Data Mining Nitrogen-Responsive Gene Expression for Source–Sink Relations and Indicators of N Status in Potato
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Trials
2.2. RNA Extraction
2.3. Transcriptome Sequencing Data Analysis
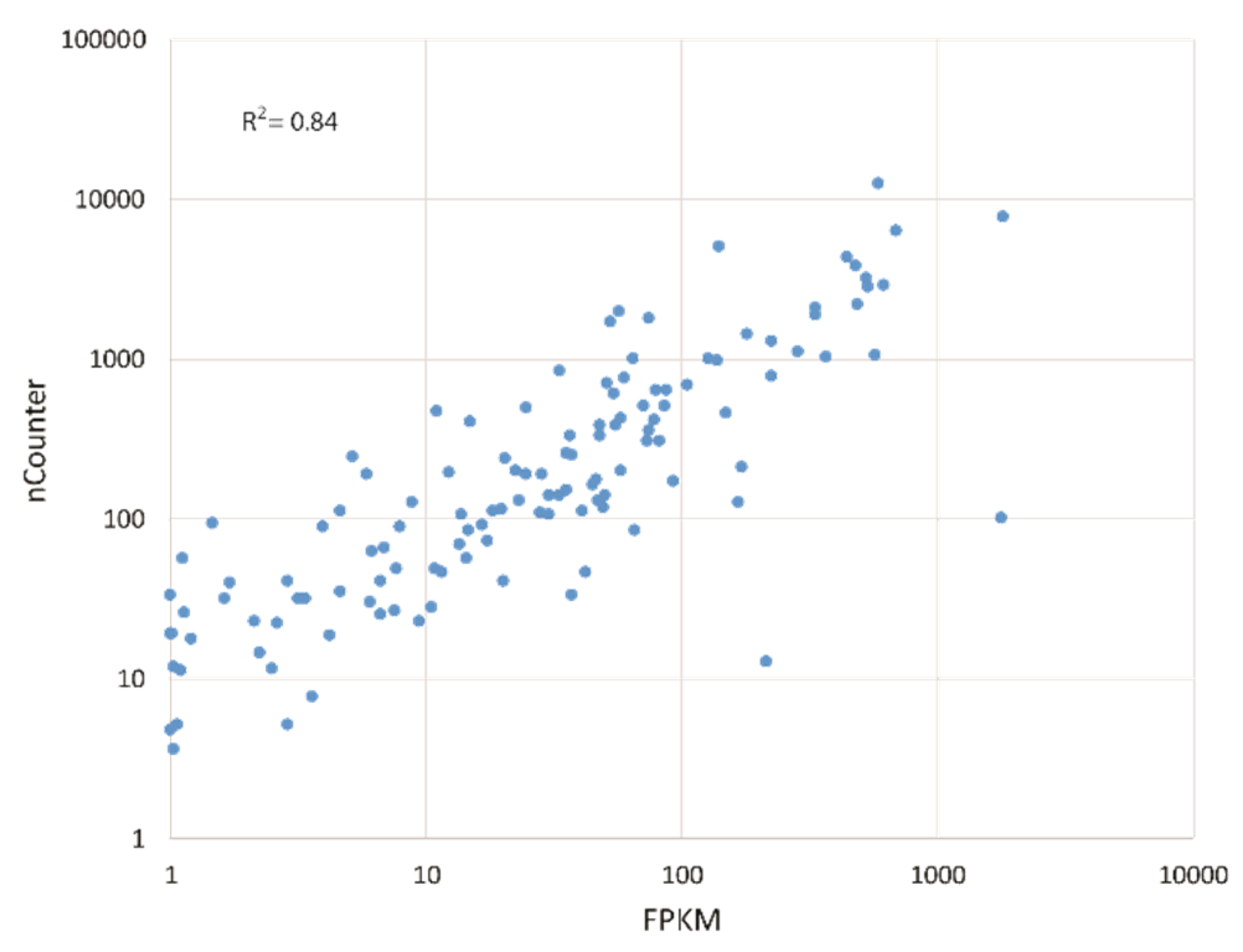
2.4. nCounter Gene Expression Analysis
2.5. Statistical Analysis
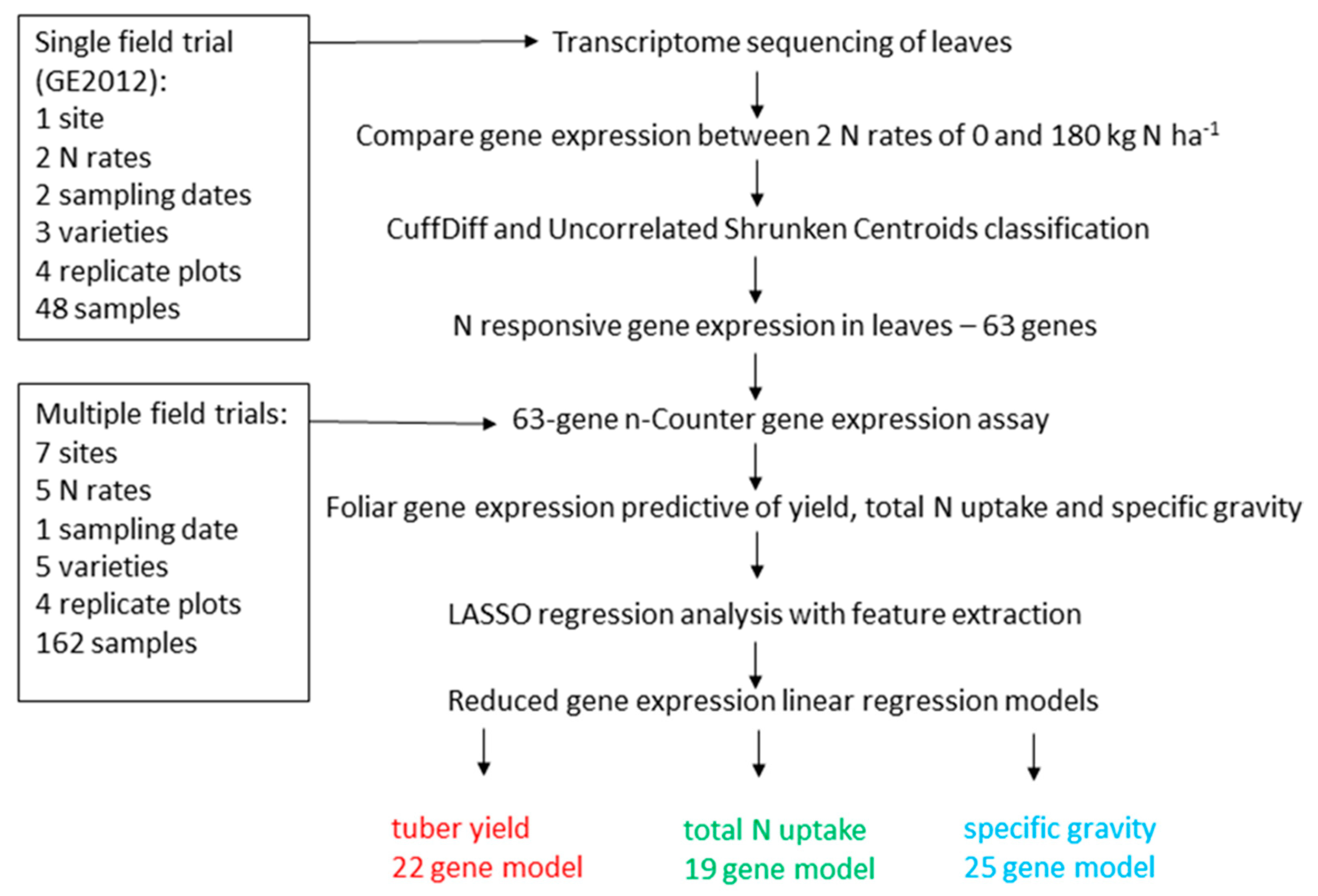
3. Results
3.1. Identification of N-Responsive Genes Using Transcriptome Sequencing
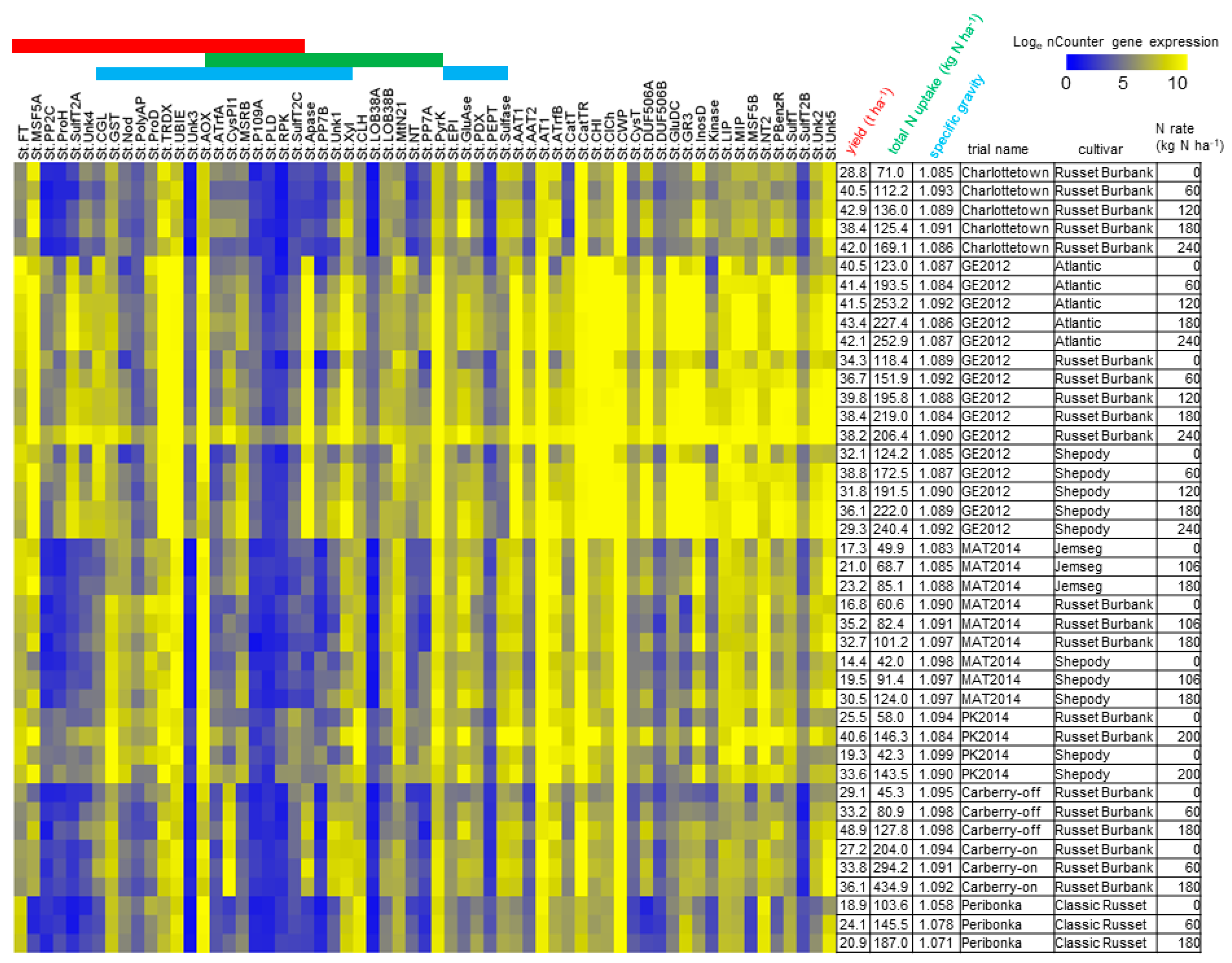
3.2. N-Responsive Gene Expression Predictive of Yield, Total N Uptake and Specific Gravity
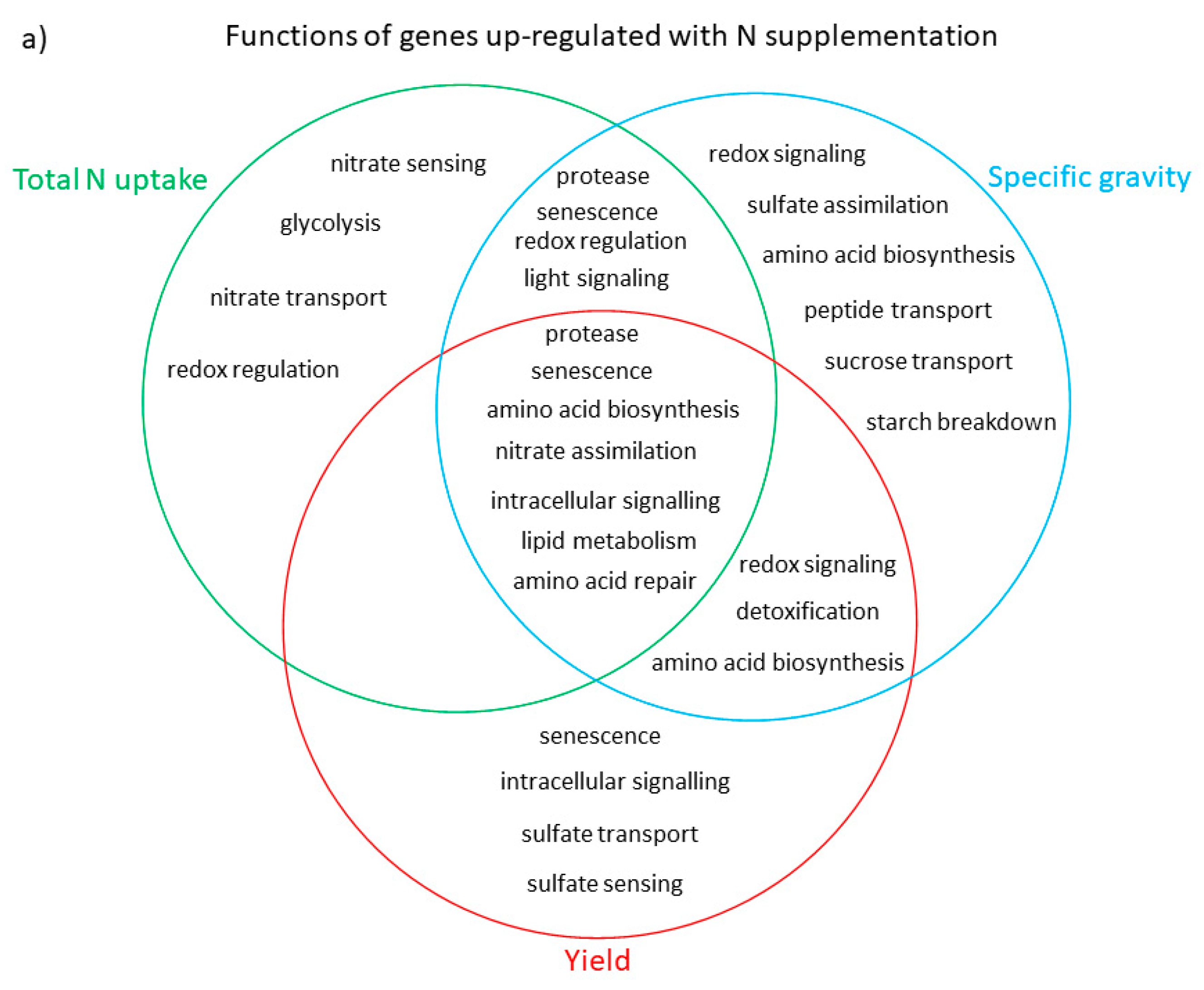
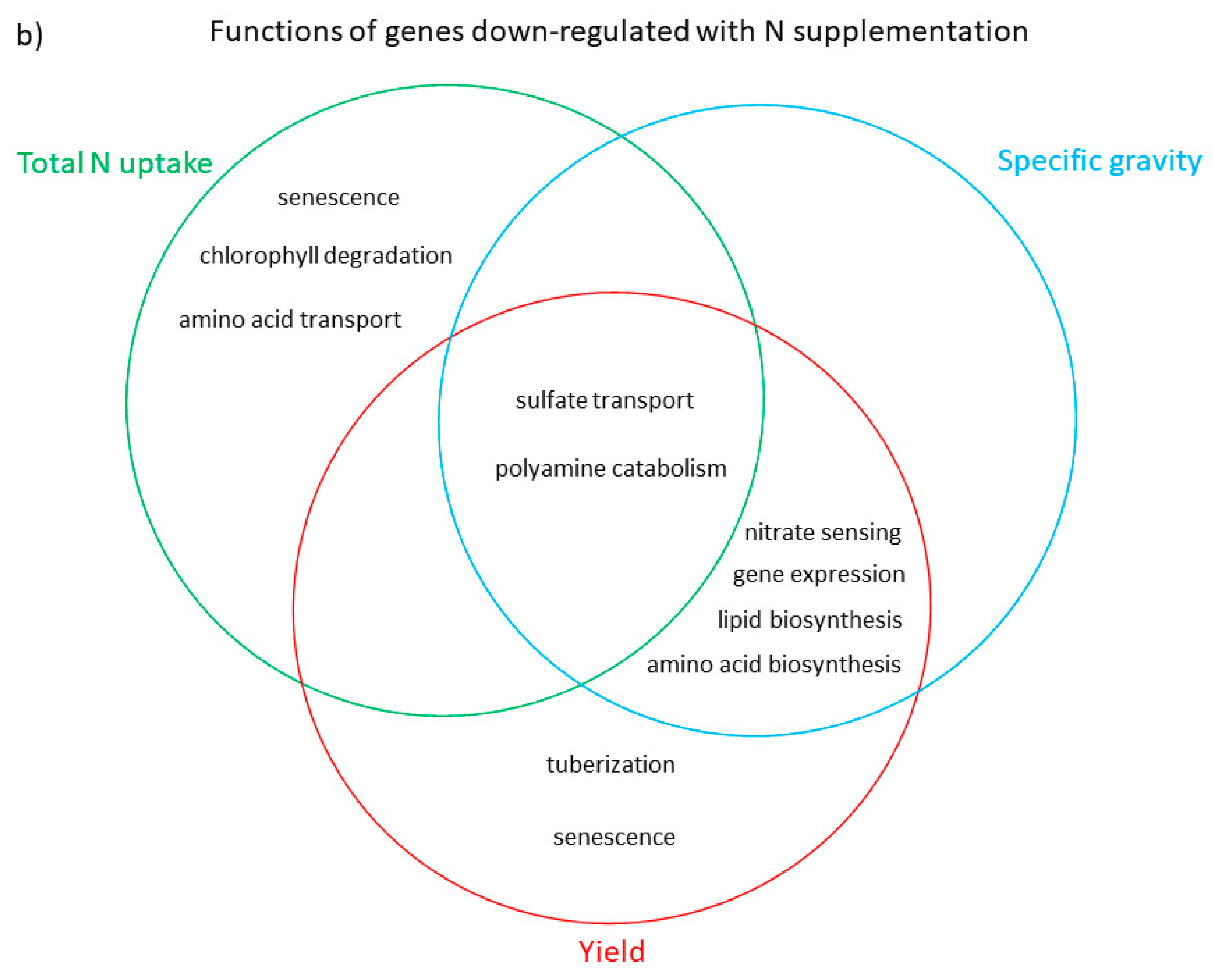
3.3. Functional Analysis of Genes in the LASSO10 Reduced Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Millard, P.; Marshall, B. Growth, nitrogen uptake and partitioning within the potato (Solatium tuberosum L.) crop, in relation to nitrogen application. J. Agric. Sci. 1986, 107, 421–429. [Google Scholar] [CrossRef]
- De la Morena, I.; Guillen, A.; del Moral, L.G. Yield development in potatoes as influenced by cultivar and the timing and level of nitrogen fertilization. Am. Potato J. 1994, 71, 165–173. [Google Scholar] [CrossRef]
- Belanger, G.; Walsh, J.; Richards, J.; Milburn, P.; Ziadi, N. Nitrogen fertilization and irrigation affects tuber characteristics of two potato cultivars. Am. J. Potato Res. 2002, 79, 269–279. [Google Scholar] [CrossRef]
- Mäck, G.; Schjoerring, J.K. Effect of no3- supply on n metabolism of potato plants (Solanum tuberosum L.) with special focus on the tubers. Plant Cell Environ. 2002, 25, 999–1009. [Google Scholar] [CrossRef]
- Zebarth, B.J.; Rosen, C.J. Research perspective on nitrogen bmp development for potato. Am. J. Potato Res. 2007, 84, 3–18. [Google Scholar] [CrossRef]
- Zebarth, B.J.; Bélanger, G.; Cambouris, A.N.; Ziadi, N. Nitrogen fertilization strategies in relation to potato tuber yield, quality, and crop n recovery. In Sustainable Potato Production: Global Case Studies; He, Z., Larkin, R., Honeycutt, W., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 165–186. [Google Scholar]
- Essah, S.Y.C.; Delgado, J.A. Nitrogen management for maximum potato yield, tuber quality, and environmental conservation. In Appropriate Technologies for Environmental Protection in the Developing World; Yanful, E.K., Ed.; Selected papers from ertep 2007, 17–19 July 2007, Ghana, Africa; Springer: Dordrecht, The Netherlands, 2009; pp. 307–315. [Google Scholar]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. of Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef] [PubMed]
- Vos, J. The effects of nitrogen supply and stem density on leaf attributes and stem branching in potato (Solanum tuberosum L.). Potato Res. 1995, 38, 271–279. [Google Scholar] [CrossRef]
- Vos, J.; Biemond, H. Effects of nitrogen on the development and growth of the potato plant. 1. Leaf appearance, expansion growth, life spans of leaves and stem branching. Ann. Bot. 1992, 70, 27–35. [Google Scholar] [CrossRef]
