Bacterial Adhesion to Natural and Synthetic Fibre-Forming Polymers: Influence of Material Properties
Abstract
1. Introduction
2. Theoretical Approach of Bacterial Adhesion
3. Textile Factors Influencing Bacterial Adhesion
3.1. Chemical and Physicochemical Properties
3.1.1. Hydrophilicity/Hydrophobicity
3.1.2. Combination of Hydrophilicity/Hydrophobicity and Surface Charge
3.1.3. Surface Free Energy
3.2. Constructional and Textural Properties
3.2.1. Porosity
3.2.2. Roughness
3.2.3. Combination of Roughness, Porosity, and Hydrophilicity/Hydrophobicity
4. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Strese, H.; Kuck, M.; Benken, R.; Schanzer, S.; Richter, H.; Fluhr, J.W.; Meinke, M.C.; Benderoth, C.; Frankowski, G.; Sterry, W.; et al. Application of Optical Methods to Characterize Textile Materials and Their Influence on the Human Skin. J. Biomed. Opt. 2011, 16, 046013. [Google Scholar] [CrossRef] [PubMed]
- Zimniewska, M.; Kozlowski, R.; Rawluk, M. Natural vs. Man-Made Fibres—Physiological Viewpoint. J. Nat. Fibers 2004, 1, 69–81. [Google Scholar] [CrossRef]
- Li, Y. The Science of Clothing Comfort. Text. Progress. 2001, 31, 1–135. [Google Scholar] [CrossRef]
- Purwar, R.; Joshi, M. Recent Developments in Antimicrobial Finishing of Textiles—A Review. AATCC Rev. 2004, 4, 22–26. [Google Scholar]
- Gao, Y.; Cranston, R. Recent Advances in Antimicrobial Treatments of Textiles. Text. Res. J. 2008, 78, 60–72. [Google Scholar] [CrossRef]
- Simončič, B.; Tomšič, B.; Orel, B.; Jerman, I. Sol-Gel Technology for Chemical Modification of Textiles; University of Twente: Enschede, The Netherlands, 2010; pp. 17–34. [Google Scholar]
- Morais, D.S.; Guedes, R.M.; Lopes, M.A. Antimicrobial Approaches for Textiles: From Research to Market. Materials 2016, 9, 498. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. How Long Can Nosocomial Pathogens Survive on Textiles? A Systematic Review. GMS Hyg. Infect. Control. 2020, 15. [Google Scholar] [CrossRef]
- Simončič, B.; Tomšič, B.; Forte-Tavčer, P.; Ivanuš, A.; Kovač, F. Biorazgradnja Tekstilnih Vlaken in Njihova Protimikrobna Zaščita; Naravoslovnotehniška fakulteta, Oddelek za tekstilstvo: Ljubljana, Slovenija, 2010; ISBN 9789616045810. [Google Scholar]
- Mallick, D.; Gupta, D.; Sharma, S. Transfer of Bacteria between Fabric and Surrogate Skin. Am. J. Infect. Control 2022, 50, 758–763. [Google Scholar] [CrossRef]
- Sattar, S.A.; Springthorpe, S.; Mani, S. Transfer of Bacteria from Fabrics to Hands and Other Fabrics: Development and Application of a Quantitative Method Using Staphylococcus Aureus as a Model. J. Appl. Microbiol. 2001, 90, 962–970. [Google Scholar] [CrossRef]
- Curtis White, W.; Bellfield, R.; Ellis, J.; Vandendaele, P. Controlling the Spread of Infections in Hospital Wards by the Use of Antimicrobials on Medical Textiles and Surfaces. In Medical and Healthcare Textiles; Anand, S.C., Kennedy, J.F., Miraftab, M., Rajendran, S., Eds.; Woodhead Publishing Series in Textiles; Woodhead Publishing: Sawston, UK, 2010; pp. 55–75. ISBN 978-1-84569-224-7. [Google Scholar]
- Whitehead, K.; Eppinger, J.; Srinivasan, V.; Ijaz, M.K.; Nims, R.W.; McKinney, J. Microbiome of Clothing Items Worn for a Single Day in a Non-Healthcare Setting. Microbiol. Res. 2023, 14, 948–958. [Google Scholar] [CrossRef]
- Ikuta, K.S.; Swetschinski, L.R.; Robles Aguilar, G.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Davis Weaver, N.; Wool, E.E.; Han, C.; Gershberg Hayoon, A.; et al. Global Mortality Associated with 33 Bacterial Pathogens in 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; ISBN 9789241564748. [Google Scholar]
- Dincer, S.; Uslu, F.M.; Delik, A. Antibiotic Resistance in Biofilm. In Bacterial Biofilms; Dincer, S., Özdenefe, M.S., Arkut, A., Eds.; IntechOpen: Rijeka, Croatia, 2020; Chapter 9; pp. 1–14. ISBN 978-1-78985-900-3. [Google Scholar]
- Simoes, M. Antimicrobial Strategies Effective against Infectious Bacterial Biofilms. Curr. Med. Chem. 2011, 18, 2129–2145. [Google Scholar] [CrossRef] [PubMed]
- Missirlis, Y.F.; Katsikogianni, M. Theoretical and Experimental Approaches of Bacteria-Biomaterial Interactions. Materwiss Werksttech 2007, 38, 983–994. [Google Scholar] [CrossRef]
- Fleming, D.; Rumbaugh, K.P. Approaches to Dispersing Medical Biofilms. Microorganisms 2017, 5, 15. [Google Scholar] [CrossRef]
- Zobell, C.E. The Effect of Solid Surfaces upon Bacterial Activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Rittman, B.E. The Effect of Shear Stress on Biofilm Loss Rate. Biotechnol. Bioeng. 1982, 24, 501–506. [Google Scholar] [CrossRef]
- Gristina, A.G. Biomaterial-Centered Infection: Microbial Adhesion Versus Tissue Integration. Science 1987, 237, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Rosan, B. Adherence of Mutans Streptococci to Other Oral Bacteria. Infect. Immun. 1990, 58, 1738–1743. [Google Scholar] [CrossRef] [PubMed]
- Neu, T.R.; Marshall, K.C. Bacterial Polymers: Physicochemical Aspects of Their Interactions at Interfaces. J. Biomater. Appl. 1990, 5, 107–133. [Google Scholar] [CrossRef]
- Ellen, R.P.; Veisman, H.; Buivids, I.A.; Rosenberg, M. Kinetics of Lactose-Reversible Coadhesion of Actinomyces Naeslundii WVU 398A and Streptococcus Oralis 34 on the Surface of Hexadecane Droplets. Oral. Microbiol. Immunol. 1994, 9, 364–371. [Google Scholar] [CrossRef]
