Two-Component Response Regulators CitT, YvcP, and YycI Differentially Control Pectin and Hemicellulose Degradation in Degumming of Ramie Fibers by Bacillus subtilis Strain 168
Abstract
1. Introduction
2. Materials and Methods
2.1. Functional Prediction of Response Regulators by Bioinformatic Analysis
2.2. Experimental Materials
2.3. Strains
2.4. Growth Conditions and Profiles
2.5. Post-Treatment and Chemical Component Analysis of Ramie Fibers
2.6. Enzymatic Activity Analysis
2.7. Statistical Analysis
3. Results and Discussion
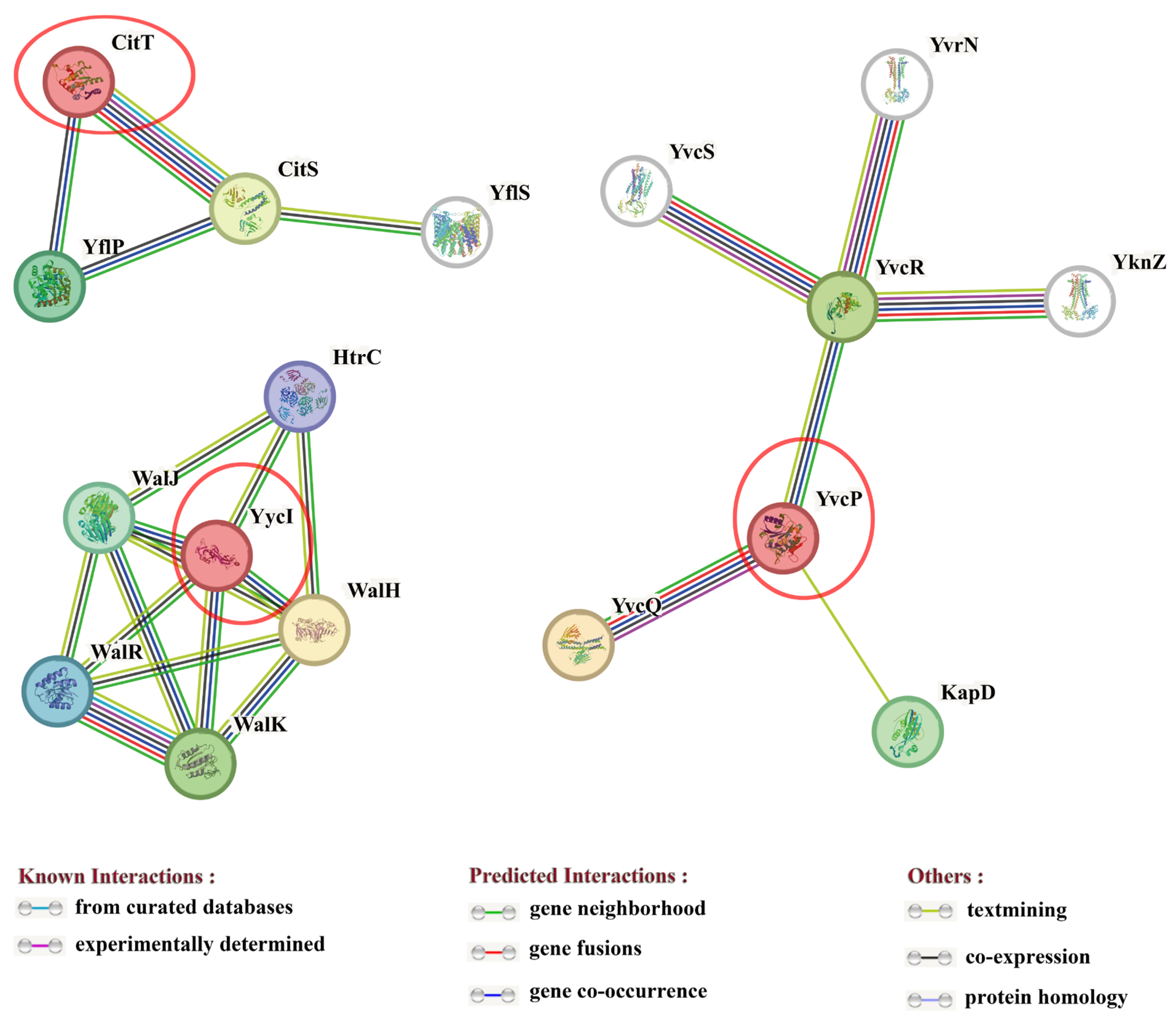
3.1. Functional Prediction of TCS Response Regulators
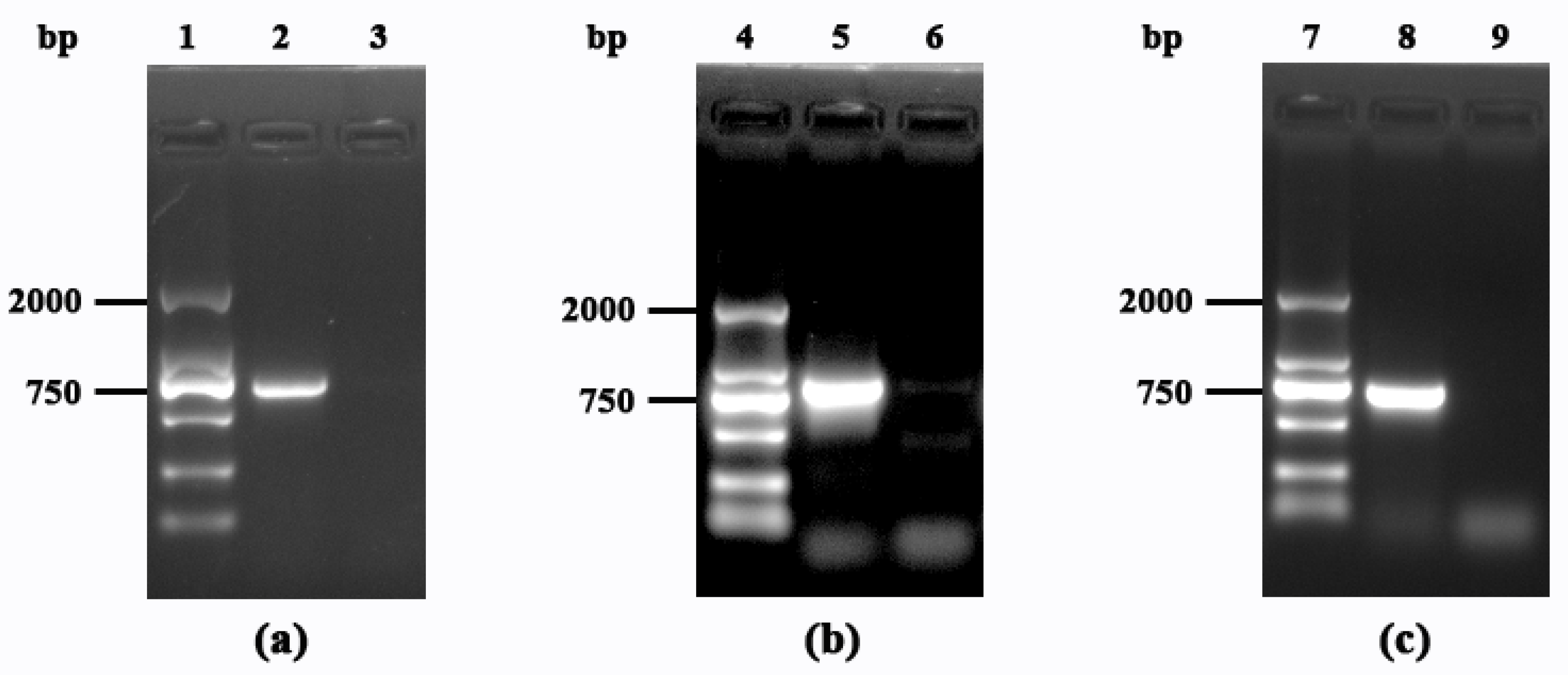
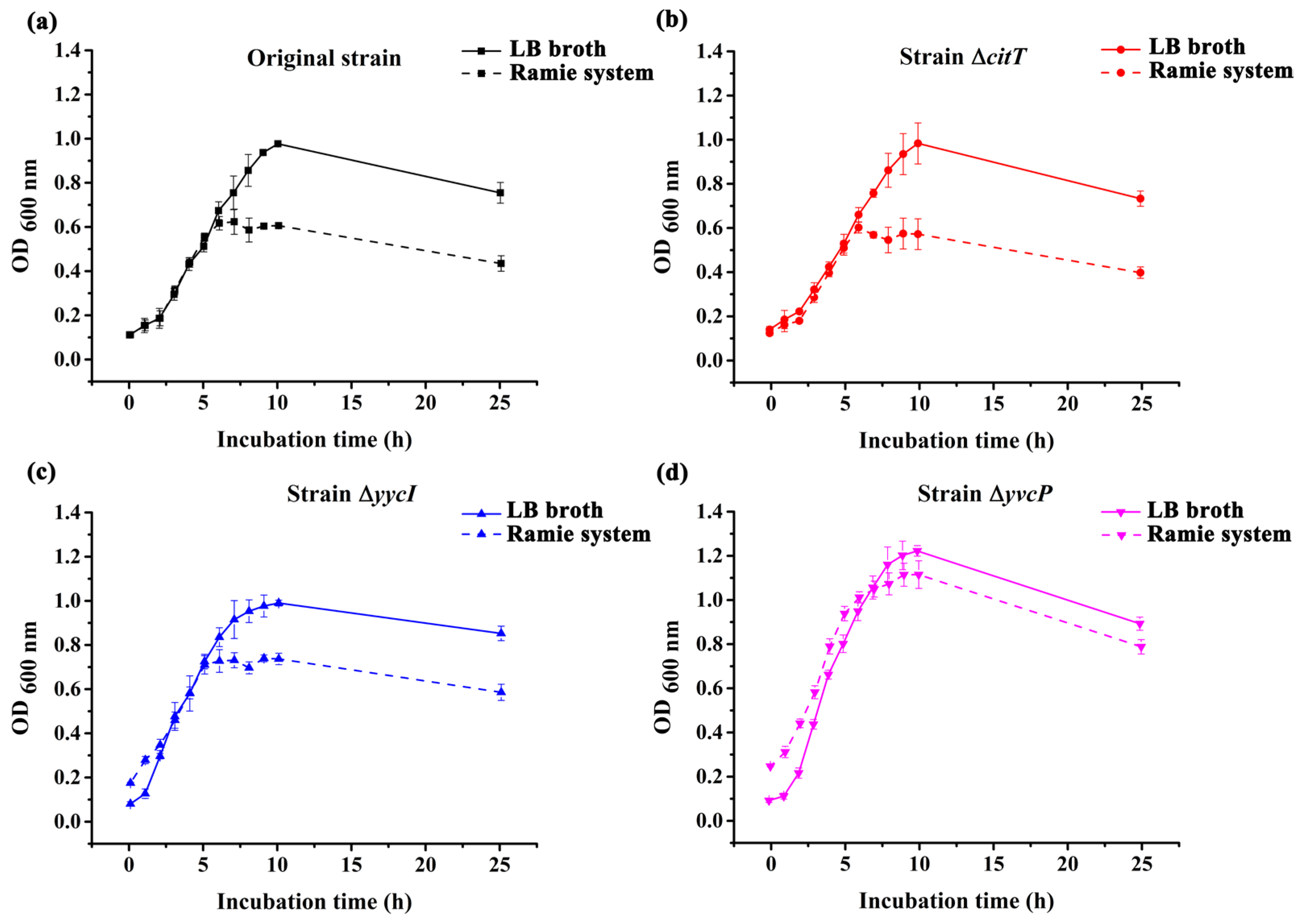
3.2. Identification of Mutant Strains and Analysis of Growth Trends
3.3. Identification of the Key TCS Response Regulator Proteins in CitT, YvcP, and YycI
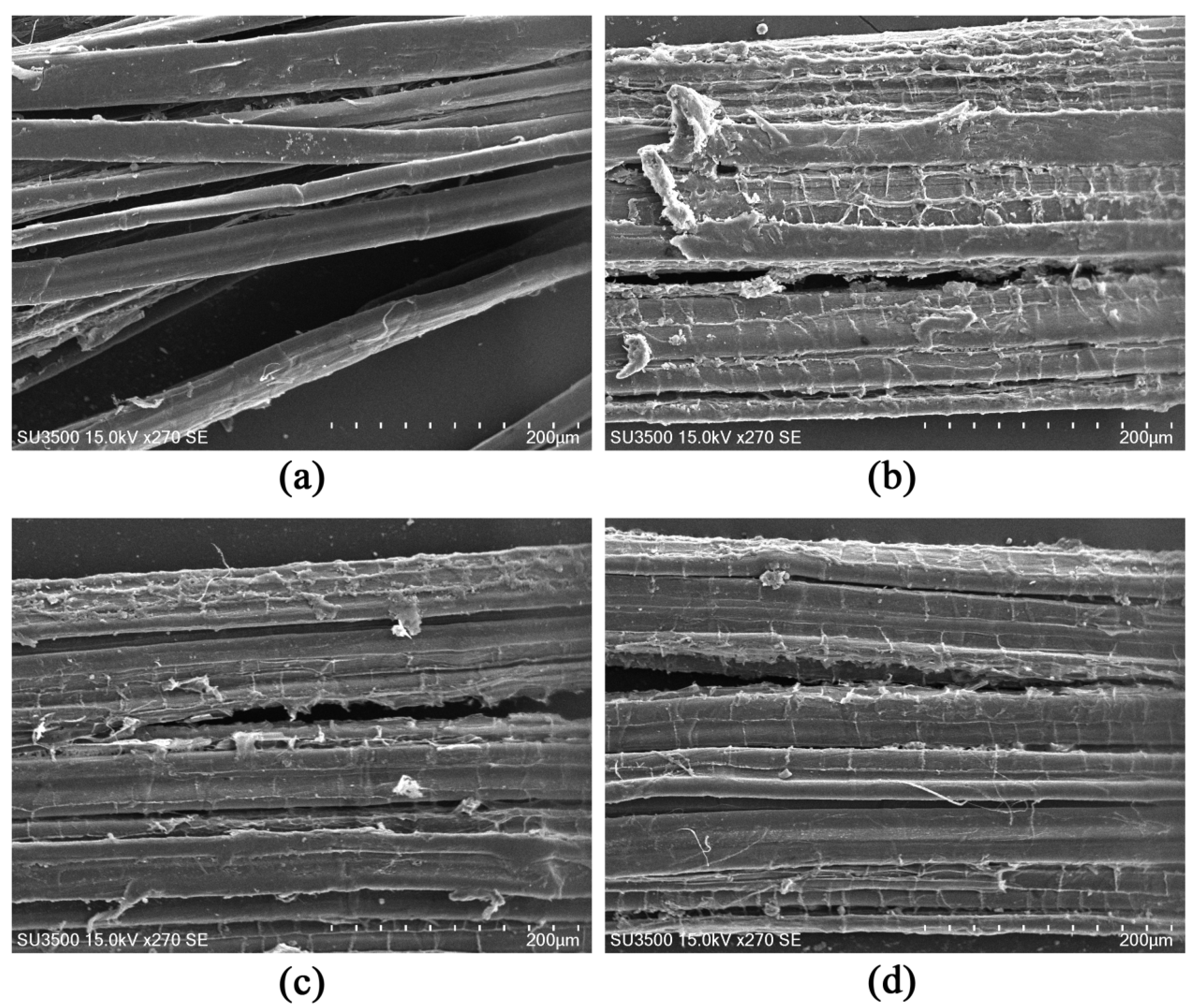
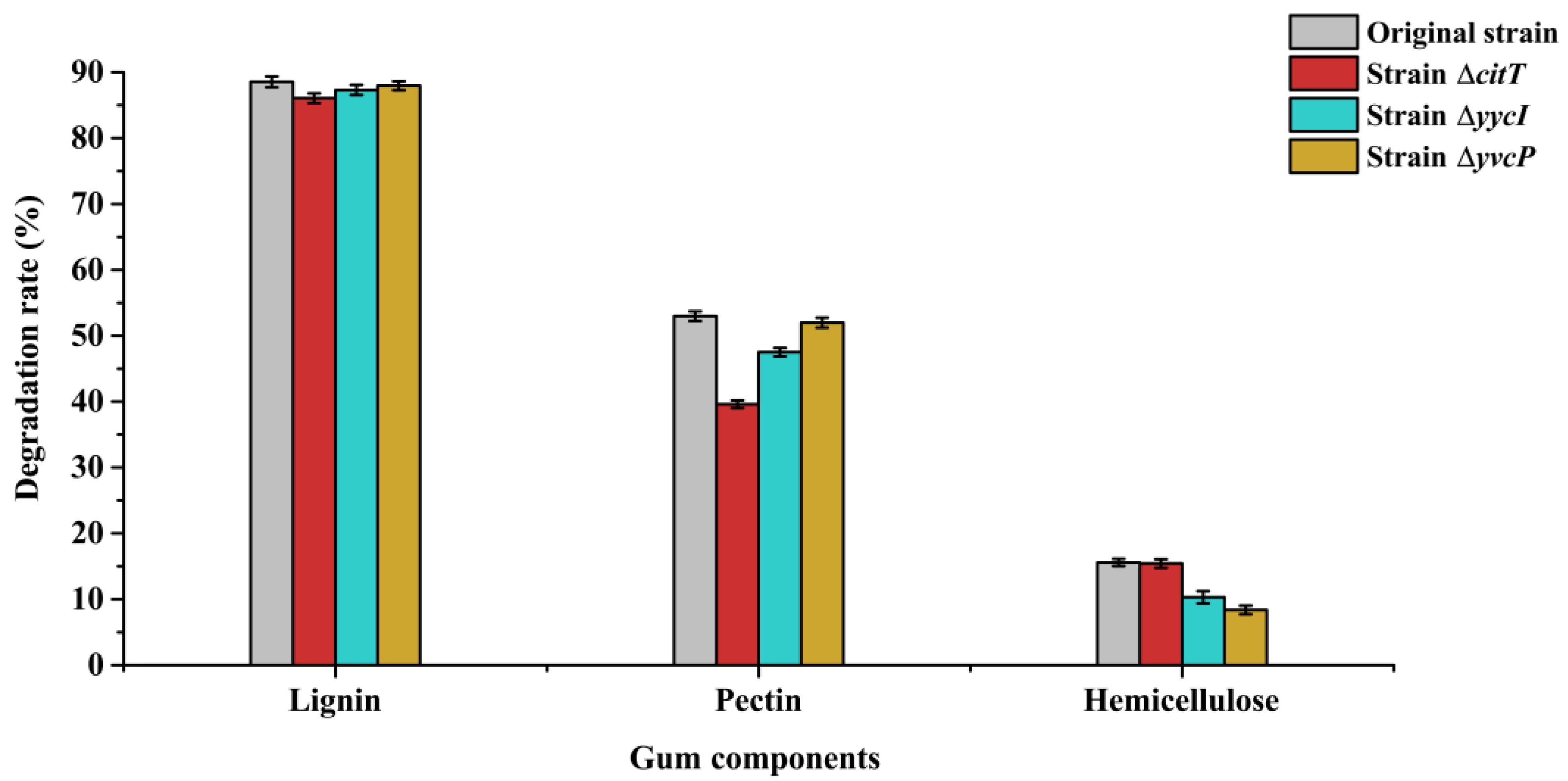
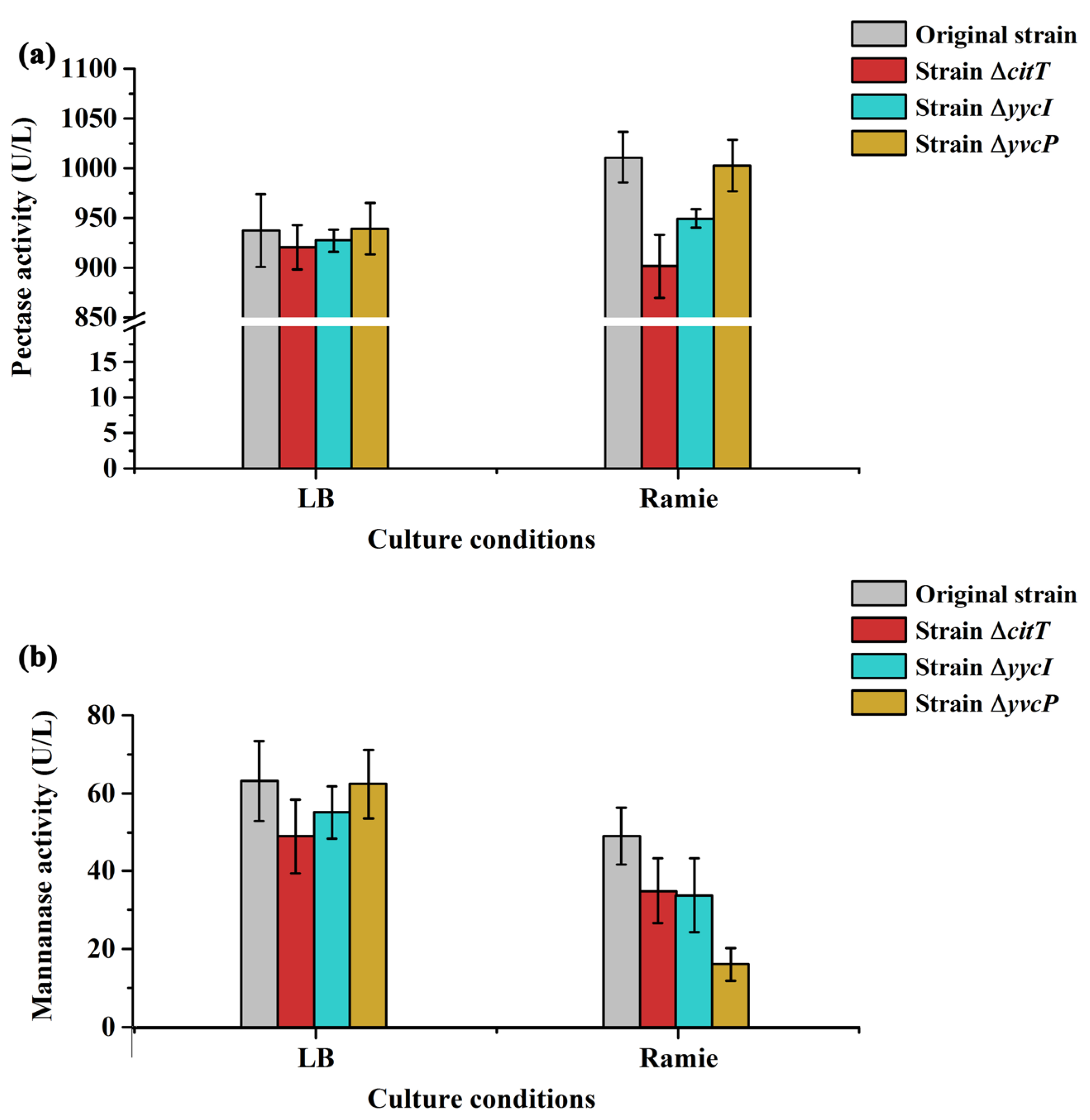
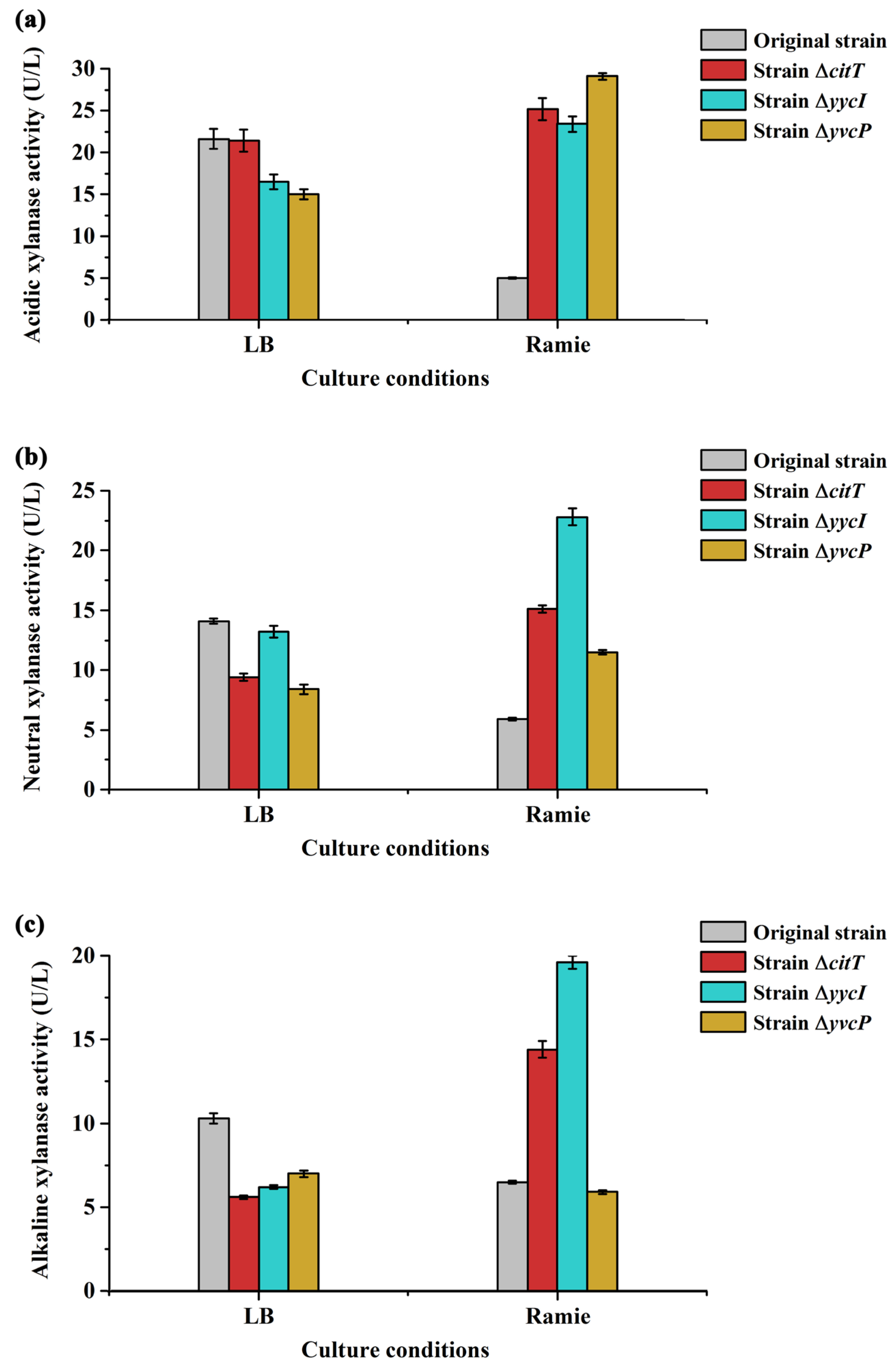
3.4. Functional Analysis of Different Regulatory Proteins