- Vos, J.; van der Putten, P.E.L. Effect of nitrogen supply on leaf growth, leaf nitrogen economy and photosynthetic capacity in potato. Field Crop. Res. 1998, 59, 63–72. [Google Scholar] [CrossRef]
- Firman, D.M.; Allen, E.J. Field measurements of the photosynthetic rate of potatoes grown with different amounts of nitrogen fertilizer. J. Agric. Sci. 1988, 111, 85–90. [Google Scholar] [CrossRef]
- Goffart, J.P.; Olivier, M.; Frankinet, M. Potato crop nitrogen status assessment to improve n fertilization management and efficiency: Past–present–future. Potato Res. 2008, 51, 355–383. [Google Scholar] [CrossRef]
- Olfs, H.-W.; Blankenau, K.; Brentrup, F.; Jasper, J.; Link, A.; Lammel, J. Soil- and plant-based nitrogen-fertilizer recommendations in arable farming. J. Plant Nutr. Soil Sci. 2005, 168, 414–431. [Google Scholar] [CrossRef]
- Ziadi, N.; Zebarth, B.J.; Bélanger, G.; Cambouris, A.N. Soil and plant tests to optimize fertilizer n management of potatoes. In Sustainable Potato Production: Global Case Studies; He, Z., Larkin, R.P., Honeycutt, C.W., Eds.; Springer: New York, NY, USA, 2012; pp. 187–207. [Google Scholar]
- Gálvez, J.H.; Tai, H.H.; Lagüe, M.; Zebarth, B.J.; Strömvik, M.V. The nitrogen responsive transcriptome in potato (Solanum tuberosum L.) reveals significant gene regulatory motifs. Sci. Rep. 2016, 6, 26090. [Google Scholar] [CrossRef]
- Li, X.-Q.; Sveshnikov, D.; Zebarth, B.; Tai, H.; de Koeyer, D.; Millard, P.; Haroon, M.; Singh, M. Detection of nitrogen sufficiency in potato plants using gene expression markers. Am. J. Potato Res. 2010, 87, 50–59. [Google Scholar] [CrossRef]
- Tai, H.H.; Zebarth, B.J. Effect of time of day of sampling on potato foliar gene expression used to assess crop nitrogen status. Am. J. Potato Res. 2015, 92, 284–293. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Dua, V.K.; Chakrabarti, S.K. Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep. 2020, 10, 1152. [Google Scholar] [CrossRef]
- Zebarth, B.J.; Tai, H.; Luo, S.; Millard, P.; De Koeyer, D.; Li, X.-Q.; Xiong, X. Differential gene expression as an indicator of nitrogen sufficiency in field-grown potato plants. Plant Soil 2011, 345, 387–400. [Google Scholar] [CrossRef]
- Zebarth, B.J.; Tai, H.; Luo, S.; Millard, P.; De Koeyer, D.; Li, X.-Q.; Xiong, X. Effect of nitrogen form on gene expression in leaf tissue of greenhouse grown potatoes during three stages of growth. Am. J. Potato Res. 2012, 89, 315–327. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Tibshirani, R.; Hastie, T.; Narasimhan, B.; Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6567–6572. [Google Scholar] [CrossRef] [PubMed]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Vasilevski, A.; Giorgi, F.M.; Bertinetti, L.; Usadel, B. Lasso modeling of the arabidopsis thaliana seed/seedling transcriptome: A model case for detection of novel mucilage and pectin metabolism genes. Mol. Biosyst. 2012, 8, 2566–2574. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Society. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- McGill, W.B.; Figueiredo, C.T. Total nitrogen. In Soil Sampling and Methods of Analysis; Gregorich, E.G., Carter, M.R., Eds.; Canadian Society of Soil Science/Lewis Publishers: Boca Raton, FL, USA, 1993; pp. 201–211. [Google Scholar]