- Schneider, R.; Marshall, K. Retention of the Gramnegative Marine Bacterium SW8 on Surfaces—Effects of Microbial Physiology, Substratum Nature and Conditioning Films. Colloids Surf. B Biointerfaces 1994, 2, 387–396. [Google Scholar] [CrossRef]
- Bloomquist, C.G.; Reilly, B.E.; Liljemark, W.F.; Liljemark, F.; Bloomquist, C.G.; Bandt, C.L.; Philstrom, B.L.; Hinrichs, J.E.; Wolff, L.F. Adherence, Accumulation, and Cell Division of a Natural Adherent Bacterial Population. J. Bacteriol. 1996, 178, 1172–1177. [Google Scholar] [CrossRef][Green Version]
- Palmer, R.J.; White, D.C. Developmental Biology of Biofilms: Implications for Treatment and Control. Trends Microbiol. 1997, 5, 435–440. [Google Scholar] [CrossRef]
- Bos, R.; Van Der Mei, H.C.; Busscher, H.J. Physico-Chemistry of Initial Microbial Adhesive Interactions—Its Mechanisms and Methods for Study. FEMS Microbiol. Rev. 1999, 23, 179–230. [Google Scholar] [CrossRef]
- Vuong, C.; Otto, M. Staphylococcus Epidermidis Infections. Microbes Infect. 2002, 4, 481–489. [Google Scholar] [CrossRef]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas Aeruginosa Displays Multiple Phenotypes during Development as a Biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, K.K. What Drives Bacteria to Produce a Biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. A Novel Signaling Network Essential for Regulating Pseudomonas Aeruginosa Biofilm Development. PLoS Pathog. 2009, 5, e1000668. [Google Scholar] [CrossRef]
- Kim, J.; Park, H.D.; Chung, S. Microfluidic Approaches to Bacterial Biofilm Formation. Molecules 2012, 17, 9818–9834. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Malic, S.; Cruz, H.; Williams, D. Introduction to Biofilms. In Biofilms and Veterinary Medicine; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6, pp. 41–68. ISBN 978-3-642-21288-8. [Google Scholar]
- Gupta, K.; Marques, C.N.H.; Petrova, O.E.; Sauer, K. Antimicrobial Tolerance of Pseudomonas Aeruginosa Biofilms Is Activated during an Early Developmental Stage and Requires the Two-Component Hybrid SagS. J. Bacteriol. 2013, 195, 4975–4987. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.; Kim, H.; Kim, G.; Lee, D. A Review of Bacterial Biofilm Formation and Growth: Rheological Characterization, Techniques, and Applications. Korea Aust. Rheol. J. 2023, 35, 267–278. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial Biofilm: Formation, Architecture, Antibiotic Resistance, and Control Strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial Biofilm: A Review on Formation, Infection, Antibiotic Resistance, Control Measures, and Innovative Treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Ashikur Rahman, M.; Akter, S.; Ashrafudoulla, M.; Anamul Hasan Chowdhury, M.; Uddin Mahamud, A.G.M.S.; Hong Park, S.; Ha, S. Do Insights into the Mechanisms and Key Factors Influencing Biofilm Formation by Aeromonas Hydrophila in the Food Industry: A Comprehensive Review and Bibliometric Analysis. Food Res. Int. 2024, 175, 113671. [Google Scholar] [CrossRef]
- Toyofuku, M.; Inaba, T.; Kiyokawa, T.; Obana, N.; Yawata, Y.; Nomura, N. Environmental Factors That Shape Biofilm Formation. Biosci. Biotechnol. Biochem. 2016, 80, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm Life Cycle: Expanding the Conceptual Model of Biofilm Formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Wood, S.R.; Kirkham, J.; Marsh, P.D.; Shore, R.C.; Nattress, B.; Robinson, C. Architecture of Intact Natural Human Plaque Biofilms Studied by Confocal Laser Scanning Microscopy. J. Dent. Res. 2000, 79, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Darmanin, T.; Guittard, F. Recent Advances in the Potential Applications of Bioinspired Superhydrophobic Materials. J. Mater. Chem. A Mater. 2014, 2, 16319–16359. [Google Scholar] [CrossRef]
- Zhou, H.; Li, Q.; Zhang, Z.; Wang, X.; Niu, H. Recent Advances in Superhydrophobic and Antibacterial Cellulose-Based Fibers and Fabrics: Bio-Inspiration, Strategies, and Applications. Adv. Fiber Mater. 2023, 5, 1555–1591. [Google Scholar] [CrossRef]
- Sotiri, I.; Overton, J.C.; Waterhouse, A.; Howell, C. Immobilized Liquid Layers: A New Approach to Anti-Adhesion Surfaces for Medical Applications. Exp. Biol. Med. 2016, 241, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Surat’man, N.E.B.; Mah, J.J.Q.; Qu, C.; Li, Z. Surface Antimicrobial Functionalization with Polymers: Fabrication, Mechanisms and Applications. J. Mater. Chem. B 2022, 10, 9349–9368. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, Z.; Chen, X.; He, C. Tissue-Adhesive, Antibacterial, Naturally-Derived Polymer Hydrogels as Wound Dressings for Infected and Chronic Wound Healing. J. Polym. Sci. 2024, 62, 2320–2339. [Google Scholar] [CrossRef]
- Dixit, S.; Varshney, S.; Gupta, D.; Sharma, S. Textiles as Fomites in the Healthcare System. Appl. Microbiol. Biotechnol. 2023, 107, 3887–3897. [Google Scholar] [CrossRef] [PubMed]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial Adhesion and Biofilms on Surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Hori, K.; Matsumoto, S. Bacterial Adhesion: From Mechanism to Control. Biochem. Eng. J. 2010, 48, 424–434. [Google Scholar] [CrossRef]
- Marshall, K.C.; Stout, R.; Mitchell, R. Mechanism of the Initial Events in the Sorption of Marine Bacteria to Surfaces. Microbiology 1971, 68, 337–348. [Google Scholar] [CrossRef]
- Marshall, K.C. Mechanisms of Bacterial Adhesion at Solid-Water Interfaces. In Bacterial Adhesion: Mechanisms and Physiological Significance; Savage Dwayne, C., Fletcher, M., Eds.; Springer: Boston, MA, USA, 1985; pp. 133–161. ISBN 978-1-4615-6514-7. [Google Scholar]
