine mapping of a major quantitative trait locus, qgnp7(t), controlling grain number per panicle in African rice (Oryza glaberrima S.). Breed. Sci. 2018, 68, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tao, Y.; Yuan, Y.; Zhang, Y.; Miao, J.; Zhang, R.; Yi, C.; Gong, Z.; Yang, Z.; Liang, G. Characterisation of a novel quantitative trait locus, GN4-1, for grain number and yield in rice (Oryza sativa L.). Theor. Appl. Genet. 2018, 131, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Ellur, R.K.; Singh, A.K.; Nagarajan, M.; Singh, B.D.; Singh, N.K. Effect of qGN4.1 QTL for grain number per panicle in genetic backgrounds of twelve different mega varieties of rice. Rice 2018, 11, 8. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, K.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Huo, X.; Wu, S.; Zhu, Z.; Liu, F.; Fu, Y.; Cai, H.; Sun, X.; Gu, P.; Xie, D.; Tan, L.; et al. NOG1 increases grain production in rice. Nat. Commun. 2017, 8, 1497. [Google Scholar] [CrossRef]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Liu, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Terao, T.; Nagata, K.; Morino, K.; Hirose, T. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 2010, 120, 875–893. [Google Scholar] [CrossRef] [PubMed]
- Ookawa, T.; Hobo, T.; Yano, M.; Murata, K.; Ando, T.; Miura, H.; Asano, K.; Ochiai, Y.; Ikeda, M.; Nishitani, R.; et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010, 1, 132. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mi, X.F.; Shan, J.X.; Li, X.M.; Xu, J.L.; Lin, H.X. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016, 12, e1006386. [Google Scholar] [CrossRef]
- Fujita, D.; Trijatmiko, K.R.; Tagle, A.G.; Sapasap, M.V.; Koide, Y.; Sasaki, K.; Tsakirpaloglou, N.; Gannaban, R.B.; Nishimura, T.; Yanagihara, Y.; et al. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 20431–20436. [Google Scholar] [CrossRef]
- Zhang, G.H.; Li, S.Y.; Wang, L.; Ye, W.J.; Zeng, D.L.; Rao, Y.C.; Peng, Y.L.; Hu, J.; Yang, Y.L.; Xu, J.; et al. LSCHL4 from Japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol. Plant 2014, 7, 1350–1364. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Weng, X.; Wang, L.; Wang, J.; Hu, Y.; Du, H.; Xu, C.; Xing, Y.; Li, X.; Xiao, J.; Zhang, Q. Grain number, plant height, and heading date 7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014, 164, 735–747. [Google Scholar] [CrossRef]
- Kinoshita, N.; Kato, M.; Koyasaki, K.; Kawashima, T.; Nishimura, T.; Hirayama, Y.; Takamure, I.; Sato, T.; Kato, K. Identification of quantitative trait loci for rice grain quality and yield-related traits in two closely related Oryza sativa L. subsp. Japonica cultivars grown near the northernmost limit for rice paddy cultivation. Breed. Sci. 2017, 67, 191–206. [Google Scholar] [CrossRef]