- Simmonds, N.W. Relations between specific gravity, dry matter content and starch content of potatoes. Potato Res. 1977, 20, 137–140. [Google Scholar] [CrossRef]
- Edgar, A.D. Determining the specific gravity of individual potatoes. Am. Potato J. 1951, 28, 729–731. [Google Scholar] [CrossRef]
- Wan, C.Y.; Wilkins, T.A. A modified hot borate method significantly enhances the yield of high-quality rna from cotton (Gossypium hirsutum L.). Anal. Biochem. 1994, 223, 7–12. [Google Scholar] [CrossRef]
- Luo, S.; Tai, H.; Zebarth, B.; Li, X.-Q.; Millard, P.; De Koeyer, D.; Xiong, X. Sample collection protocol effects on quantification of gene expression in potato leaf tissue. Plant Mol. Biol. Report. 2010, 29, 369–378. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. Tm4: A free, open-source system for microarray data management and analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- The Potato Genome Sequencing Consortium; Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time rt-pcr normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Tai, H.; Conn, G.; Davidson, C.; Platt, H. Arbitrary multi-gene reference for normalization of real-time pcr gene expression data. Plant Mol. Biol. Report. 2009, 27, 315–320. [Google Scholar] [CrossRef]
- Nakane, E.; Kawakita, K.; Doke, N.; Yoshioka, H. Elicitation of primary and secondary metabolism during defense in the potato. J. Gen. Plant Pathol. 2003, 69, 378–384. [Google Scholar] [CrossRef]
- Quinones, V.; Zanlungo, S.; Holuigue, L.; Litvak, S.; Jordana, X. The cox1 initiation codon is created by rna editing in potato mitochondria. Plant Physiol. 1995, 108, 1327–1328. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, M.R.; de Bastiani, D.; Gaeta, M.L.; de Araujo Mariath, J.E.; de Costa, F.; Retallick, J.; Nolan, L.; Tai, H.H.; Stromvik, M.V.; Fett-Neto, A.G. Comparative transcriptional analysis provides new insights into the molecular basis of adventitious rooting recalcitrance in eucalyptus. Plant Sci. Int. J. Exp. Plant Biol. 2015, 239, 155–165. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Hirsch, C.D.; Hamilton, J.P.; Childs, K.L.; Cepela, J.; Crisovan, E.; Vaillancourt, B.; Hirsch, C.N.; Habermann, M.; Neal, B.; Buell, C.R. Spud db: A resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. Plant Genome 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Huala, E.; Dickerman, A.W.; Garcia-Hernandez, M.; Weems, D.; Reiser, L.; LaFond, F.; Hanley, D.; Kiphart, D.; Zhuang, M.; Huang, W.; et al. The arabidopsis information resource (tair): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001, 29, 102–105. [Google Scholar] [CrossRef]
- Zebarth, B.; Drury, C.; Tremblay, N.; Cambouris, A. Opportunities for improved fertilizer nitrogen management in production of arable crops in eastern Canada: A review. Can. J. Soil Sci. 2009, 89, 113–132. [Google Scholar] [CrossRef]