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hamad, S.F.; Stehling, N.; Hayes, S.A.; Foreman, J.P.; Rodenburg, C. Exploiting plasma exposed, natural surface nanostructures in ramie fibers for polymer composite applications. Materials 2019, 12, 1631. [Google Scholar] [CrossRef]
- Kandimalla, R.; Kalita, S.; Choudhury, B.; Devi, D.; Kalita, D.; Kalita, K.; Dash, S.; Kotoky, J. Fiber from ramie plant (Boehmeria nivea): A novel suture biomaterial. Mat. Sci. Eng. C-Mater. 2016, 62, 816–822. [Google Scholar] [CrossRef]
- Cheng, L.; Duan, S.; Feng, X.; Zheng, K.; Yang, Q.; Xu, H.; Luo, W.; Peng, Y. Ramie-degumming methodologies: A short review. J. Eng. Fibers Fabr. 2020, 15, 1558925020940105. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, Y.; Du, M.; Yu, J.; Li, Z.; Ding, B. Research progress in Ramie fiber extraction: Degumming method, working mechanism, and fiber performance. Ind. Crops Prod. 2024, 222, 119876. [Google Scholar] [CrossRef]
- Liu, Z.C.; Duan, S.W.; Sun, Q.X.; Peng, Y.D.; Feng, X.Y.; Zheng, K.; Hu, Z.X.; Zhang, Y.X. A rapid process of ramie bio-degumming by Pectobacterium sp. CXJZU-120. Text Res. J. 2012, 82, 1553–1559. [Google Scholar] [CrossRef]