- Yamamoto, T.; Yonemaru, J.; Yano, M. Towards the understanding of complex traits in rice: Substantially or superficially? DNA Res. 2009, 16, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Ando, T.; Nonoue, Y.; Mizubayashi, T.; Kitazawa, N.; Shomura, A.; Matsubara, K.; Ono, N.; Mizobuchi, R.; Shibaya, T.; et al. Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica × indica cross. Breed. Sci. 2015, 65, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Guo, L.; Ma, H.; Chen, Y.Y.; Zhang, H.W.; Ying, J.Z.; Zhuang, J.Y. Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.). Theor. Appl. Genet. 2014, 127, 2515–2524. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.J.; Fan, Y.Y.; Wang, K.; Huang, D.R.; Liu, W.Z.; Ying, J.Z.; Zhuang, J.Y. Rice flowering locus T1 plays an important role in heading date influencing yield traits in rice. Sci. Rep. 2017, 7, 4918. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Zhu, Y.; Fan, Y.; Zhuang, J. Fine-mapping of qTGW1.2a, a quantitative trait locus for 1000-grain weight in rice. Rice Sci. 2019, 26, 220–228. [Google Scholar]
- Dong, Q.; Zhang, Z.H.; Wang, L.L.; Zhu, Y.J.; Fan, Y.Y.; Mou, T.M.; Ma, L.Y.; Zhuang, J.Y. Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice. Rice 2018, 11, 44. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.Y.; Zhu, Y.J.; Fan, Y.Y.; Zhuang, J.Y. Validation of qGS10, a quantitative trait locus for grain size on the long arm of chromosome 10 in rice (Oryza sativa L.). J. Integr. Agric. 2017, 16, 16–26. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Huang, D.R.; Fan, Y.Y.; Zhang, Z.H.; Ying, J.Z.; Zhuang, J.Y. Detection of QTLs for yield heterosis in rice using a RIL population and its testcross population. Int. J. Genomics 2016, 2016, 2587823. [Google Scholar] [CrossRef]
- Mei, D.Y.; Zhu, Y.J.; Yu, Y.H.; Fan, Y.Y.; Huang, D.R.; Zhuang, J.Y. Quantitative trait loci for grain chalkiness and endosperm transparency detected in three recombinant inbred line populations of indica rice. J. Integr. Agric. 2013, 12, 1–11. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Zhang, Z.H.; Chen, J.Y.; Fan, Y.Y.; Mou, T.M.; Tang, S.Q.; Zhuang, J.Y. Fine mapping of qTGW10-20.8, a QTL having important contribution to grain weight variation in rice. Crop J. 2019, 1, 587–597. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, Y.J.; Fan, Y.Y.; Huang, T.X.; Zhang, J.F.; Xie, H.A.; Zhuang, J.Y. Identification and verification of quantitative trait loci for eating and cooking quality of rice (Oryza sativa). Plant Breed. 2019, 138, 568–576. [Google Scholar] [CrossRef]