- Bélanger, G.; Walsh, J.R.; Richards, J.E.; Milburn, P.H.; Ziadi, N. Critical nitrogen curve and nitrogen nutrition index for potato in eastern Canada. Am. J. Potato Res. 2001, 78, 355–364. [Google Scholar] [CrossRef]
- Zebarth, B.J.; Leclerc, Y.; Moreau, G.; Botha, E. Rate and timing of nitrogen fertilization of russet burbank potato: Yield and processing quality. Can. J. Plant Sci. 2004, 84, 855–863. [Google Scholar] [CrossRef]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [PubMed]
- Canales, J.; Moyano, T.C.; Gutiérrez, R.A.; Vidal, E.A. Nitrogen control of developmental phase transitions in arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5611–5618. [Google Scholar]
- Ewing, E.E.; Struik, P.C. Tuber formation in potato: Induction, initiation, and growth. Hortic. Rev. 1992, 14, 89. [Google Scholar]
- Rodriguez-Falcon, M.; Bou, J.; Prat, S. Seasonal control of tuberization in potato: Conserved elements with the flowering response. Annu. Rev. Plant Biol. 2006, 57, 151–180. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, Y.; Zhao, J.; Shi, X.; Ma, Z.; Fan, M. Tuber formation as influenced by the c: N ratio in potato plants. J. Plant Nutr. Soil Sci. 2018, 181, 686–693. [Google Scholar] [CrossRef]
- Navarro, C.; Abelenda, J.A.; Cruz-Oro, E.; Cuellar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by flowering locus t. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. Fd, a bzip protein mediating signals from the floral pathway integrator ft at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.G.F.; Sonnewald, U.; Bachem, C.W.B. Source-sink regulation is mediated by interaction of an ft homolog with a sweet protein in potato. Curr. Biol. 2019, 29, 1178–1186.e6. [Google Scholar] [CrossRef]
- Viola, R.; Roberts, A.G.; Haupt, S.; Gazzani, S.; Hancock, R.D.; Marmiroli, N.; Machray, G.C.; Oparka, K. Tuberization in potato involves a switch from apoplastic to symplastic phloem unloading. Plant Cell 2001, 13, 385–398. [Google Scholar] [CrossRef]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-G.; Zhu, X.-G. Source–sink interaction: A century old concept under the light of modern molecular systems biology. J. Exp. Bot. 2017, 68, 4417–4431. [Google Scholar] [CrossRef] [PubMed]
- Bowden, R.L.; Rouse, D.I.; Sharkey, T.D. Mechanism of photosynthesis decrease by verticillium dahliae in potato. Plant Physiol. 1990, 94, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Tai, H.H.; de Koeyer, D.; Sønderkær, M.; Hedegaard, S.; Lagüe, M.; Goyer, C.; Nolan, L.; Davidson, C.; Gardner, K.; Neilson, J.; et al. Verticillium dahliae disease resistance and the regulatory pathway for maturity and tuberization in potato. Plant Genome 2018, 11, 1–5. [Google Scholar] [CrossRef]
- Kloosterman, B.; Abelenda, J.A.; Gomez, M.d.M.C.; Oortwijn, M.; de Boer, J.M.; Kowitwanich, K.; Horvath, B.M.; van Eck, H.J.; Smaczniak, C.; Prat, S.; et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 2013, 495, 246. [Google Scholar] [CrossRef]