- Deng, Q.; Li, N.; Bai, S.N.; Cao, J.Q.; Jin, Y.L.; Zhang, H.E.; Wang, J.K.; Wang, Q. SbPL1CE8 from Segatella bryantii combines with SbGH28GH105 in a multi-enzyme cascade for pectic biomass utilization. Int. J. Biol. Macromol. 2024, 279, 135217. [Google Scholar] [CrossRef]
- Liu, S.J.; Qin, Y.; Wang, Q.Y.; Zhang, J.; Zhou, J.; He, B.X.; Liang, X.Q.; Xian, L.; Wu, J.H. A novel pectate lyase with high specific activity from Bacillus sp. B58-2: Gene cloning, heterologous expression and use in ramie degumming. Enzyme Microb. Tech. 2024, 175, 110395. [Google Scholar] [CrossRef]
- Guo, G.; Liu, Z.C.; Xu, J.F.; Liu, J.P.; Dai, X.Y.; Xie, D.P.; Peng, K.Q.; Feng, X.Y.; Duan, S.W.; Zheng, K.; et al. Purification and characterization of a xylanase from Bacillus subtilis isolated from the degumming line. J. Basic Microb. 2012, 52, 419–428. [Google Scholar] [CrossRef]
- Wang, Y.W.; Shu, T.; Fan, P.; Zhang, H.S.; Turunen, O.; Xiong, H.R.; Yu, L.J. Characterization of a recombinant alkaline thermostable β-mannanase and its application in eco-friendly ramie degumming. Process Biochem. 2017, 61, 73–79. [Google Scholar] [CrossRef]
- Yang, Q.; Gao, M.Q.; Cheng, L.F.; Feng, X.Y.; Zheng, K.; Peng, Z.H.; Peng, Y.D.; Duan, S.W. Genomic scanning and extracellular proteomic analysis of Dickeya dadantii DCE-01 Reveal its excellent performance on Ramie degumming. Fiber Polym. 2023, 24, 1517–1525. [Google Scholar] [CrossRef]
