Computational Analysis of the Tripartite Interaction of Phasins (PhaP4 and 5)-Sigma Factor (σ24)-DNA of Azospirillum brasilense Sp7
Abstract
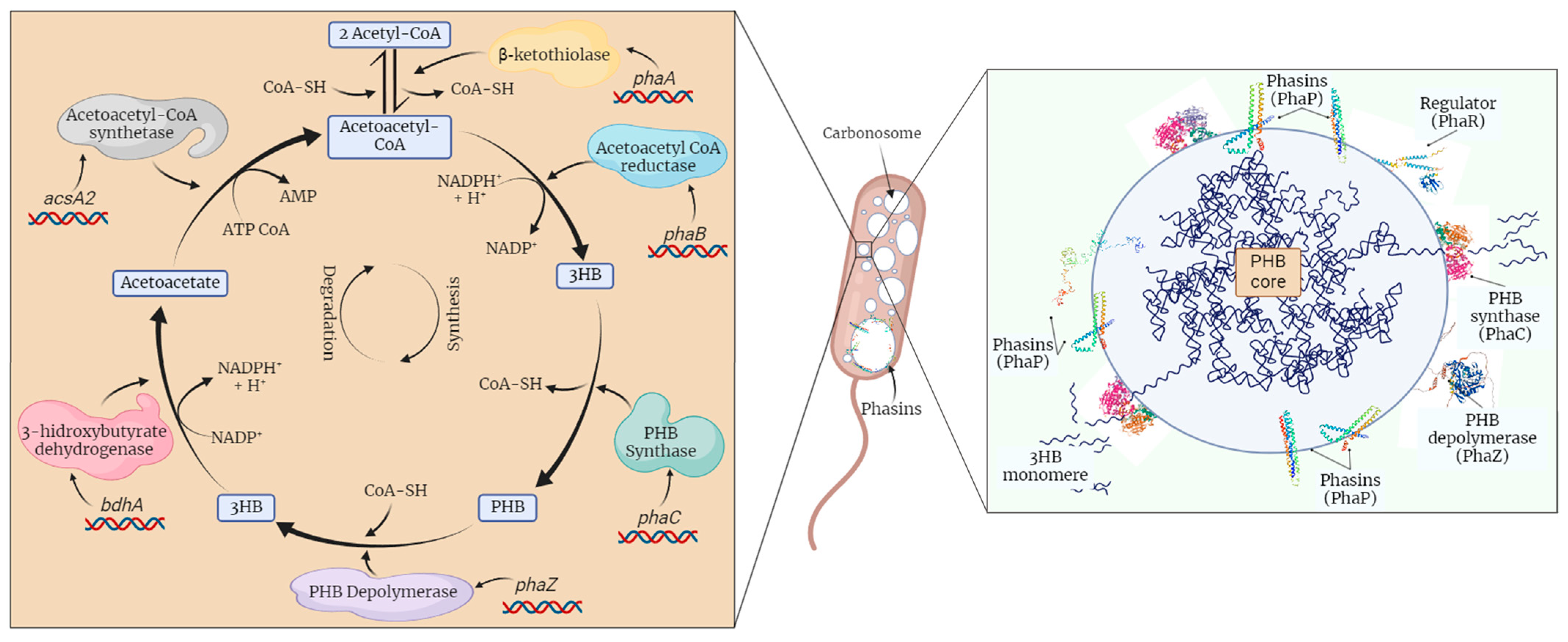
1. Introduction
2. Materials and Methods
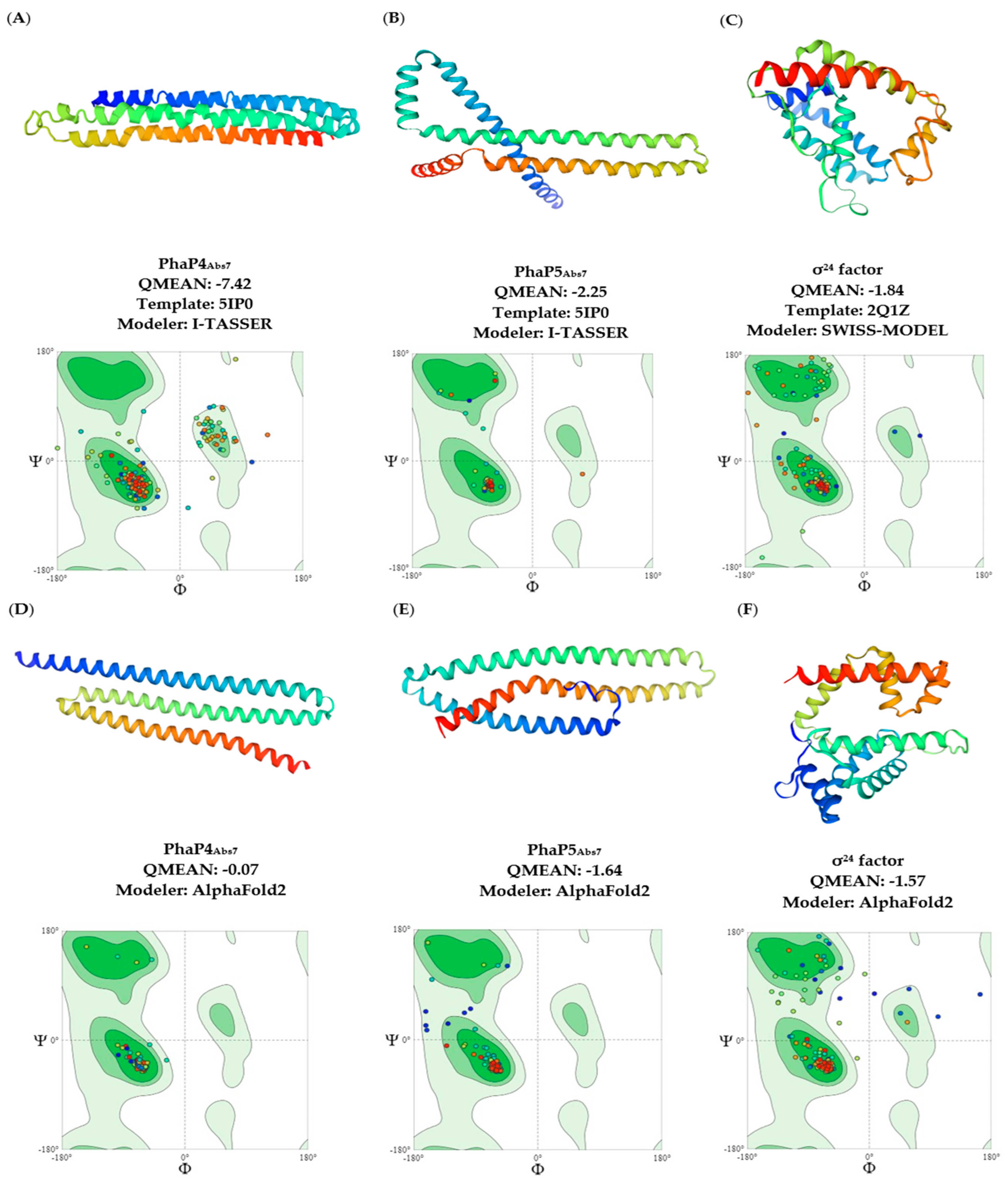
2.1. Three-Dimensional Analysis of the Structures of PhaP4Abs7, PhaP5Abs7, and σ24
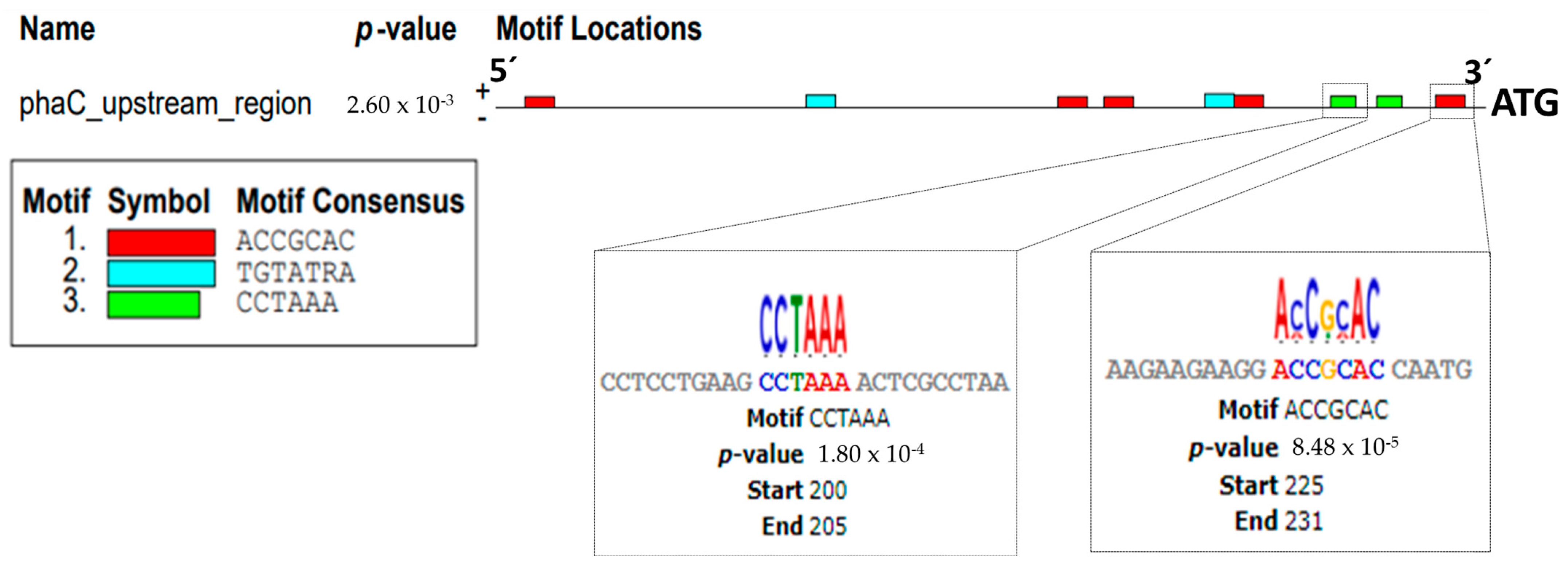
2.2. Determination of the Promoter Regions (−10 and −35) Upstream of the phaC Gene
2.3. Molecular Docking to Form the Phasin-σ24 Factor Protein Complex and Individual Docking of Phasins and the σ24 Factor with DNA
2.4. Tripartite Molecular Docking between PhaP4Abs7-σ24 and PhaP5Abs7-σ24 Protein Complexes with the Promoter Region of the phaC Gene
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S.; Rai, A.K.; Mishra, M.N.; Shukla, M.; Singh, P.K.; Tripathi, A.K. RpoH2 sigma factor controls the photooxidative stress response in a non-photosynthetic rhizobacterium, Azospirillum brasilense Sp7. Microbiology 2012, 158, 2891–2902. [Google Scholar] [CrossRef]
- McAdam, B.; Fournet, M.B.; McDonald, P.; Mojicevic, M. Production of Polyhydroxybutyrate (PHB) and Factors Impacting Its Chemical and Mechanical Characteristics. Polymers 2020, 12, 2908. [Google Scholar] [CrossRef]
- Khatami, K.; Perez-Zabaleta, M.; Owusu-Agyeman, I.; Cetecioglu, Z. Waste to bioplastics: How close are we to sustainable polyhydroxyalkanoates production? Waste Manag. 2020, 119, 374–388. [Google Scholar] [CrossRef]
- Martínez-Martínez, M.D.L.A.; González-Pedrajo, B.; Dreyfus, G.; Soto-Urzúa, L.; Martínez-Morales, L.J. Phasin PhaP1 is involved in polyhydroxybutyrate granules morphology and in controlling early biopolymer accumulation in Azospirillum brasilense Sp7. AMB Express 2019, 9, 155. [Google Scholar] [CrossRef]
- Martínez, M.D.L.M.; Urzúa, L.S.; Carrillo, Y.A.; Ramírez, M.B.; Morales, L.J.M. Polyhydroxybutyrate Metabolism in Azospirillum brasilense and Its Applications, a Review. Polymers 2023, 15, 3027. [Google Scholar] [CrossRef]
- Maestro, B.; Sanz, J.M. Polyhydroxyalkanoate-associated phasins as phylogenetically heterogeneous, multipurpose proteins. Microb. Biotechnol. 2017, 10, 1323–1337. [Google Scholar] [CrossRef]