- Pearce, D.; Bazin, M.J.; Lynch, J.M. The Rhizosphere as a Biofilm. In Microbial Biofilms; Lappin-Scott, H.M., Costerton, J.W., Eds.; Biotechnology Research; Cambridge University Press: Cambridge, UK, 1995; pp. 207–220. ISBN 9780521542128. [Google Scholar]
- An, Y.H.; Friedman, R.J. Concise Review of Mechanisms of Bacterial Adhesion to Biomaterial Surfaces. J. Biomed. Mater. Res. 1998, 43, 338–348. [Google Scholar] [CrossRef]
- An, Y.H.; Friedman, R.J. Handbook of Bacterial Adhesion: Principles, Methods, and Applications; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2000; Volume 204, ISBN 1592592244. [Google Scholar]
- Boland, T.; Latour, R.A.; Stutzenberger, F.J. Molecular Basis of Bacterial Adhesion. In Handbook of Bacterial Adhesion: Principles, Methods, and Applications; An, Y.H., Friedman, R.J., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 29–41. ISBN 978-1-59259-224-1. [Google Scholar]
- Dunne, W.M. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin. Microbiol. Rev. 2002, 15, 155–166. [Google Scholar] [CrossRef]
- Berne, C.; Ellison, C.K.; Ducret, A.; Brun, Y.V. Bacterial Adhesion at the Single-Cell Level. Nat. Rev. Microbiol. 2018, 16, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wei, C.; Cheng, H.; Chen, G. Adhesion of Colloids and Bacteria to Porous Media: A Critical Review. Rev. Adhes. Adhes. 2019, 7, 417–460. [Google Scholar] [CrossRef]
- Uzoma, P.C.; Etim, I.I.N.; Okonkwo, B.O.; Olanrele, O.S.; Njoku, D.I.; Kolawole, S.K.; Emori, W.; Ikeuba, A.I.; Njoku, C.N.; Ekerenam, O.O.; et al. Recent Design Approaches, Adhesion Mechanisms, and Applications of Antibacterial Surfaces. Chem. Eng. J. Adv. 2023, 16, 100563. [Google Scholar] [CrossRef]
- Ashok, D.; Cheeseman, S.; Wang, Y.; Funnell, B.; Leung, S.F.; Tricoli, A.; Nisbet, D. Superhydrophobic Surfaces to Combat Bacterial Surface Colonization. Adv. Mater. Interfaces 2023, 10, 2300324. [Google Scholar] [CrossRef]
- Bian, X.; Kim, C.; Karniadakis, G.E. 111 Years of Brownian Motion. Soft Matter 2016, 12, 6331–6346. [Google Scholar] [CrossRef]
- Adler, J. Chemotaxis in Bacteria. In Surface Membrane Receptors: Interface Between Cells and Their Environment; Bradshaw, R.A., Frazier, W.A., Merrell, R.C., Gottlieb, D.I., Hogue-Angeletti, R.A., Eds.; Springer: Boston, MA, USA, 1976; pp. 419–435. ISBN 978-1-4684-2772-1. [Google Scholar]
- Preedy, V.R. Adhesion Molecules; CRC Press: Boca Raton, FL, USA, 2016; ISBN 9781439840603. [Google Scholar]
- Kumar, A.S.; Mody, K.; Jha, B. Bacterial Exopolysaccharides—A Perception. J. Basic. Microbiol. 2007, 47, 103–117. [Google Scholar] [CrossRef]
- Absolom, D.R.; Lamberti, F.V.; Policova, Z.; Zingg, W.; Van Oss, C.J.; Wilhelm Neumann, A.A. Surface Thermodynamics of Bacterial Adhesion. Appl. Environ. Microbiol. 1983, 46, 90–97. [Google Scholar] [CrossRef]
- Van Oss, C.J.; Good, R.J.; Chaudhury, M.K. The Role of van Der Waals Forces and Hydrogen Bonds in “Hydrophobic Interactions” between Biopolymers and Low Energy Surfaces. J. Colloid Interface Sci. 1986, 111, 378–390. [Google Scholar] [CrossRef]
- Adamson, A.W. Physical Chemistry of Surfaces, 5th ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1990. [Google Scholar]
- Hermansson, M. The DLVO Theory in Microbial Adhesion. Colloids Surf. B Biointerfaces 1999, 14, 105–119. [Google Scholar] [CrossRef]
- Busscher, H.J.; Weerkamp, A.H.; Van, H.C.; Mei, D.; Van Pelt, A.W.J.; De Jong, H.P.; Arends, J. Measurement of the Surface Free Energy of Bacterial Cell Surfaces and Its Relevance for Adhesion. Appl. Environ. Microbiol. 1984, 48, 980–983. [Google Scholar] [CrossRef]
- Rutter, P.R.; Vincent, B. The Adhesion of microorganisms to Surfaces, Physico-Chemical Aspects; Berkeley, R.C.W., Lynch, J.M., Melling, J., Rutter, P.R., Vincent, B., Eds.; Ellis Horwood: London, UK, 1980. [Google Scholar]
- Katsikogianni, M.; Missirlis, Y.F.; Harris, L.; Douglas, J. Concise Review of Mechanisms of Bacterial Adhesion to Biomaterials and of Techniques Used in Estimating Bacteria-Material Interactions. Eur. Cell Mater. 2004, 8, 37–57. [Google Scholar] [CrossRef]
- Varshney, S.; Sain, A.; Gupta, D.; Sharma, S. Factors Affecting Bacterial Adhesion on Selected Textile Fibres. Indian. J. Microbiol. 2021, 61, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, H.; Wang, P.; Zhang, D.; Sun, Y.; Li, E. Oxygen Reduction Reaction Affected by Sulfate-Reducing Bacteria: Different Roles of Bacterial Cells and Metabolites. Indian. J. Microbiol. 2017, 57, 344–350. [Google Scholar] [CrossRef]
- Flath, H.; Saleh, N. Zur Bedeutung Des Zetapotentials von Faserstoffen Für Den Färbeprozeß. Acta Polym. 1980, 31, 510–517. [Google Scholar] [CrossRef]
- Jacobasch, H.; Baubock, G.; Schurz, J. Problems and Results of Zeta-Potential Measurements on Fibers. Colloid Polym. Sci. 1985, 263, 3–24. [Google Scholar] [CrossRef]
- Grancaric, A.M.; Tarbuk, A.; Pusic, T. Electrokinetic Properties of Textile Fabrics. Color. Technol. 2005, 121, 221–227. [Google Scholar] [CrossRef]
- Oh, J.K.; Yegin, Y.; Yang, F.; Zhang, M.; Li, J.; Huang, S.; Verkhoturov, S.V.; Schweikert, E.A.; Perez-Lewis, K.; Scholar, E.A.; et al. The Influence of Surface Chemistry on the Kinetics and Thermodynamics of Bacterial Adhesion. Sci. Rep. 2018, 8, 17247. [Google Scholar] [CrossRef]