- Qin, Y.; Xie, X.; Mo, H.; Li, Y.; Chen, X.; Ma, Y.; Zhao, Z.; Zheng, H.; Sun, Y.; Li, D.; et al. Comparative analysis of anaerobic degumming effects on different bast fibers from the angle of enzyme activity and microbial community structure. J. Clean. Prod. 2024, 480, 144147. [Google Scholar] [CrossRef]
- Shu, T.; Li, P.; Hou, Z.; Wang, H.; Yu, T.; Chen, Y.; Fu, C.; Yu, L. Improving microbial degumming efficiency for ramie fibers: Synergistic action of enhanced xylanase activity and optimized pretreatment. Bioresour. Technol. 2025, 443, 132747. [Google Scholar] [CrossRef]
- Basu, S.; Saha, M.N.; Chattopadhyay, D.; Chakrabarti, K. Large-scale degumming of ramie fibre using a newly isolated Bacillus pumilus DKS1 with high pectate lyase activity. J. Ind. Microbiol. Biot. 2009, 36, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Ye, J.T.; Xue, Y.F.; Ma, Y.H. Directed evolution and structural analysis of alkaline pectate lyase from the alkaliphilic bacterium Bacillus sp. Strain N16-5 to improve its thermostability for efficient Ramie degumming. Appl. Environ. Microb. 2015, 81, 5714–5723. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.F.; Zou, M.Y.; Li, X.Z.; Zhao, J.; Qu, Y.B. An effective degumming enzyme from Bacillus sp. Y1 and synergistic action of hydrogen peroxide and protease on enzymatic degumming of Ramie fibers. Biomed. Res. Int. 2013, 2013, 212315. [Google Scholar] [CrossRef]
- Wang, X.W.; Lu, Z.H.; Xu, T.; Selvaraj, J.N.; Yi, L.; Zhang, G.M. Improving the specific activity and thermo-stability of alkaline pectate lyase from Bacillus subtilis 168 for bioscouring. Biochem. Eng. J. 2018, 129, 74–83. [Google Scholar] [CrossRef]
- Zhang, C.J.; Yao, J.; Zhou, C.; Mao, L.W.; Zhang, G.M.; Ma, Y.H. The alkaline pectate lyase PEL168 of Bacillus subtilis heterologously expressed in Pichia pastorisis more stable and efficient for degumming ramie fiber. BMC Biotechnol. 2013, 13, 26. [Google Scholar] [CrossRef]
- Niu, T.; Lv, X.; Liu, Z.; Li, J.; Du, G.; Liu, L. Synergetic engineering of central carbon and nitrogen metabolism for the production of N-acetylglucosamine in Bacillus subtilis. Biotechnol. Appl. Biochem. 2020, 67, 123–132. [Google Scholar] [CrossRef]
- Park, Y.C. Xylose metabolism and transport in Bacillus subtilis and its application to D-ribose production. J. Microbiol. Biotechnol. 2025, 35, e2504021. [Google Scholar] [CrossRef]