- Ewing, E.E. Heat stress and the tuberization stimulus. Am. Potato J. 1981, 58, 31. [Google Scholar] [CrossRef]
- Lehretz, G.G.; Sonnewald, S.; Hornyik, C.; Corral, J.M.; Sonnewald, U. Post-transcriptional regulation of flowering locus t modulates heat-dependent source-sink development in potato. Curr. Biol. 2019, 29, 1614–1624.e3. [Google Scholar] [CrossRef]
- Hancock, R.D.; Morris, W.L.; Ducreux, L.J.M.; Morris, J.A.; Usman, M.; Verrall, S.R.; Fuller, J.; Simpson, C.G.; Zhang, R.; Hedley, P.E.; et al. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plant Cell Environ. 2014, 37, 439–450. [Google Scholar]
- Hastilestari, B.R.; Lorenz, J.; Reid, S.; Hofmann, J.; Pscheidt, D.; Sonnewald, U.; Sonnewald, S. Deciphering source and sink responses of potato plants (Solanum tuberosum L.) to elevated temperatures. Plant Cell Environ. 2018, 41, 2600–2616. [Google Scholar] [CrossRef]
- Sonnewald, S.; van Harsselaar, J.; Ott, K.; Lorenz, J.; Sonnewald, U. How potato plants take the heat? Procedia Environ. Sci. 2015, 29, 97. [Google Scholar] [CrossRef]
- Tang, R.; Niu, S.; Zhang, G.; Chen, G.; Haroon, M.; Yang, Q.; Rajora, O.P.; Li, X.-Q. Physiological and growth responses of potato cultivars to heat stress. Botany 2018, 96, 897–912. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Ewing, E.E.; Owens, T.G. Photosynthesis at high temperature in tuber-bearing solanum species: A comparison between accessions of contrasting heat tolerance. Plant Physiol. 1990, 93, 791–797. [Google Scholar] [CrossRef]
- Dahal, K.; Li, X.-Q.; Tai, H.; Creelman, A.; Bizimungu, B. Improving potato stress tolerance and tuber yield under a climate change scenario—A current overview. Front. Plant Sci. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth Res. 2015, 123, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Liepman, A.H.; Olsen, L.J. Peroxisomal alanine: Glyoxylate aminotransferase (agt1) is a photorespiratory enzyme with multiple substrates in arabidopsis thaliana. Plant J. 2001, 25, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Cambouris, A.N.; Ziadi, N.; Bertrand, A.; Khelifi, M. Nitrogen fertilization effects on the composition of foliar amino acids of russet burbank potato. Am. J. Potato Res. 2019, 96, 541–551. [Google Scholar] [CrossRef]
- Brunold, C.; Suter, M. Regulation of sulfate assimilation by nitrogen nutrition in the duckweed Lemna minor L. Plant Physiol. 1984, 76, 579–583. [Google Scholar] [CrossRef]
- Laboski, C.A.; Kelling, K.A. Influence of fertilizer management and soil fertility on tuber specific gravity: A review. Am. J. Potato Res. 2007, 84, 283–290. [Google Scholar] [CrossRef]

| Trial Name | Location (Decimal Degrees) | Planting Date m/d/y | Second Application m/d/y (DAP) | Harvest Date m/d/y (DAP) | Soil Type | Avg ℃ | Precipitation (mm) |
|---|---|---|---|---|---|---|---|
| Carberry-Off | Carberry, MB (49.932–99.388) | 5/16/2014 | 6/25/2014 (40) | 9/19/2014 (126) | loamy fine sand | 13.7 | 337 |