- Ong, Y.L.; Razatos, A.; Georgiou, G.; Sharma, M.M. Adhesion Forces between E. c Oli Bacteria and Biomaterial Surfaces. Langmuir 1999, 15, 2719–2725. [Google Scholar] [CrossRef]
- Van Loosdrecht, M.C.M.; Lyklema, J.; Norde, W.; Schraa, G.; Zehnder1, A.J.B. Electrophoretic Mobility and Hydrophobicity as a Measure To Predict the Initial Steps of Bacterial Adhesion. Appl. Enviorn. Microbiol. 1987, 53, 1898–1901. [Google Scholar] [CrossRef]
- van Loosdrecht, M.C.M.; Lyklema, J.; Norde, W.; Zehnder, A.J.B. Bacterial Adhesion: A Physicochemical Approach. Microb. Ecol. 1989, 17, 1–15. [Google Scholar] [CrossRef]
- Faraudo, J.; Andreu, J.S.; Camacho, J. Understanding Diluted Dispersions of Superparamagnetic Particles under Strong Magnetic Fields: A Review of Concepts, Theory and Simulations. Soft Matter 2013, 9, 6654–6664. [Google Scholar] [CrossRef]
- Alam, F.; Kumar, S.; Varadarajan, K.M. Quantification of Adhesion Force of Bacteria on the Surface of Biomaterials: Techniques and Assays. ACS Biomater. Sci. Eng. 2019, 5, 2093–2110. [Google Scholar] [CrossRef]
- Carniello, V.; Peterson, B.W.; van der Mei, H.C.; Busscher, H.J. Physico-Chemistry from Initial Bacterial Adhesion to Surface-Programmed Biofilm Growth. Adv. Colloid Interface Sci. 2018, 261, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M. Effects of Electrolytes on Attachment of Aquatic Bacteria to Solid Surfaces. Estuaries 1988, 11, 226–230. [Google Scholar] [CrossRef]
- Nguyen, V.; Karunakaran, E.; Collins, G.; Biggs, C.A. Physicochemical Analysis of Initial Adhesion and Biofilm Formation of Methanosarcina Barkeri on Polymer Support Material. Colloids Surf. B Biointerfaces 2016, 143, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.; Flint, S.; Brooks, J. Bacterial Cell Attachment, the Beginning of a Biofilm. J. Ind. Microbiol. Biotechnol. 2007, 34, 577–588. [Google Scholar] [CrossRef]
- Pereni, C.I.; Zhao, Q.; Liu, Y.; Abel, E. Surface Free Energy Effect on Bacterial Retention. Colloids Surf. B Biointerfaces 2006, 48, 143–147. [Google Scholar] [CrossRef]
- Jan Van Oss, C. Use of the Combined Lifshitz-Van Der Waals and Lewis Acid-Base Approaches in Determining the Apolar and Polar Contributions to Surface and Interfacial Tensions and Free Energies. J. Adhes. Sci. Technol. 2002, 16, 669–677. [Google Scholar] [CrossRef]
- Clint, J.H. Adhesion and Components of Solid Surface Energies. Curr. Opin. Colloid Interface Sci. 2001, 6, 28–33. [Google Scholar] [CrossRef]
- Morra, M.; Cassinelli, C. Bacterial Adhesion to Polymer Surfaces: A Critical Review of Surface Thermodynamic Approaches. J. Biomater. Sci. 1998, 9, 55–74. [Google Scholar] [CrossRef]
- Owens, D.K.; Wendt, R.C. Estimation of the Surface Free Energy of Polymers. J. Appl. Polym. Sci. 1969, 13, 1741–1747. [Google Scholar] [CrossRef]
- Fowkes, F.M. Determination of Interfacial Tensions, Contact Angles, and Dispersion Forces in Surfaces by Assuming Additivity of Intermolecular Interactions in Surfaces. J. Phys. Chem. 1962, 66, 382. [Google Scholar] [CrossRef]
- Fowkes, F.M. Attractive Forces at Interfaces. Ind. Eng. Chem. 1964, 56, 40–52. [Google Scholar] [CrossRef]
- Fowkes, F.M. Interface Acid-Base/Charge-Transfer Properties. In Surface and Interfacial Aspects of Biomedical Polymers: Volume 1 Surface Chemistry and Physics; Andrade, J.D., Ed.; Springer: Boston, MA, USA, 1985; pp. 337–372. ISBN 978-1-4684-8610-0. [Google Scholar]
- Van Oss, C.J.; Ju, L.; Chaudhury, M.K.; Good, R.J. Estimation of the Polar Parameters of the Surface Tension of Liquids by Contact Angle Measurements on Gels. J. Colloid Interface Sci. 1989, 128, 313–319. [Google Scholar] [CrossRef]
- Van Oss, C.J. Long-Range and Short-Range Mechanisms of Hydrophobic Attraction and Hydrophilic Repulsion in Specific and Aspecific Interactions. J. Mol. Recognit. 2003, 16, 177–190. [Google Scholar] [CrossRef]
- van Oss, C.J.; Chaudhury, M.K.; Good, R.J. Monopolar Surfaces. Adv. Colloid Interface Sci. 1987, 28, 35–64. [Google Scholar] [CrossRef]
- van Oss, C.J. Development and Applications of the Interfacial Tension between Water and Organic or Biological Surfaces. Colloids Surf. B Biointerfaces 2007, 54, 2–9. [Google Scholar] [CrossRef]
- Chen, H.; Li, L.; Jones, P.M.; Hsia, Y.T.; Jhon, M.S. Lifshitz-van Der Waals and Lewis Acid-Base Approach for Analyzing Surface Energy of Molecularly Thin Lubricant Films. IEEE Trans. Magn. 2007, 43, 2226–2228. [Google Scholar] [CrossRef]
- Van Oss, C.J. Hydrophobic, Hydrophilic and Other Interactions in Epitope-Paratope Binding. Mol. Immunol. 1995, 32, 199–211. [Google Scholar] [CrossRef]
- Bayoudh, S.; Othmane, A.; Mora, L.; Ben Ouada, H. Assessing Bacterial Adhesion Using DLVO and XDLVO Theories and the Jet Impingement Technique. Colloids Surf. B Biointerfaces 2009, 73, 1–9. [Google Scholar] [CrossRef]
- Chen, G.; Flury, M. Retention of Mineral Colloids in Unsaturated Porous Media as Related to Their Surface Properties. Colloids Surf. A Physicochem. Eng. Asp. 2005, 256, 207–216. [Google Scholar] [CrossRef]
- van Oss, C.J. Interfacial Forces in Aqueous Media; Taylor & Francis: Boca Raton, FL, USA, 1994; ISBN 9780824791681. [Google Scholar]
- ISO 6938; Textiles-Natural Fibres-Generic Names and Definitions Textiles-Fibres Naturelles-Noms Génériques et Définitions. International Organization for Standardization: Geneva, Switzerland, 2012.
- ISO 2076; Textiles-Man-Made Fibres-Generic Names. International Organization for Standardization: Geneva, Switzerland, 2021.