- Meyer, H.; Weidmann, H.; Mäder, U.; Hecker, M.; Völker, U.; Lalk, M. A time resolved metabolomics study: The influence of different carbon sources during growth and starvation of Bacillus subtilis. Mol. Biosyst. 2014, 10, 1812–1823. [Google Scholar] [CrossRef]
- Handtke, S.; Albrecht, D.; Otto, A.; Becher, D.; Hecker, M.; Voigt, B. The proteomic response of Bacillus pumilus cells to glucose starvation. Proteomics 2018, 18, 1700109. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Duan, S.W.; Cheng, L.F.; Feng, X.Y.; Zheng, K.; Xu, C.; Peng, Y.D. Proteomic characterization of Bacillus Subtilis on bio-degumming of Ramie bast. J. Nat. Fibers 2022, 19, 9886–9903. [Google Scholar] [CrossRef]
- Kim, I.M.; Szurmant, H. Two-component signaling pathways: A bacterial goldilocks mechanism. Elife 2020, 9, e54244. [Google Scholar] [CrossRef]
- Gao, R.; Stock, A.M. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009, 63, 133–154. [Google Scholar] [CrossRef]
- Quandt, E.M.; Deatherage, D.E.; Ellington, A.D.; Georgiou, G.; Barrick, J.E. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, 2217–2222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.M.; Li, X.F.; Wang, X.; Li, Z.; He, J. The two-component signal transduction system YvcPQ regulates the bacterial resistance to bacitracin in Bacillus thuringiensis. Arch. Microbiol. 2016, 198, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Takada, H.; Yoshikawa, H. Essentiality and function of WalK/WalR two-component system: The past, present, and future of research. Biosci. Biotech. Bioch. 2018, 82, 741–751. [Google Scholar] [CrossRef]
- Yang, Q.; Duan, S.W.; Cheng, L.F.; Feng, X.Y.; Zheng, K.; Xie, C.L.; Liu, Z.Y.; Pen, Y.D. Engineering of a Bacillus subtilis strain deficient in cellulase: Application in degumming of Ramie. Fiber Polym. 2019, 20, 57–62. [Google Scholar] [CrossRef]
- Altenbuchner, J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microb. 2016, 82, 5421–5427. [Google Scholar] [CrossRef]
- Duan, S.W.; Cheng, L.F.; Liu, Z.C.; Feng, X.Y.; Zheng, K.; Peng, Y.D. Diversity and characteristics of kenaf bast degumming microbial resources. J. Nat. Fibers. 2018, 15, 799–807. [Google Scholar] [CrossRef]
- GB 5889-1986; Method of Quantitative Analysis of Ramie Chemical Components. China Standard Press: Beijing, China, 1986.