| Carberry-On | Carberry, MB (49.904–99.353) | 5/15/2014 | 6/24/2014 (40) | 9/22/2014 (130) | clay loam | 13.7 | 337 |
| Peribonka | Péribonka, QC (48.766–72.053) | 6/11/2014 | 7/21/2014 (45) | 9/24/2014 (106) | silty loam | 12.0 | 500 |
| Charlottetown | Harrington, PE (46.352–63.154) | 5/21/2014 | N/A | 10/6/2014 (138) | sandy loam | 15.0 | 461 |
| MAT2014 | Fredericton, NB (45.923–66.616) | 5/25/2014 | N/A | 9/29/2014 (127) | loam | 14.6 | 549 |
| GE2012 | Fredericton, NB (45.925–66.612) | 5/23/2012 | N/A | 10/3/2012 (105) | loam | 14.6 | 549 |
| PK2014 | Fredericton, NB (45.925 -66.615) | 2/6/2014 | N/A | 9/30/2014 (120) | loam | 14.6 | 549 |
| iTAG | nCounter | Description | FPKM Control | FPKM Treated | nCounter Control | nCounter Treated | LASSO |
|---|---|---|---|---|---|---|---|
| Sotub05g028860 | St.FT | Flowering locus T protein | 56.76 | 23.52 | 431.5 | 188.6 | Y |
| Sotub06g008080 | St.MSF5A | Male sterility 5 family protein (Fragment) | 90.98 | 617.84 | 173.38 | 2920.28 | Y |
| Sotub10g020080 | St.PP2C | Protein phosphatase 2C | 0.02 | 0.2 | 10.87 | 16.74 | Y |
| Sotub10g025870 | St.ProH | DUF584 domain containing protein; S40 senescence regulator | 1.13 | 6.61 | 22.23 | 48.45 | Y |
| Sotub04g027100 | St.SulfT2A | High affinity sulfate transporter 2 | 2.35 | 18.66 | 31.03 | 114.11 | Y |
| Sotub12g011840 | St.Unk4 | Hypothetical gene of unknown function | 213.13 | 1771.26 | 11.82 | 101.41 | Y |
| Sotub08g027190 | St.CGL | Cystathionine gamma-synthase | 6.44 | 21.9 | 25.82 | 128.98 | Y, S |
| Sotub10g024560 | St.GST | Glutathione S-transferase | 76.76 | 331.91 | 419.04 | 1913.08 | Y, S |
| Sotub01g049920 | St.Nod | Nodule inception protein; RWP-RK domain-containing protein | 29.26 | 12.47 | 140.67 | 68.78 | Y, S |
| Sotub12g031130 | St.PolyAP | Poly(A) polymerase | 5.07 | 1.61 | 62.26 | 21.28 | Y, S |
| Sotub02g033320 | St.ProD | Proline dehydrogenase | 35.88 | 12.61 | 252.32 | 105.8 | Y, S |
| Sotub12g027600 | St.TRDX | DSBA-like thioredoxin domain containing protein | 53.79 | 478.31 | 390.79 | 3815.08 | Y, S |
| Sotub12g020880 | St.UBIE | Phosphoethanolamine N-methyltransferase | 281.83 | 125.14 | 1108.19 | 1014.9 | Y, S |
| Sotub04g006310 | St.Unk3 | Unknown Protein | 0 | 2.14 | 18.52 | 30.83 | Y, S |
| Sotub08g025870 | St.AOX | Primary amine oxidase | 72.65 | 32.27 | 1795.55 | 843.27 | Y, N, S |
| Sotub10g018540 | St.ATrfA | Aminotransferase like protein; alanine-glyoxylate aminotransferase | 2.23 | 56.51 | 30.74 | 202.64 | Y, N, S |
| Sotub03g016050 | St.CysPI1 | Cysteine protease inhibitor 1 | 0.11 | 0.45 | 56.44 | 94.26 | Y, N, S |
| Sotub01g022620 | St.MSRB | Peptide methionine sulfoxide reductase msrB | 36.19 | 164.77 | 32.43 | 128.18 | Y, N, S |
| Sotub03g011570 | St.P109A | DUF607 domain containing protein/Ca2+ uniporter | 0 | 0.69 | 32.66 | 38.72 | Y, N, S |
| Sotub10g013730 | St.PLD | Phospholipase D | 0 | 0.07 | 3.84 | 4.25 | Y, N, S |
| Sotub08g007230 | St.RPK | Receptor-like protein kinase; ATP binding protein | 0.02 | 1.87 | 2.69 | 4.23 | Y, N, S |