- Møllebjerg, A.; Palmén, L.G.; Gori, K.; Meyer, R.L. The Bacterial Life Cycle in Textiles is Governed by Fiber Hydrophobicity. Microbiol. Spectr. 2021, 9, e0118521. [Google Scholar] [CrossRef]
- Roy, S.; Ghosh, S.; Bhowmick, N.; Choudhury, P. Role of Physicochemical Factors on Bacterial Attachment in Textile Fibrous Media. Water Environ. J. 2018, 33, 21–30. [Google Scholar] [CrossRef]
- Rebenfeld, L. Fibers. Encyclopedia of Polymer Science and Engineering; Wiley-Interscience: Hoboken, NJ, USA, 1986; Volume 6, pp. 647–733. [Google Scholar]
- Çaykara, T.; Sande, M.G.; Azoia, N.; Rodrigues, L.R.; Silva, C.J. Exploring the Potential of Polyethylene Terephthalate in the Design of Antibacterial Surfaces. Med. Microbiol. Immunol. 2020, 209, 363–372. [Google Scholar] [CrossRef]
- Suzawa, T. Electrical Phenomena at Interfaces; Marcel Dekker, INC: New York, NY, USA, 1984; p. 299. [Google Scholar]
- Matsumoto, S.; Ohtaki, A.; Hori, K. Carbon Fiber as an Excellent Support Material for Wastewater Treatment Biofilms. Environ. Sci. Technol. 2012, 46, 10175–10181. [Google Scholar] [CrossRef]
- Wu, S.; Altenried, S.; Zogg, A.; Zuber, F.; Maniura-Weber, K.; Ren, Q. Role of the Surface Nanoscale Roughness of Stainless Steel on Bacterial Adhesion and Microcolony Formation. ACS Omega 2018, 3, 6456–6464. [Google Scholar] [CrossRef]
- Koziarz, J. Adhesion and Immobilization of Bacteria on Hydrophobic Cloths. Master’s Thesis, Ottawa-Carleton Institute of Biology, Ottawa, ON, Canada, 1998. [Google Scholar]
- Takashima, M.; Shirai, F.; Sageshima, M.; Ikeda, N.; Okamoto, Y.; Dohi, Y. Distinctive Bacteria-Binding Property of Cloth Materials. Am. J. Infect. Control 2004, 32, 27–30. [Google Scholar] [CrossRef]
- Gu, P.; Fan, N.; Wang, Y.; Wang, J.; Müller-Buschbaum, P.; Zhong, Q. Linear Control of Moisture Permeability and Anti-Adhesion of Bacteria in a Broad Temperature Region Realized by Cross-Linking Thermoresponsive Microgels onto Cotton Fabrics. ACS Appl. Mater. Interfaces 2019, 11, 30269–30277. [Google Scholar] [CrossRef]
- Rochex, A.; Lecouturier, D.; Pezron, I.; Lebeault, J.M. Adhesion of a Pseudomonas Putida Strain Isolated from a Paper Machine to Cellulose Fibres. Appl. Microbiol. Biotechnol. 2004, 65, 727–733. [Google Scholar] [CrossRef]
- Bajpai, V.; Dey, A.; Ghosh, S.; Bajpai, S.; Jha, M.K. Quantification of Bacterial Adherence on Different Textile Fabrics. Int. Biodeterior. Biodegrad. 2011, 65, 1169–1174. [Google Scholar] [CrossRef]
- Bajpai, S.; Bajpai, V.; Dey, A.; Ghosh, S.; Jha, M.K. Study of Adherence Kinetics of Escherichia Coli on Cotton Knitted Fabrics. Indian. Chem. Eng. 2019, 61, 296–308. [Google Scholar] [CrossRef]
- Hemmatian, T.; Lee, H.; Kim, J. Bacteria Adhesion of Textiles Influenced by Wettability and Pore Characteristics of Fibrous Substrates. Polymers 2021, 13, 223. [Google Scholar] [CrossRef]
- Wang, H.; Newby, B.Z. Applicability of the Extended Derjaguin–Landau–Verwey–Overbeek Theory on the Adsorption of Bovine Serum Albumin on Solid Surfaces. Biointerphases 2014, 9, 041006. [Google Scholar] [CrossRef]
- Yuan, Y.; Hays, M.P.; Hardwidge, P.R.; Kim, J. Surface Characteristics Influencing Bacterial Adhesion to Polymeric Substrates. RSC Adv. 2017, 7, 14254–14261. [Google Scholar] [CrossRef]
- Jiang, Y.; Yin, Y.J.; Zha, X.C.; Dou, X.Q.; Feng, C.L. Wettability Regulated Gram-Negative Bacterial Adhesion on Biomimetic Hierarchical Structures. Chin. Chem. Lett. 2017, 28, 813–817. [Google Scholar] [CrossRef]
- Roy, S.; Ghosh, S.; Bhowmick, N. Mechanism of Bacterial Attachment on Textile Fibrous Media. J. Text. Inst. 2019, 110, 916–923. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Xi, H.; Sun, J.; Liang, X.; Wei, J.; Xiao, X.; Liu, Z.; Li, S.; Liang, Z.; et al. Interpretation of Adhesion Behaviors between Bacteria and Modified Basalt Fiber by Surface Thermodynamics and Extended DLVO Theory. Colloids Surf. B Biointerfaces 2019, 177, 454–461. [Google Scholar] [CrossRef]
- Van Oss, C.; Giese, R.; Li, Z.; Murphy, K.; Norris, J.; Chaudhury, M.; Good, R. Determination of Contact Angles and Pore Sizes of Porous Media by Column and Thin Layer Wicking. J. Adhes. Sci. Technol. 1992, 6, 413–428. [Google Scholar] [CrossRef]
- Chibowski, E.; González-Caballero, F. Theory and Practice of Thin-Layer Wicking. Langmuir 1993, 9, 330–340. [Google Scholar] [CrossRef]
- Chibowski, E.; Holysz, L. Use of the Washburn Equation for Surface Free Energy Determination. Langmuir 1992, 8, 710–716. [Google Scholar] [CrossRef]