- Sun, Z.C.; Zhu, Y.J.; Chen, J.Y.; Zhang, H.; Zhang, Z.H.; Niu, X.J.; Fan, Y.Y.; Zhuang, J.Y. Minor-effect QTL for heading date detected in crosses between indica rice cultivar Teqing and near isogenic lines of IR24. Crop J. 2018, 6, 291–298. [Google Scholar] [CrossRef]
- Zheng, K.; Huang, N.; Bennett, J.; Khush, G.S. PCR-based marker-assisted selection in rice breeding. In IRRI Discussion Paper Series No.12; International Rice Research Institute: Los Banos, Philippines, 1995. [Google Scholar]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.G.; McCouch, S.R. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 1997, 95, 553–567. [Google Scholar] [CrossRef]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newberg, L.A. MAPMARKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- McCouch, S.R.; CGSNL (Committee on Gene Symbolization, Nomenclature and Linkage, Rice Genetics Cooperative). Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef]
- Wang, S.C.; Basten, J.; Zeng, Z.B. Windows QTL Cartographer 2.5, Department of Statistics; North Carolina State University: Raleigh, NC, USA, 2012. [Google Scholar]
- Tian, F.; Li, D.J.; Fu, Q.; Zhu, Z.F.; Fu, Y.C.; Wang, X.K.; Sun, C.Q. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 2006, 112, 570–580. [Google Scholar] [CrossRef]
- Thomson, M.J.; Tai, T.H.; McClung, A.M.; Lai, X.H.; Hinga, M.E.; Lobos, K.B.; Xu, Y.; Martinez, C.P.; McCouch, S.R. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 2003, 107, 479–493. [Google Scholar] [CrossRef]
- Zhu, A.; Sun, Z.; Zhu, Y.; Zhang, H.; Niu, X.; Fan, Y.; Zhang, Z.; Zhuang, J. Identification of QTL for grain weight and grain shape using populations derived from residual heterozygous lines of indica rice. Chin. J. Rice Sci. 2019, 33, 144–151. [Google Scholar]
- Wu, W.; Zheng, X.M.; Luo, G.; Zhong, Z.; Gao, H.; Chen, L.; Wu, C.; Wang, H.J.; Wang, Q.; Zhou, K.; et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 2013, 110, 2775–2780. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Liu, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.W.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.Y.; Chen, C.; Wu, J.R.; Cheng, S.H.; Zhuang, J.Y. Quantitative trait loci for yield traits located between Hd3a and Hd1 on short arm of chromosome 6 in rice. Rice Sci. 2011, 18, 257–264. [Google Scholar] [CrossRef]

| Trait a | Population b | Year | Mean | SD | CV | Range | Skew | Kurt | Parental Mean | pd | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male c | ||||||||||
| NGP | TI | 2009 | 107.6 | 20.9 | 0.194 | 65.1–166.8 | 0.34 | −0.16 | 187.8 | 80.9 | |
| 2010 | 150.7 | 26.6 | 0.177 | 92.5–259.9 | 0.57 | 0.81 | 172.7 | 122.7 | |||
| 2011 | 166.5 | 25.0 | 0.150 | 112.4–232.4 | 0.35 | −0.05 | 212.9 | 124.2 | |||
| 2016 | 161.5 | 30.5 | 0.189 | 54.3–308.3 | 0.35 | 2.35 | 187.7 | 131.3 | 0.011 | ||