- Yang, Q.; Cheng, L.F.; Feng, X.Y.; Zheng, K.; Liu, C.J.; Liu, Z.Y.; Peng, Y.D.; Duan, S.W. Response regulator CitT from CitST two-component system specifically regulates the pectin degradation in bio-degumming of Ramie fibers by Bacillus subtilis. J. Nat. Fibers 2022, 19, 15488–15500. [Google Scholar] [CrossRef]
- Tanaka, K.; Kobayashi, K.; Ogasawara, N. The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiol.-Sgm 2003, 149, 2317–2329. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Murata, M.; Sekiguchi, J. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 2000, 37, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Bernhards, C.B.; Chen, Y.; Toutkoushian, H.; Popham, D.L. HtrC is involved in proteolysis of YpeB during germination of Bacillus anthracis and Bacillus subtilis spores. J. Bacteriol. 2015, 197, 326–336. [Google Scholar] [CrossRef]
- Biller, S.J.; Wayne, K.J.; Winkler, M.E.; Burkholder, W.F. The putative hydrolase YycJ (WalJ) affects the coordination of cell division with DNA replication in Bacillus subtilis and may play a conserved role in cell wall metabolism. J. Bacteriol. 2011, 193, 896–908. [Google Scholar] [CrossRef]
- Huang, W.Z.; Wang, J.J.; Chen, H.J.; Chen, J.T.; Shaw, G.C. The heat-inducible essential response regulator WalR positively regulates transcription of sigI, mreBH and lytE in Bacillus subtilis under heat stress. Res. Microbiol. 2013, 164, 998–1008. [Google Scholar] [CrossRef]
- Dobihal, G.S.; Brunet, Y.R.; Flores-Kim, J.; Rudner, D.Z. Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. eLife 2019, 8, e52088. [Google Scholar] [CrossRef]
- Yamada, Y.; Tikhonova, E.B.; Zgurskaya, H.I. YknWXYZ is an unusual four-component transporter with a role in protection against sporulation-delaying-protein-induced killing of Bacillus subtilis. J. Bacteriol. 2012, 194, 4386–4394. [Google Scholar] [CrossRef]
- Nam, D.H.; Lim, S.K.; Chung, M.H.; Lee, E.S.; Sohn, Y.S.; Ryu, D.D.Y. Membrane transporter genes in Cephabacin biosynthetic gene cluster of Lysobacter lactamgenus. J. Microbiol. Biotechn. 2001, 11, 153–159. [Google Scholar]
- Asina, F.N.U.; Brzonova, I.; Kozliak, E.; Kubátová, A.; Ji, Y. Microbial treatment of industrial lignin: Successes, problems and challenges. Renew. Sust. Energ. Rev. 2017, 77, 1179–1205. [Google Scholar] [CrossRef]
- Yang, Q.; Hu, Y.Q.; Feng, X.Y.; Xi, G.G.; Zheng, K.; Peng, Z.H.; Han, F.B.; Cheng, L.F.; Duan, S.W. Proteomic insights into citT-deletion induced metabolic sensitivity in bio-degumming of Ramie fibers by Bacillus subtilis. J. Nat. Fibers 2024, 21, 2334414. [Google Scholar] [CrossRef]
- Cruz-Magalhaes, V.; Nieto-Jacobo, M.F.; Rostás, M.; Echaide-Aquino, J.F.; Esquivel-Naranjo, E.U.; Stewart, A.; Loguercio, L.L.; Mendoza-Mendoza, A. Histidine kinase two-component response regulators Ssk1, Skn7 and Rim15 differentially control growth, developmental and volatile organic compounds emissions as stress responses in Trichoderma atroviride. Curr. Res. Microb. Sci. 2022, 3, 100139. [Google Scholar] [CrossRef]
- Ruller, R.; Rosa, J.C.; Faça, V.M.; Greene, L.J.; Ward, R.J. Efficient constitutive expression of Bacillus subtilis xylanase A in Escherichia coli DH5α under the control of the Bacillus BsXA promoter. Biotechnol. Appl. Biochem. 2006, 43, 9–15. [Google Scholar] [CrossRef]
- John, F.J.S.; Rice, J.D.; Preston, J.F. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J. Bacteriol. 2006, 188, 8617–8626. [Google Scholar] [CrossRef] [PubMed]
- Abena, T.; Simachew, A. A review on xylanase sources, classification, mode of action, fermentation processes, and applications as a promising biocatalyst. Biotechnologia 2024, 105, 273. [Google Scholar] [CrossRef]
- Yang, Q.; Cheng, L.; Feng, X.; Zheng, K.; Liu, Z.; Duan, S.; Peng, Y. Analysis of the relationship between enzymatic activity and microbial degumming effect of kenaf bast. J. Nat. Fibers 2021, 18, 1217–1228. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Ma, S.; Cheng, L.; Zhou, X.; Xi, G.; Chen, C.; Peng, Z.; Hu, Y.; Tan, S.; Duan, S. Two-Component Response Regulators CitT, YvcP, and YycI Differentially Control Pectin and Hemicellulose Degradation in Degumming of Ramie Fibers by Bacillus subtilis Strain 168. Polymers 2025, 17, 2473. https://doi.org/10.3390/polym17182473
Yang Q, Ma S, Cheng L, Zhou X, Xi G, Chen C, Peng Z, Hu Y, Tan S, Duan S. Two-Component Response Regulators CitT, YvcP, and YycI Differentially Control Pectin and Hemicellulose Degradation in Degumming of Ramie Fibers by Bacillus subtilis Strain 168. Polymers. 2025; 17(18):2473. https://doi.org/10.3390/polym17182473
Chicago/Turabian StyleYang, Qi, Shihang Ma, Lifeng Cheng, Xiang Zhou, Guoguo Xi, Chen Chen, Zhenghong Peng, Yuqin Hu, Si Tan, and Shengwen Duan. 2025. "Two-Component Response Regulators CitT, YvcP, and YycI Differentially Control Pectin and Hemicellulose Degradation in Degumming of Ramie Fibers by Bacillus subtilis Strain 168" Polymers 17, no. 18: 2473. https://doi.org/10.3390/polym17182473
APA StyleYang, Q., Ma, S., Cheng, L., Zhou, X., Xi, G., Chen, C., Peng, Z., Hu, Y., Tan, S., & Duan, S. (2025). Two-Component Response Regulators CitT, YvcP, and YycI Differentially Control Pectin and Hemicellulose Degradation in Degumming of Ramie Fibers by Bacillus subtilis Strain 168. Polymers, 17(18), 2473. https://doi.org/10.3390/polym17182473