- Pezron, I.; Rochex, A.; Lebeault, J.M.; Clausse, D. Determination of Cellulose Surface Energy by Imbibition Experiments in Relation to Bacterial Adhesion. J. Dispers. Sci. Technol. 2004, 25, 781–787. [Google Scholar] [CrossRef]
- Grancarić, A.M.; Tarbuk, A.; Hadžić, S.; Simončič, B. From Raw to Finished Cotton—Characterization by Interface Phenomena. Autex Res. J. 2023, 23, 184–192. [Google Scholar] [CrossRef]
- Tian, G.; Chen, B.; Qi, S.; Niu, H.; Han, E.; Wu, D. Enhanced Surface Free Energy of Polyimide Fibers by Alkali Treatment and Its Interfacial Adhesion Behavior to Epoxy Resins. Compos. Interfaces 2016, 23, 145–155. [Google Scholar] [CrossRef]
- Lewin, M.; Sello, S.B. (Eds.) Handbook of Fiber Science and Technology: Volume II: Chemical Processing of Fibers and Fabrics. Functional Finishes: Part B; Marcel Dekker, Inc.: New York, NY, USA; Basel, Switzerland, 1984. [Google Scholar]
- Zhang, X.; Zhang, Q.; Yan, T.; Jiang, Z.; Zhang, X.; Zuo, Y.Y. Quantitatively Predicting Bacterial Adhesion Using Surface Free Energy Determined with a Spectrophotometric Method. Environ. Sci. Technol. 2015, 49, 6164–6171. [Google Scholar] [CrossRef]
- Fletcher, M.; Pringle, J.H. The Effect of Surface Free Energy and Medium Surface Tension on Bacterial Attachment to Solid Surfaces. J. Colloid Interface Sci. 1985, 104, 5–14. [Google Scholar] [CrossRef]
- Vilčnik, A.; Jerman, I.; Vuk, A.Š.; Koželj, M.; Orel, B.; Tomšič, B.; Simončič, B.; Kovač, J. Structural Properties and Antibacterial Effects of Hydrophobic and Oleophobic Sol-Gel Coatings for Cotton Fabrics. Langmuir 2009, 25, 5869–5880. [Google Scholar] [CrossRef]
- Tomšič, B.; Simončič, B.; Orel, B.; Černe, L.; Tavčer, P.F.; Zorko, M.; Jerman, I.; Vilčnik, A.; Kovač, J. Sol-Gel Coating of Cellulose Fibres with Antimicrobial and Repellent Properties. J. Solgel Sci. Technol. 2008, 47, 44–57. [Google Scholar] [CrossRef]
- Swar, S.; Máková, V.; Stibor, I. The Covalent Tethering of Poly(Ethylene Glycol) to Nylon 6 Surface via N,N′-Disuccinimidyl Carbonate Conjugation: A New Approach in the Fight against Pathogenic Bacteria. Polymers 2020, 12, 2181. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Baggerman, J.; Paulusse, J.M.J.; Van Rijn, C.J.M.; Zuilhof, H. Stable Protein-Repellent Zwitterionic Polymer Brushes Grafted from Silicon Nitride. Langmuir 2011, 27, 2587–2594. [Google Scholar] [CrossRef]
- Zhang, X.; Brodus, D.S.; Hollimon, V.; Hu, H. A Brief Review of Recent Developments in the Designs That Prevent Bio-Fouling on Silicon and Silicon-Based Materials. Chem. Cent. J. 2017, 11, 1. [Google Scholar] [CrossRef]
- Hsieh, Y.L.; Timm, D.A.; Merry, J. Bacterial Adherence on Fabrics by a Radioisotope Labeling Method. Text. Res. J. 1987, 57, 20–28. [Google Scholar] [CrossRef]
- Baier, R. Substrata Influences on the Adhesion of Microorganisms and Their Resultant New Surface Properties. In Adsorption of Micro-Organisms to Surface; Bitton, G., Marshall, K., Eds.; Wiley Interscience: New York, NY, USA, 1980; pp. 59–104. [Google Scholar]
- Becker, K. Detachment Studies on Microfouling in Natural Biofilms on Substrata with Different Surface Tensions. Int. Biodeterior. Biodegrad. 1998, 41, 93–100. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, S.; Müller-Steinhagen, H. Tailored Surface Free Energy of Membrane Diffusers to Minimize Microbial Adhesion. Appl. Surf. Sci. 2004, 230, 371–378. [Google Scholar] [CrossRef]
- Salta, M.; Wharton, J.A.; Stoodley, P.; Dennington, S.P.; Goodes, L.R.; Werwinski, S.; Mart, U.; Wood, R.J.K.; Stokes, K.R. Designing Biomimetic Antifouling Surfaces. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2010, 368, 4729–4754. [Google Scholar] [CrossRef]
- Baier, R.E.; DePalma, V.A.; Dale, W.A. Management of Occlusive Arterial Disease; Yearbook Medical, Chicago, IL, USA, 1971; p. 147.
- Su, X.J.; Zhao, Q.; Wang, S.; Bendavid, A. Modification of Diamond-like Carbon Coatings with Fluorine to Reduce Biofouling Adhesion. Surf. Coat. Technol. 2010, 204, 2454–2458. [Google Scholar] [CrossRef]
- Bayoudh, S.; Ponsonnet, L.; Ben Ouada, H.; Bakhrouf, A.; Othmane, A. Bacterial Detachment from Hydrophilic and Hydrophobic Surfaces Using a Microjet Impingement. Colloids Surf. A Physicochem. Eng. Asp. 2005, 266, 160–167. [Google Scholar] [CrossRef]
- Dobnik Dubrovski, P. Konstrukcija Tekstilij; Univerza v Mariboru, Fakulteta za strojništvo, Oddelek za tekstilne materiale in oblikovanje: Maribor, Slovenia, 2007. [Google Scholar]