- Fibach-Paldi, S.; Burdman, S.; Okon, Y. Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol. Lett. 2012, 326, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Sagong, H.-Y.; Son, H.F.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural Insights into Polyhydroxyalkanoates Biosynthesis. Trends Biochem. Sci. 2018, 43, 790–805. [Google Scholar] [CrossRef] [PubMed]
- Jendrossek, D.; Pfeiffer, D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate): A simple molecule with multiple functions. Environ. Microbiol. 2014, 16, 2357–2373. [Google Scholar] [CrossRef] [PubMed]
- Sznajder, A.; Pfeiffer, D.; Jendrossek, D. Comparative Proteome Analysis Reveals Four Novel Polyhydroxybutyrate (PHB) Granule-Associated Proteins in Ralstonia eutropha H16. Appl. Environ. Microbiol. 2015, 81, 1847–1858. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ushimaru, K.; Motoda, Y.; Numata, K.; Tsuge, T. Phasin Proteins Activate Aeromonas caviae Polyhydroxyalkanoate (PHA) Synthase but Not Ralstonia eutropha PHA Synthase. Appl. Environ. Microbiol. 2014, 80, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Hauf, W.; Watzer, B.; Roos, N.; Klotz, A.; Forchhammer, K. Photoautotrophic Polyhydroxybutyrate Granule Formation Is Regulated by Cyanobacterial Phasin PhaP in Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2015, 81, 4411–4422. [Google Scholar] [CrossRef] [PubMed]
- Mezzina, M.P.; Pettinari, M.J. Phasins, Multifaceted Polyhydroxyalkanoate Granule-Associated Proteins. Appl. Environ. Microbiol. 2016, 82, 5060–5067. [Google Scholar] [CrossRef] [PubMed]
- Müller-Santos, M.; Koskimäki, J.J.; Alves, L.P.S.; de Souza, E.M.; Jendrossek, D.; Pirttilä, A.M. The protective role of PHB and its degradation products against stress situations in bacteria. FEMS Microbiol. Rev. 2021, 45, fuaa058. [Google Scholar] [CrossRef] [PubMed]
- Tugarova, A.V.; Dyatlova, Y.A.; Kenzhegulov, O.A.; Kamnev, A.A. Poly-3-hydroxybutyrate synthesis by different Azospirillum brasilense strains under varying nitrogen deficiency: A comparative in-situ FTIR spectroscopic analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 252, 119458. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Singh, S.; Dwivedi, S.K.; Srivastava, A.; Pandey, P.; Kumar, S.; Singh, B.N.; Tripathi, A.K. Catalase Expression in Azospirillum brasilense Sp7 Is Regulated by a Network Consisting of OxyR and Two RpoH Paralogs and Including an RpoE1→RpoH5 Regulatory Cascade. Appl. Environ. Microbiol. 2018, 84, e01787-18. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Gupta, A.; Kumar, S.; Mishra, R.; Singh, C.; Tripathi, A.K. Cross-Talk Between Cognate and Noncognate RpoE Sigma Factors and Zn2+-Binding Anti-Sigma Factors Regulates Photooxidative Stress Response in Azospirillum brasilense. Antioxidants Redox Signal. 2014, 20, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.D.A.E.; Echeverrigaray, S.; Gerhardt, G.J. BacPP: Bacterial promoter prediction—A tool for accurate sigma-factor specific assignment in enterobacteria. J. Theor. Biol. 2011, 287, 92–99. [Google Scholar] [CrossRef]
- Gupta, N.; Kumar, S.; Mishra, M.N.; Tripathi, A.K. A constitutively expressed pair of rpoE2–chrR2 in Azospirillum brasilense Sp7 is required for survival under antibiotic and oxidative stress. Microbiology 2013, 159, 205–218. [Google Scholar] [CrossRef]
- Alvarez-Gonzalez, S.; Erill, I. Design of Machine Learning Models for the Prediction of Transcription Factor Binding Regions in Bacterial DNA. Eng. Proc. 2021, 7, 59. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wei, H.; Liu, X.; Yao, Z.; Xu, M.; Wei, D.; Wang, J.; Wang, X.; Chen, G.-Q. Structural Insights on PHA Binding Protein PhaP from Aeromonas hydrophila. Sci. Rep. 2016, 6, 39424. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.A.; Greenwell, R.; Anthony, J.R.; Wang, S.; Lim, L.; Das, K.; Sofia, H.J.; Donohue, T.J.; Darst, S.A. A Conserved Structural Module Regulates Transcriptional Responses to Diverse Stress Signals in Bacteria. Mol. Cell 2007, 27, 793–805. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins 2016, 85, 435–444. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2012, 9, 173–175. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- BIOVIA; Dassault Systèmes. Discovery Studio Visualizer, v21.1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2021; Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 21 December 2021).