| ZM | 1999 | 90.5 | 25.1 | 0.278 | 39.5–185.5 | 0.68 | 0.70 | 114.4 | 117.2 | ||
| 2000 | 71.2 | 16.7 | 0.235 | 26.0–130.4 | 0.20 | 0.15 | 88.7 | 88.0 | |||
| 2003 | 73.7 | 16.1 | 0.219 | 22.8–117.7 | −0.12 | 0.26 | 59.6 | 70.8 | |||
| 2016 | 104.0 | 26.9 | 0.259 | 33.9–183.1 | 0.17 | −0.06 | 70.6 | 88.0 | 0.157 | ||
| XM | 1999 | 84.2 | 22.3 | 0.265 | 39.4–173.2 | 0.76 | 1.09 | 101.3 | 117.2 | ||
| 2000 | 71.0 | 19.8 | 0.279 | 21.3–131.2 | 0.22 | 0.10 | 72.2 | 88.0 | |||
| 2003 | 79.9 | 15.3 | 0.191 | 41.4–122.9 | 0.34 | 0.08 | 70.2 | 70.8 | 0.170 | ||
| NSP | TI | 2009 | 121.2 | 24.2 | 0.200 | 73.3–190.0 | 0.38 | −0.18 | 205.8 | 95.1 | |
| 2010 | 194.0 | 32.5 | 0.168 | 128.4–318.3 | 0.75 | 1.00 | 226.7 | 159.3 | |||
| 2011 | 204.2 | 30.7 | 0.150 | 141.8–298.8 | 0.59 | 0.01 | 248.4 | 165.0 | |||
| 2016 | 188.6 | 35.9 | 0.191 | 87.6–351.2 | 0.65 | 1.88 | 225.9 | 141.6 | 0.002 | ||
| ZM | 1999 | 129.7 | 31.6 | 0.244 | 61.9–242.2 | 0.66 | 0.75 | 126.2 | 138.5 | ||
| 2000 | 119.4 | 22.0 | 0.185 | 68.7–191.2 | 0.33 | −0.03 | 118.4 | 123.3 | |||
| 2003 | 105.9 | 21.6 | 0.204 | 57.1–175.4 | 0.46 | 0.28 | 88.3 | 88.3 | |||
| 2016 | 132.0 | 28.2 | 0.213 | 62.5–226.0 | 0.40 | 0.18 | 105.4 | 112.9 | 0.094 | ||
| XM | 1999 | 121.8 | 27.5 | 0.226 | 56.9–203.4 | 0.46 | 0.20 | 117.4 | 138.5 | ||
| 2000 | 113.6 | 20.9 | 0.184 | 56.0–193.6 | 0.32 | 0.97 | 101.2 | 123.3 | |||
| 2003 | 105.8 | 20.2 | 0.191 | 60.4–164.0 | 0.40 | −0.17 | 99.2 | 88.3 | 0.423 | ||
| SF | TI | 2009 | 89.0 | 4.9 | 0.055 | 70.1–96.2 | −1.05 | 1.28 | 91.4 | 88.7 | |
| 2010 | 77.9 | 7.3 | 0.093 | 58.8–91.4 | −0.55 | −0.10 | 75.8 | 79.1 | |||
| 2011 | 81.8 | 7.0 | 0.085 | 60.5–94.7 | −0.46 | −0.23 | 85.7 | 79.8 | |||
| 2016 | 85.9 | 6.5 | 0.076 | 63.4–96.3 | −0.95 | 0.82 | 82.9 | 86.4 | 0.856 | ||
| ZM | 1999 | 69.9 | 10.1 | 0.145 | 40.7–92.6 | −0.12 | −0.26 | 90.7 | 84.6 | ||
| 2000 | 59.9 | 10.5 | 0.175 | 29.6–89.1 | −0.33 | 0.22 | 73.4 | 70.8 | |||
| 2003 | 69.8 | 9.4 | 0.134 | 38.4–92.0 | −0.43 | 0.17 | 67.5 | 80.1 | |||
| 2016 | 78.4 | 9.1 | 0.116 | 42.0–94.5 | −0.99 | 1.01 | 75.7 | 79.1 | 0.682 | ||
| XM | 1999 | 69.7 | 11.7 | 0.168 | 34.8–90.3 | −0.37 | −0.52 | 86.3 | 84.6 | ||
| 2000 | 62.1 | 11.9 | 0.191 | 28.7–89.4 | −0.36 | 0.00 | 71.6 | 70.8 | |||
| 2003 | 75.9 | 7.6 | 0.100 | 53.7–93.9 | −0.34 | −0.30 | 70.6 | 80.1 | 0.579 | ||
| Trait | QTL a | TI Population | ZM Population | XM Population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interval | LOD | Ab | R2 (%) c | Interval | LOD | A | R2 (%) | Interval | LOD | A | R2 (%) | ||
| NGP | qNGP1.1 | RG532-RM151 | 29.1 | 6.13 | 15.2 | RG532-RM1195 | 10.1 | 4.32 | 6.6 | ||||
| qNGP1.2 | Wn34352-RM11869 | 4.9 | −3.17 | 1.9 | RM315-RZ538 | 7.4 | 2.66 | 2.5 | |||||