- Ho, C.P.; Fan, J.; Newton, E.; Au, R. 7—Improving Thermal Comfort in Apparel. In Improving Comfort in Clothing; Song, G., Ed.; Woodhead Publishing Series in Textiles; Woodhead Publishing: Sawston, UK, 2011; pp. 165–181. ISBN 978-1-84569-539-2. [Google Scholar]
- Burleigh, E.G., Jr.; Wakeham, H.; Honold, E.; Skau, E.L. Pore-Size Distribution in Textiles. Text. Res. J. 1949, 19, 547–555. [Google Scholar] [CrossRef]
- Biglary, F.; Dashtizadeh, M. Surface Metrology and Contact Roughness Measurement; Amirkabir University of Technology: Tehran, Iran, 2005. [Google Scholar]
- Militky, J.; Bajzik, V. Characterization of Protective Clothing Surface Roughness. In Proceedings of the 2nd International Congress on Technical textiles, Greenville, SC, USA, 21 July 2004. [Google Scholar]
- Mooneghi, S.A.; Saharkhiz, S.; Mohammad, S.; Varkiani, H. Surface Roughness Evaluation of Textile Fabrics: A Literature Review. J. Eng. Fibers Fabr. 2014, 9, 155892501400900201. [Google Scholar] [CrossRef]
- Mao, N.; Wang, Y.; Qu, J. Smoothness and Roughness: Characteristics of Fabric-to-Fabric Self-Friction Properties. In Proceedings of the 90th Textile Institute World Conference, Poznan, Polan, 25–28 April 2016. [Google Scholar]
- Whitehouse, D. Surfaces and Their Measurement; Butterworth-Heinemann: Boston, MA, USA, 2012. [Google Scholar]
- Ajayi, J.O. Elder HM Effects of Surface Geometry on Fabric Friction. J. Test. Eval. 1997, 25, 182–188. [Google Scholar] [CrossRef]
- Akgun, M. Effect of Yarn Filament Fineness on the Surface Roughness of Polyester Woven Fabrics. J. Eng. Fibers Fabr. 2015, 10, 155892501501000214. [Google Scholar] [CrossRef]
- Banerjee, P.K. Principles of Fabric Formation; Taylor & Francis: Abingdon, UK, 2014; ISBN 9781466554443. [Google Scholar]
- Bos, R.; Mei, H.C.; Gold, J.; Busscher, H.J. Retention of Bacteria on a Substratum Surface with Micro-Patterned Hydrophobicity. FEMS Microbiol. Lett. 2000, 189, 311–315. [Google Scholar] [CrossRef]
- Scheuerman, T.R.; Camper, A.K.; Hamilton, M.A. Effects of Substratum Topography on Bacterial Adhesion. J. Colloid Interface Sci. 1998, 208, 23–33. [Google Scholar] [CrossRef]
- Medilanski, E.; Kaufmann, K.; Wick, L.Y.; Wanner, O.; Harms, H. Influence of the Surface Topography of Stainless Steel on Bacterial Adhesion. Biofouling 2002, 18, 193–203. [Google Scholar] [CrossRef]
- Edwards, K.J.; Rutenberg, A.D. Microbial Response to Surface Microtopography: The Role of Metabolism in Localized Mineral Dissolution. Chem. Geol. 2001, 180, 19–32. [Google Scholar] [CrossRef]
- Hong, H.R.; Kim, J. Nanoroughness-Mediated Bacterial Adhesion on Fabrics. ACS Appl. Nano Mater. 2023, 6, 18518–18530. [Google Scholar] [CrossRef]
- Roach, P.; Farrar, D.; Perry, C.C. Surface Tailoring for Controlled Protein Adsorption: Effect of Topography at the Nanometer Scale and Chemistry. J. Am. Chem. Soc. 2006, 128, 3939–3945. [Google Scholar] [CrossRef] [PubMed]
- Shiu, J.Y.; Chen, P. Addressable Protein Patterning via Switchable Superhydrophobic Microarrays. Adv. Funct. Mater. 2007, 17, 2680–2686. [Google Scholar] [CrossRef]
- Caykara, T.; Fernandes, S.; Braga, A.; Rodrigues, J.; Rodrigues, L.R.; Silva, C.J. Can Superhydrophobic PET Surfaces Prevent Bacterial Adhesion? Nanomaterials 2023, 13, 1117. [Google Scholar] [CrossRef]
- Boyd, R.D.; Verran, J.; Jones, M.V.; Bhakoo, M. Use of the Atomic Force Microscope to Determine the Effect of Substratum Surface Topography on Bacterial Adhesion. Langmuir 2002, 18, 2343–2346. [Google Scholar] [CrossRef]
- Craighead, H.G.; James, C.D.; Turner, A.M.P. Chemical and Topographical Patterning for Directed Cell Attachment. Curr. Opin. Solid. State Mater. Sci. 2001, 5, 177–184. [Google Scholar] [CrossRef]
- Tang, H.; Cao, T.; Liang, X.; Wang, A.; Salley, S.O.; McAllister, J.; Ng, K.Y.S. Influence of Silicone Surface Roughness and Hydrophobicity on Adhesion and Colonization of Staphylococcus Epidermidis. J. Biomed. Mater. Res. A 2009, 88, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Hizal, F.; Rungraeng, N.; Jun, S.; Choi, C.-H. Nano-Engineered Alumina Surfaces for Prevention of Bacteria Adhesions. In Proceedings of the 9th IEEE International Conference on Nano/Micro Engineered and Molecular Systems (NEMS), Waikiki Beach, HI, USA, 13–16 April 2014; pp. 17–22. [Google Scholar]
- Perera-Costa, D.; Bruque, J.M.; González-Martín, M.L.; Gómez-García, A.C.; Vadillo-Rodríguez, V. Studying the Influence of Surface Topography on Bacterial Adhesion Using Spatially Organized Microtopographic Surface Patterns. Langmuir 2014, 30, 4633–4641. [Google Scholar] [CrossRef]