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Wiltgen, M. Algorithms for Structure Comparison and Analysis: Homology Modelling of Proteins. Encycl. Bioinforma. Comput. Biol. 2019, 1, 38–61. [Google Scholar] [CrossRef]
- García Ordaz, D.M. Identificacion de Secuencias Reguladoras Mediante Agrupamiento, Tesis de Maestría, INAOE. 2011. Available online: http://inaoe.repositorioinstitucional.mx/jspui/handle/1009/682 (accessed on 3 February 2024).
- Pötter, M.; Steinbüchel, A. Poly(3-hydroxybutyrate) Granule-Associated Proteins: Impacts on Poly(3-hydroxybutyrate) Synthesis and Degradation. Biomacromolecules 2005, 6, 552–560. [Google Scholar] [CrossRef]
- Wahl, A.; Schuth, N.; Pfeiffer, D.; Nussberger, S.; Jendrossek, D. PHB granules are attached to the nucleoid via PhaM in Ralstonia eutropha. BMC Microbiol. 2012, 12, 262. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.-J.; Lai, W.-J.; Zheng, Z.; Wang, H.-X.; Chen, G.-Q. Effect of over-expression of phasin gene from Aeromonas hydrophilaon biosynthesis of copolyesters of 3-hydroxybutyrate and 3-hydroxyhexanoate. FEMS Microbiol. Lett. 2005, 244, 19–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Galán, B.; Dinjaski, N.; Maestro, B.; de Eugenio, L.I.; Escapa, I.F.; Sanz, J.M.; García, J.L.; Prieto, M.A. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol. Microbiol. 2011, 79, 402–418. [Google Scholar] [CrossRef] [PubMed]
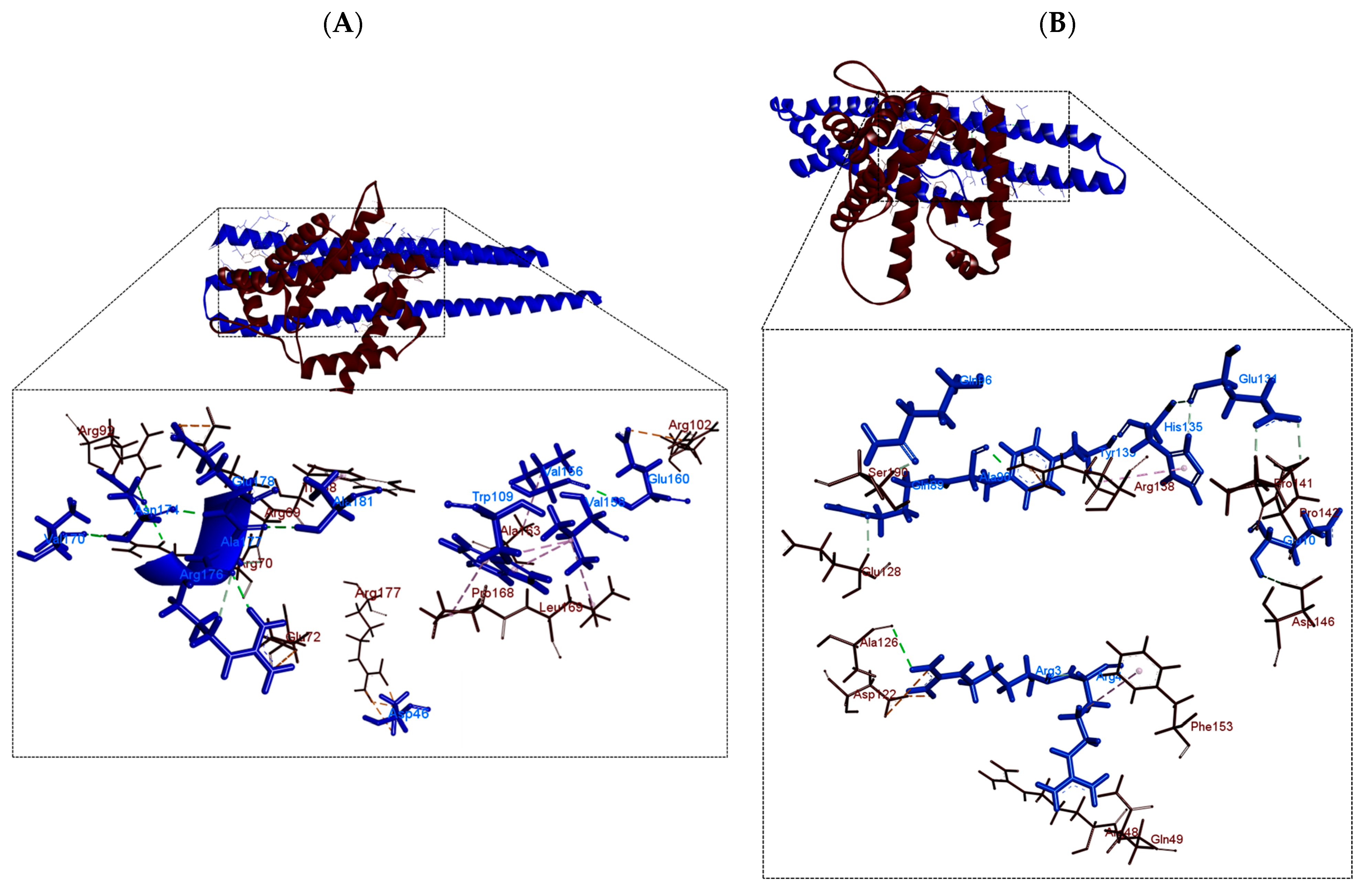

| Receptor | Ligand | Number of Hydrogen Bonds | Amino Acids with Hydrogen Interactions | ||
|---|---|---|---|---|---|
| Biovia | LigPlot | Biovia | LigPlot | ||
| Factor σ24 | PhaP4Abs7 | 4 | 5 | Gln83, Val170, Asn174 | Asp46, Gln83, Val170, Asn174 |