| Sotub09g023510 | St.SulfT2C | Sulfate/bicarbonate/oxalate exchanger and transporter sat-1; group 3 sulfate transporter | 5.78 | 1.85 | 65.77 | 40.66 | Y, N, S |
| Sotub12g007850 | St.Apase | Cytosol aminopeptidase family protein; leucine aminopeptidase | 9.44 | 41.07 | 26.88 | 45.69 | N, S |
| Sotub03g018710 | St.PP7B | Serine/threonine-protein phosphatase 7 long form homolog | 0.09 | 2.93 | 10.41 | 88.73 | N, S |
| Sotub04g023170 | St.Unk1 | Phytochrome-interacting factor 3.1 | 3.61 | 53.37 | 34.04 | 607.97 | N, S |
| Sotub01g023000 | St.Xyl | Aspartic peptidase | 21.13 | 69.85 | 198.5 | 515.82 | N, S |
| Sotub06g016810 | St.CLH | Chlorophyllase | 58.61 | 49.38 | 770.14 | 703.2 | N |
| Sotub04g031010 | St.LOB38A | LOB domain protein 38 | 0.12 | 0.63 | 25.45 | 30.7 | N |
| Sotub02g031040 | St.LOB38B | LOB domain protein 38 | 13.59 | 31.9 | 83.58 | 141.23 | N |
| Sotub05g012720 | St.MtN21 | Nodulin MtN21 family protein | 84.96 | 27.2 | 508.49 | 192.39 | N |
| Sotub08g005390 | St.NT | Nitrate transporter | 1.48 | 9.72 | 10.65 | 47.64 | N |
| Sotub09g012260 | St.PP7A | Serine/threonine-protein phosphatase 7 long form homolog | 0.02 | 4.82 | 18.1 | 192.28 | N |
| Sotub11g010490 | St.PyrK | Pyruvate kinase | 104.48 | 223.18 | 686.65 | 1302.26 | N |
| Sotub02g033060 | St.EPI | PQ-loop repeat family protein / transmembrane family protein; sucrose transporter | 6.83 | 48.14 | 88.05 | 118.25 | S |
| Sotub03g018720 | St.GluAse | Alpha-glucosidase-like | 45.81 | 523.27 | 129.69 | 3215.26 | S |
| Sotub04g026530 | St.PDX | Peroxidase | 16.23 | 43.55 | 72.72 | 164.05 | S |
| Sotub07g009860 | St.PEPT | Peptide transporter | 2.59 | 18.9 | 6.7 | 40.64 | S |
| Sotub09g024290 | St.Sulfase | Sulfate adenylyltransferase | 170.6 | 566.95 | 210.07 | 1051.94 | S |
| Sotub05g024960 | St.AAT1 | Amino acid transporter | 46.46 | 177.1 | 335.53 | 1427.92 | |
| Sotub11g012150 | St.AAT2 | Transmembrane amino acid transporter | 5.57 | 39.63 | 24.62 | 112.82 | |
| Sotub04g019200 | St.AT1 | Ammonium transporter 1, member 2 (LeAMT1;2) | 683.45 | 443.32 | 6349.83 | 4392.36 | |
| Sotub12g011100 | St.ATrfB | Gamma aminobutyrate transaminase isoform3 | 73.48 | 532.05 | 354.92 | 2851.1 | |
| Sotub09g009440 | St.CatT | Cation/H+ antiporter | 13.85 | 4.16 | 402.58 | 245.55 | |
| Sotub08g007240 | St.CatTR | Cation transport regulator-like protein 2; gamma-glutamylclyclotransferase | 364.05 | 1806.56 | 1044.62 | 7793.81 | |
| Sotub08g014020 | St.CHI | Chalcone isomerase | 11.17 | 63.56 | 196.46 | 1021.25 | |
| Sotub02g014150 | St.ClCh | Voltage-gated chloride channel; CLC-Nt2 protein | 34.33 | 71.91 | 150.63 | 306.12 | |
| Sotub09g029950 | St.CWP | Cell wall protein | 586.33 | 138.66 | 12701.21 | 5091.67 | |
| Sotub02g036900 | St.CysT | Lysosomal Cystine Transporter family protein | 4.98 | 17.18 | 29.55 | 112.29 | |
| Sotub11g007090 | St.DUF506A | DUF506 domain containing protein | 7.73 | 56.04 | 128.44 | 1994.76 | |
| Sotub11g007110 | St.DUF506B | DUF506 domain containing protein | 8.33 | 64.64 | 21.95 | 83.52 | |