| qNGP2.1 | A5-RM71 | 5.3 | −2.85 | 2.5 | |||||||||
| qNGP2.2 | RM6-RM240 | 48.9 | −10.78 | 20.6 | |||||||||
| qNGP3.1 | RM15303-RM16 | 25.9 | −7.90 | 11.4 | |||||||||
| qNGP3.2 | RM85-RG418A | 5.7 | −3.60 | 4.6 | |||||||||
| qNGP4 | RM349-RM3333 | 8.7 | 4.00 | 3.4 | RG776A-RG620 | 5.4 | −2.60 | 2.9 | |||||
| qNGP5.1 | CDO82-RG182 | 4.6 | −2.78 | 2.6 | |||||||||
| qNGP5.2 | RM146-RM164 | 7.5 | 3.25 | 2.9 | RG13-RM164 | 5.5 | −2.72 | 2.6 | |||||
| qNGP5.3 | RG573-RG470 | 5.8 | −2.70 | ||||||||||
| qNGP5.4 | RM274-RM334 | 9.3 | 3.81 | 3.2 | |||||||||
| qNGP6.1 | RM508-RM190 | 6.8 | 2.19 | 4.4 | |||||||||
| qNGP6.2 | RZ398-RM204 | 19.2 | −4.46 | 12.4 | RZ398-RM217 | 6.5 | −3.16 | 3.6 | |||||
| qNGP6.3 | RM276-RM549 | 9.2 | −4.43 | 3.9 | |||||||||
| qNGP6.4 | RM3827-RM20361 | 4.7 | −2.69 | 1.5 | |||||||||
| qNGP7.1 | RM3859-RG678 | 9.3 | 2.54 | 5.4 | |||||||||
| qNGP7.2 | RM70-RM18 | 7.8 | −3.97 | 2.7 | |||||||||
| qNGP9.1 | RM8206-RM219 | 5.8 | −3.00 | 1.6 | |||||||||
| qNGP9.2 | RM242-RM108 | 5.4 | 2.58 | 2.5 | |||||||||
| qNGP11 | RG167-RM287 | 6.5 | 2.99 | 2.9 | |||||||||
| qNGP12.1 | RM511-RM28313 | 9.5 | 4.35 | 3.9 | |||||||||
| qNGP12.2 | RM28597-RM17 | 4.9 | −3.09 | 2.1 | |||||||||
| NSP | qNSP1 | RG532-RM151 | 33.6 | 7.41 | 15.0 | RG532-RM1195 | 15.4 | 6.60 | 11.1 | ||||
| qNSP2.1 | RM3732-RM71 | 4.8 | 3.67 | 1.8 | RZ742-RZ512 | 7.4 | −4.58 | 5.2 | |||||
| qNSP2.2 | RM6-RM240 | 53.7 | −13.37 | 22.1 | RM240-RZ123 | 4.8 | −3.07 | 2.1 | |||||
| qNSP2.3 | Tw35293-RM207 | 5.2 | −3.63 | 1.9 | |||||||||
| qNSP3 | RM15303-RM16 | 9.0 | −5.19 | 3.3 | |||||||||
| qNSP4 | RM3474-RM6992 | 8.7 | 4.36 | 3.0 | RG776A-RG620 | 7.4 | −3.41 | 2.9 | |||||
| qNSP5.1 | RM164-RM18927 | 5.4 | 3.13 | 1.9 | RM164-RM163 | 6.7 | −3.28 | 2.9 | RM163-RG470 | 4.5 | −3.44 | 2.9 | |
| qNSP5.2 | RG573-RG470 | 5.1 | −2.93 | 2.7 | |||||||||
| qNSP5.3 | RM274-RM334 | 7.5 | 4.27 | 2.5 | |||||||||
| qNSP6.1 | RZ398-RM217 | 7.5 | −3.79 | 6.5 | |||||||||
| qNSP6.2 | RM276-RM549 | 22.6 | −8.35 | 8.6 | RM253-RM276 | 24.3 | −7.30 | 13.6 | |||||
| qNSP7.1 | RM1243-RM3859 | 7.2 | 2.93 | 4.6 | RM1243-RM3859 | 7.9 | 3.72 | 6.4 | |||||
| qNSP7.2 | RM70-RM18 | 11.2 | −5.86 | 4.1 | RZ264-RZ626 | 5.4 | −4.03 | 3.4 | |||||
| qNSP10 | RM1859-RM184 | 5.0 | −3.42 | 4.2 | |||||||||
| qNSP11.1 | RG118-RM202 | 5.1 | 2.85 | 2.0 | |||||||||
| qNSP11.2 | RZ797-RG103 | 6.9 | 4.09 | 5.5 | |||||||||
| qNSP12 | RM511-RM28313 | 18.4 | 7.24 | 6.8 | |||||||||
| SF | qSF1.1 | RM294A-RM294B | 6.6 | 1.31 | 4.8 | ||||||||
| qSF1.2 | RM12178-RM12210 | 7.0 | −0.95 | 3.1 | |||||||||
| qSF2.1 | RZ318-RM263 | 6.4 | −0.74 | 3.6 | |||||||||
| qSF2.2 | RM6-RM240 | 6.5 | 0.87 | 3.3 | |||||||||
| qSF3.1 | RM15303-RM16 | 13.1 | −1.45 | 8.2 | |||||||||