| PhaP5Abs7 | 3 | 3 | Arg3, Glu10, Ala90 | Arg3, Ala90, Thr93 | |
| Receptor | Ligand | Number of Hydrogen Bonds | Amino Acids with Hydrogen Interactions | ||
|---|---|---|---|---|---|
| Biovia | LigPlot | Biovia | LigPlot | ||
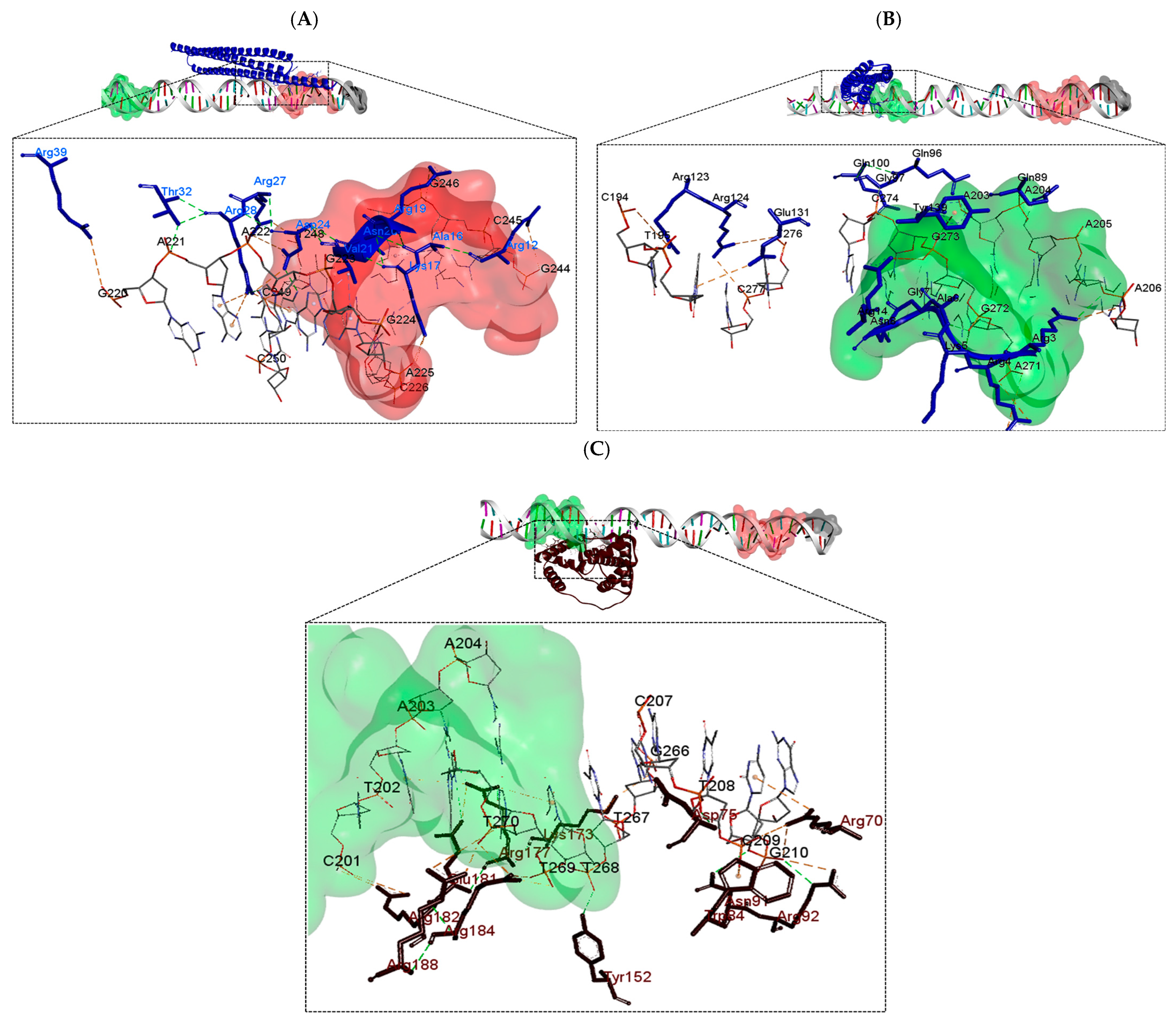
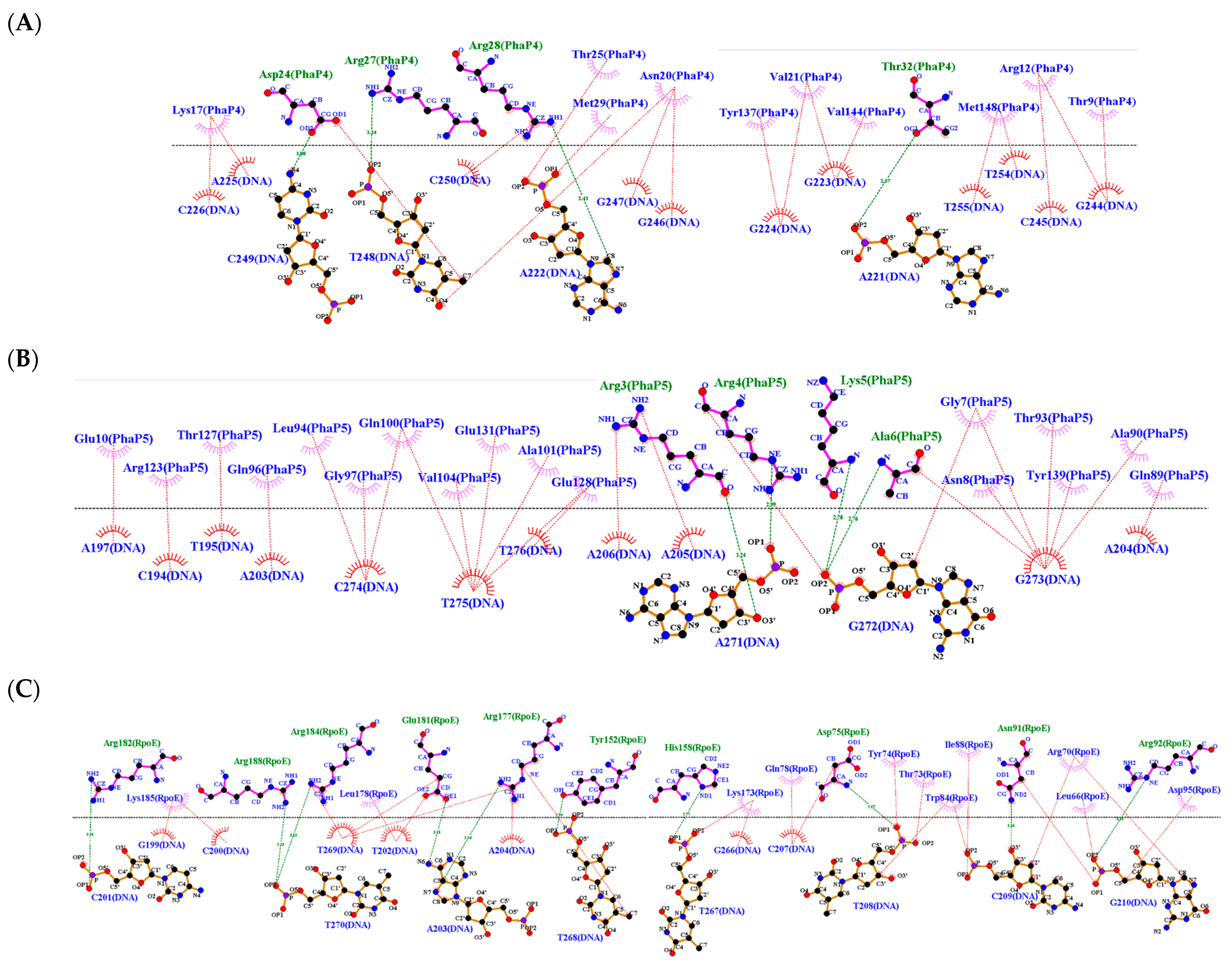
| DNA (upstream region of phaC) | PhaP4Abs7 | 3 | 4 | Asp24, Arg28, Thr32 | Asp24, Arg27, Arg28, Thr32 |
| PhaP5Abs7 | 5 | 4 | Arg3, Arg4, Lys5, Ala6, Asn8 | Arg3, Arg4, Lys5, Ala6 | |
| Factor σ24 | 6 | 10 | Asp75, Asn91, Arg92, Tyr152, Arg177, Glu181 | Asp75, Asn91, Arg92, Tyr152, His158, Arg177, Glu181 Arg182, Arg184, Arg188 | |
| Receptor | Ligand | Number of Hydrogen Bonds | Amino Acids with Hydrogen Interactions | ||
|---|---|---|---|---|---|
| Biovia | LigPlot | Biovia | LigPlot | ||