| Sotub01g005580 | St.GluDC | Glutamate decarboxylase isozyme 3 | 10.4 | 26.87 | 45.43 | 110.42 | |
| Sotub04g010640 | St.GR3 | Glutaredoxin | 3.58 | 23.35 | 112.2 | 495.13 | |
| Sotub08g024220 | St.InosD | Oxidoreductase | 28.88 | 329.76 | 105.2 | 2087.67 | |
| Sotub01g049270 | St.Kinase | Protein kinase | 3.2 | 44.89 | 17.8 | 178.44 | |
| Sotub12g012740 | St.LIP | Chloroplast lipocalin | 77.39 | 35.31 | 641.61 | 329.74 | |
| Sotub08g028270 | St.MIP | Methanol inducible protein | 80.14 | 221.33 | 306.61 | 791.64 | |
| Sotub09g018850 | St.MSF5B | Male sterility 5 family protein; Pollenless3 | 13.37 | 135.3 | 56 | 984.12 | |
| Sotub08g024820 | St.NT2 | Low-affinity nitrate transporter | 46.84 | 34.42 | 385.18 | 257.99 | |
| Sotub10g014450 | St.PBenzR | Isoflavone reductase homolog | 15.41 | 487.58 | 90.58 | 2220.52 | |
| Sotub04g021910 | St.SulfT | Plasma membrane sulfate transporter | 19.26 | 85.77 | 238.24 | 645.07 | |
| Sotub10g013960 | St.SulfT2B | High affinity sulfate transporter type 1 | 1.24 | 5.6 | 13.63 | 40.54 | |
| Sotub03g017290 | St.Unk2 | UP-9A; Response to low sulfur 1 protein | 49.28 | 148.11 | 139.56 | 465.3 | |
| Sotub10g017020 | St.Unk5 | Unknown Protein | 9.92 | 51.13 | 478.09 | 1714.8 |
| LASSO Regression Model | Yield | Specific Gravity | Total N Uptake | |||
|---|---|---|---|---|---|---|
| Adjusted R2 | MSE | Adjusted R2 | MSE | Adjusted R2 | MSE | |
| Full | 0.30 | 65.24 | −0.08 | 6.68 × 10−5 | 0.52 | 2825.11 |
| Factors | 0.63 | 34.49 | 0.33 | 4.18 × 10−5 | 0.71 | 1693.08 |
| All genes | 0.35 | 61.40 | 0.08 | 5.70 × 10−5 | 0.35 | 3858.10 |
| LASSO10 genes | 0.57 | 40.58 | 0.35 | 4.06 × 10−5 | 0.58 | 2489.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parenteau, M.T.; Gu, H.; Zebarth, B.J.; Cambouris, A.N.; Lafond, J.; Nelson, A.; Nyiraneza, J.; Davidson, C.; Lagüe, M.; Galvez, J.H.; et al. Data Mining Nitrogen-Responsive Gene Expression for Source–Sink Relations and Indicators of N Status in Potato. Agronomy 2020, 10, 1617. https://doi.org/10.3390/agronomy10101617
Parenteau MT, Gu H, Zebarth BJ, Cambouris AN, Lafond J, Nelson A, Nyiraneza J, Davidson C, Lagüe M, Galvez JH, et al. Data Mining Nitrogen-Responsive Gene Expression for Source–Sink Relations and Indicators of N Status in Potato. Agronomy. 2020; 10(10):1617. https://doi.org/10.3390/agronomy10101617
Chicago/Turabian StyleParenteau, Mia T., Hong Gu, Bernie J. Zebarth, Athyna N. Cambouris, Jean Lafond, Alison Nelson, Judith Nyiraneza, Charlotte Davidson, Martin Lagüe, José Héctor Galvez, and et al. 2020. "Data Mining Nitrogen-Responsive Gene Expression for Source–Sink Relations and Indicators of N Status in Potato" Agronomy 10, no. 10: 1617. https://doi.org/10.3390/agronomy10101617
APA StyleParenteau, M. T., Gu, H., Zebarth, B. J., Cambouris, A. N., Lafond, J., Nelson, A., Nyiraneza, J., Davidson, C., Lagüe, M., Galvez, J. H., Strömvik, M. V., & Tai, H. H. (2020). Data Mining Nitrogen-Responsive Gene Expression for Source–Sink Relations and Indicators of N Status in Potato. Agronomy, 10(10), 1617. https://doi.org/10.3390/agronomy10101617