| qSF3.2 | R1927-RM143 | 4.9 | −1.25 | 3.2 | |||||||||
| qSF3.3 | RZ613-RM85 | 11.4 | −1.80 | 7.4 | RM85-RG418A | 6.1 | −1.99 | 7.6 | |||||
| qSF4.1 | RM551-RM261 | 4.8 | 0.93 | 3.3 | |||||||||
| qSF4.2 | Fo13346-RM303 | 8.6 | −1.07 | 5.1 | |||||||||
| qSF5.1 | RM13-RM267 | 6.0 | −1.99 | 5.8 | |||||||||
| qSF5.2 | RM18038-RM18189 | 22.2 | 1.96 | 13.9 | |||||||||
| qSF6.1 | RM190-RZ516 | 7.3 | 1.83 | 7.9 | |||||||||
| qSF6.2 | RM6119-RM276 | 7.5 | 0.93 | 3.1 | RG138-RM111 | 5.3 | 0.88 | 4.2 | |||||
| qSF6.3 | RM340-RM20731 | 9.0 | 1.19 | 5.4 | |||||||||
| qSF8 | RM23001-RM210 | 6.4 | 0.95 | 3.2 | |||||||||
| qSF9 | RM105-RM3700 | 7.0 | 1.35 | 4.0 | |||||||||
| qSF10.1 | RM3229B-RM1376 | 5.0 | 1.13 | 5.6 | |||||||||
| qSF10.1 | RM3773-RM3123 | 5.4 | 0.83 | 2.9 | |||||||||
| qSF12 | RM28313-RM28597 | 6.8 | −0.94 | 2.9 |
| Trait | QTL | Interval | LOD | Aa | R2 (%) b |
|---|---|---|---|---|---|
| NGP | qNGP3.3 | RM232 | 3.2 | 2.29 | 4.1 |
| qNGP5.5 | RM18927-RM3321 | 4.1 | −2.46 | 4.9 | |
| qNGP6.1 | RM469-RM589 | 5.8 | 2.31 | 9.9 | |
| qNGP9.1 | RM5688-RM219 | 3.5 | −2.18 | 5.6 | |
| qNGP12.1 | Pita-RM511 | 3.1 | 2.19 | 4.8 | |
| NSP | qNSP3.2 | RM232 | 2.6 | 1.89 | 2.2 |
| qNSP5.4 | RM18927-RM3321 | 5.5 | −3.44 | 5.3 | |
| qNSP12 | RM3246-Pita | 6.8 | 3.73 | 7.8 | |
| SF | qSF3.4 | RM14303-RM14383 | 3.8 | 0.53 | 7.3 |
| qSF6.1 | RM587-RM584 | 7.8 | 1.08 | 16.3 | |
| qSF9.2 | RM5688-RM219 | 6.1 | −0.98 | 11.3 | |
| qSF11 | RM224-RM5926 | 6.0 | 0.97 | 9.0 |
| QTL | Population | Interval | LOD | Aa | D b | R2 (%) c |
|---|---|---|---|---|---|---|
| qNGP5.5 | ZC5 S1:2 | RM18927-RM3321 | 2.72 | −1.94 | 3.42 | 6.3 |
| qNGP9.1 | ZC9 S1:2 | RM219-RM1896 | 2.22 | −2.96 | 1.00 | 5.1 |
| qNGP12.1 | ZC12 S1:2 | RM3246-Pita | 9.06 | 5.68 | 0.43 | 16.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, X.; Zhu, Y.; Sun, Z.; Yu, S.; Zhuang, J.; Fan, Y. Identification and Validation of Quantitative Trait Loci for Grain Number in Rice (Oryza sativa L.). Agronomy 2020, 10, 180. https://doi.org/10.3390/agronomy10020180
Niu X, Zhu Y, Sun Z, Yu S, Zhuang J, Fan Y. Identification and Validation of Quantitative Trait Loci for Grain Number in Rice (Oryza sativa L.). Agronomy. 2020; 10(2):180. https://doi.org/10.3390/agronomy10020180
Chicago/Turabian StyleNiu, Xiaojun, Yujun Zhu, Zhichao Sun, Sibin Yu, Jieyun Zhuang, and Yeyang Fan. 2020. "Identification and Validation of Quantitative Trait Loci for Grain Number in Rice (Oryza sativa L.)" Agronomy 10, no. 2: 180. https://doi.org/10.3390/agronomy10020180
APA StyleNiu, X., Zhu, Y., Sun, Z., Yu, S., Zhuang, J., & Fan, Y. (2020). Identification and Validation of Quantitative Trait Loci for Grain Number in Rice (Oryza sativa L.). Agronomy, 10(2), 180. https://doi.org/10.3390/agronomy10020180