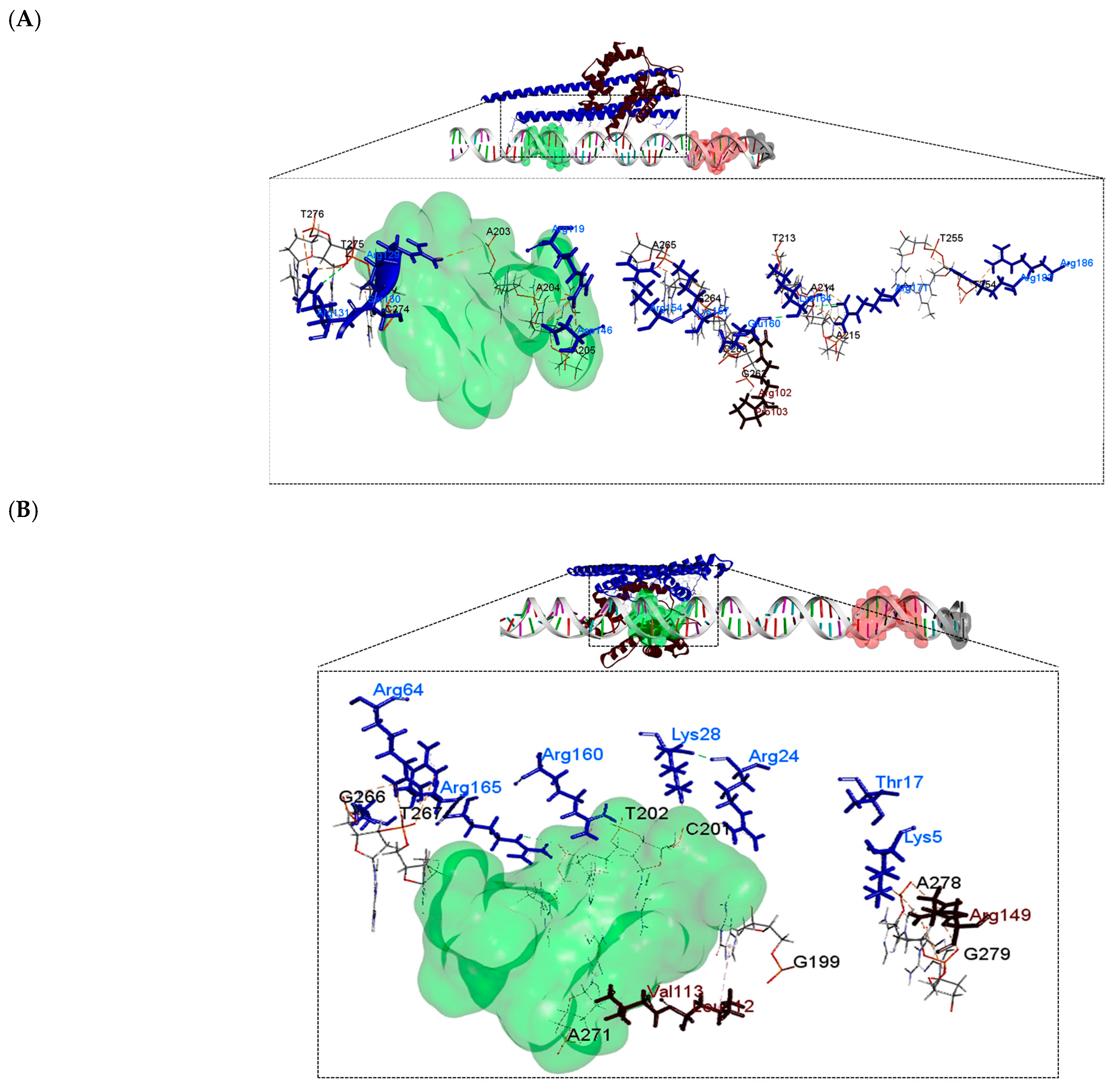
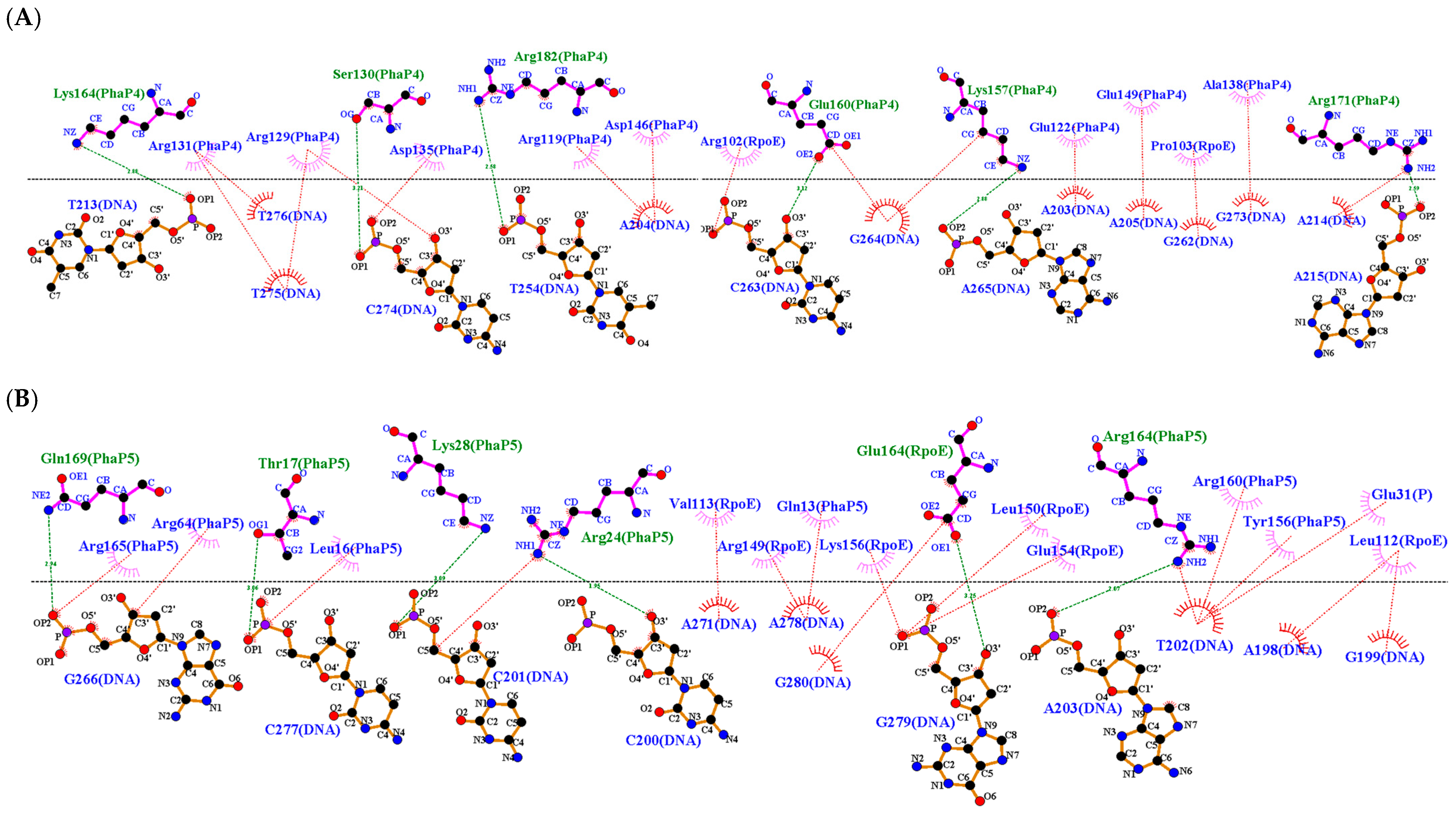
| DNA (upstream region of phaC) | PhaP4Abs7-σ24 | 3 | 6 | Ser130 *, Arg131 *, Arg171 * | Ser130 *, Lys157 *, Glu160 *, Lys164 *, Arg171 *, Arg182 * |
| PhaP5Abs7-σ24 | 3 | 6 | Thr17 *, Arg164 *, Gln169 * | Thr17 *, Arg24 *, Lys28 *, Gln169 *, Arg164 *, Glu164 ** | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilar-Carrillo, Y.; Soto-Urzúa, L.; Martínez-Martínez, M.D.L.Á.; Becerril-Ramírez, M.; Martínez-Morales, L.J. Computational Analysis of the Tripartite Interaction of Phasins (PhaP4 and 5)-Sigma Factor (σ24)-DNA of Azospirillum brasilense Sp7. Polymers 2024, 16, 611. https://doi.org/10.3390/polym16050611
Aguilar-Carrillo Y, Soto-Urzúa L, Martínez-Martínez MDLÁ, Becerril-Ramírez M, Martínez-Morales LJ. Computational Analysis of the Tripartite Interaction of Phasins (PhaP4 and 5)-Sigma Factor (σ24)-DNA of Azospirillum brasilense Sp7. Polymers. 2024; 16(5):611. https://doi.org/10.3390/polym16050611
Chicago/Turabian StyleAguilar-Carrillo, Yovani, Lucía Soto-Urzúa, María De Los Ángeles Martínez-Martínez, Mirian Becerril-Ramírez, and Luis Javier Martínez-Morales. 2024. "Computational Analysis of the Tripartite Interaction of Phasins (PhaP4 and 5)-Sigma Factor (σ24)-DNA of Azospirillum brasilense Sp7" Polymers 16, no. 5: 611. https://doi.org/10.3390/polym16050611
APA StyleAguilar-Carrillo, Y., Soto-Urzúa, L., Martínez-Martínez, M. D. L. Á., Becerril-Ramírez, M., & Martínez-Morales, L. J. (2024). Computational Analysis of the Tripartite Interaction of Phasins (PhaP4 and 5)-Sigma Factor (σ24)-DNA of Azospirillum brasilense Sp7. Polymers, 16(5), 611. https://doi.org/10.3390/polym16050611