- Vadillo-Rodríguez, V.; Guerra-García-Mora, A.I.; Perera-Costa, D.; Gónzalez-Martín, M.L.; Fernández-Calderón, M.C. Bacterial Response to Spatially Organized Microtopographic Surface Patterns with Nanometer Scale Roughness. Colloids Surf. B Biointerfaces 2018, 169, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Gao, Y.; Jin, T.; Luo, X.; Zeng, Q.; Shang, Z. Effects of Surface Roughness and Texture on the Bacterial Adhesion on the Bearing Surface of Bio-Ceramic Joint Implants: An In Vitro Study. Ceram. Int. 2020, 46, 6550–6559. [Google Scholar] [CrossRef]
- Gao, Q.; Li, J.; Ding, C.; Wang, J.; Chen, Z.; Yang, X. In Situ Real-Time Investigation of Staphylococcus Aureus on Hemisphere-Patterned Polyurethane Films. Colloids Surf. B Biointerfaces 2022, 216, 112577. [Google Scholar] [CrossRef]
- Hočevar, M.; Jenko, M.; Godec, M.; Drobne, D. An Overview of the Influence of Stainless-Steel Surface Properties on Bacterial Adhesion. Mater. Tehnol. 2014, 48, 609–617. [Google Scholar]
- Fink, R.; Okanovič, D.; Dražič, G.; Abram, A.; Oder, M.; Jevšnik, M.; Bohinc, K. Bacterial Adhesion Capacity on Food Service Contact Surfaces. Int. J. Environ. Health Res. 2017, 27, 169–178. [Google Scholar] [CrossRef]
- Liu, Q.; Li, R.; Qu, W.; Tian, X.; Zhang, Y.; Wang, W. Influence of Surface Properties on the Adhesion of Bacteria onto Different Casings. Food Res. Int. 2023, 164, 112463. [Google Scholar] [CrossRef]
- Chung, K.K.; Schumacher, J.F.; Sampson, E.M.; Burne, R.A.; Antonelli, P.J.; Brennan, A.B. Impact of Engineered Surface Microtopography on Biofilm Formation of Staphylococcus Aureus. Biointerphases 2007, 2, 89–94. [Google Scholar] [CrossRef]
- Reddy, S.T.; Chung, K.K.; McDaniel, C.J.; Darouiche, R.O.; Landman, J.; Brennan, A.B. Micropatterned Surfaces for Reducing the Risk of Catheter-Associated Urinary Tract Infection: An In Vitro Study on the Effect of Sharklet Micropatterned Surfaces to Inhibit Bacterial Colonization and Migration of Uropathogenic Escherichia Coli. J. Endourol. 2011, 25, 1547–1552. [Google Scholar] [CrossRef]
- Xu, L.C.; Siedlecki, C.A. Staphylococcus Epidermidis Adhesion on Hydrophobic and Hydrophilic Textured Biomaterial Surfaces. Biomed. Mater. 2014, 9, 035003. [Google Scholar] [CrossRef]
- Vasudevan, R.; Kennedy, A.J.; Merritt, M.; Crocker, F.H.; Baney, R.H. Microscale Patterned Surfaces Reduce Bacterial Fouling-Microscopic and Theoretical Analysis. Colloids Surf. B Biointerfaces 2014, 117, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Leng, Y.; Lu, X.; Ren, F.; Wang, K.; Ding, Y.; Yang, M. Bacterial Responses to Periodic Micropillar Array. J. Biomed. Mater. Res. A 2015, 103, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Tang, P.; Zhang, W.; Wang, Y.; Zhang, B.; Wang, H.; Zhang, L. Effect of Superhydrophobic Surface of Titanium on Staphylococcus Aureus Adhesion. J. Nanomater. 2011, 2011, 1–8. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Levänen, E. Superhydrophobic Surfaces for the Reduction of Bacterial Adhesion. RSC Adv. 2013, 3, 12003–12020. [Google Scholar] [CrossRef]
- Oh, J.K.; Lu, X.; Min, Y.; Cisneros-Zevallos, L.; Akbulut, M. Bacterially Antiadhesive, Optically Transparent Surfaces Inspired from Rice Leaves. ACS Appl. Mater. Interfaces 2015, 7, 19274–19281. [Google Scholar] [CrossRef]
- Falde, E.J.; Yohe, S.T.; Colson, Y.L.; Grinstaff, M.W. Superhydrophobic Materials for Biomedical Applications. Biomaterials 2016, 104, 87–103. [Google Scholar] [CrossRef]
- Yang, K.; Shi, J.; Wang, L.; Chen, Y.; Liang, C.; Yang, L.; Wang, L.N. Bacterial Anti-Adhesion Surface Design: Surface Patterning, Roughness and Wettability: A Review. J. Mater. Sci. Technol. 2022, 99, 82–100. [Google Scholar] [CrossRef]
- Fink, R.; Oder, M.; Rangus, D.; Raspor, P.; Bohinc, K. Microbial Adhesion Capacity. Influence of Shear and Temperature Stress. Int. J. Environ. Health Res. 2015, 25, 656–669. [Google Scholar] [CrossRef]
- Tamayo, L.; Melo, F.; Caballero, L.; Hamm, E.; Díaz, M.; Leal, M.S.; Guiliani, N.; Urzúa, M.D. Does Bacterial Elasticity Affect Adhesion to Polymer Fibers? ACS Appl. Mater. Interfaces 2020, 12, 14507–14517. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of Surface Properties, Bacterial Motility, and Hydrodynamic Conditions on Bacterial Surface Sensing and Their Initial Adhesion. Front. Bioeng. Biotechnol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]


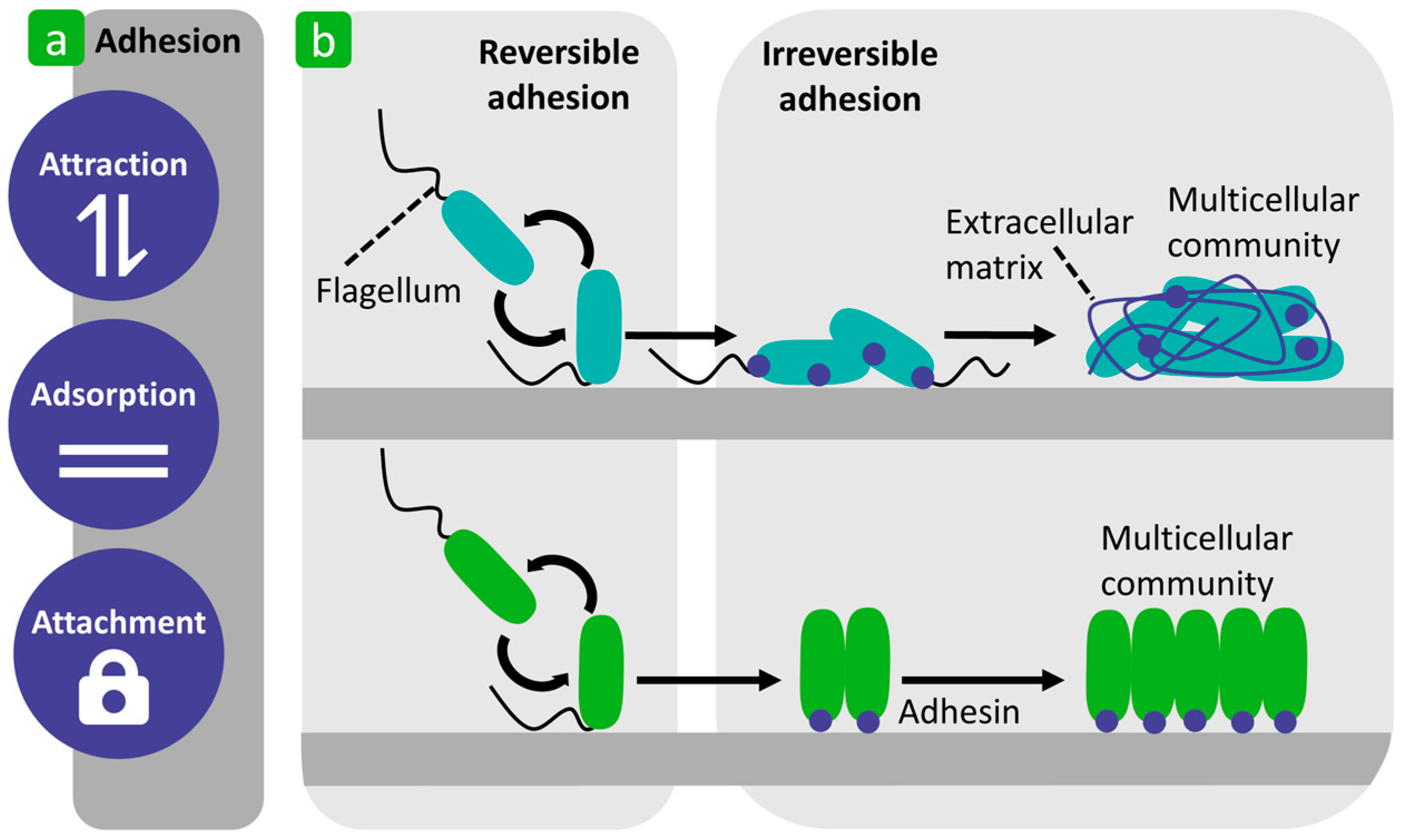
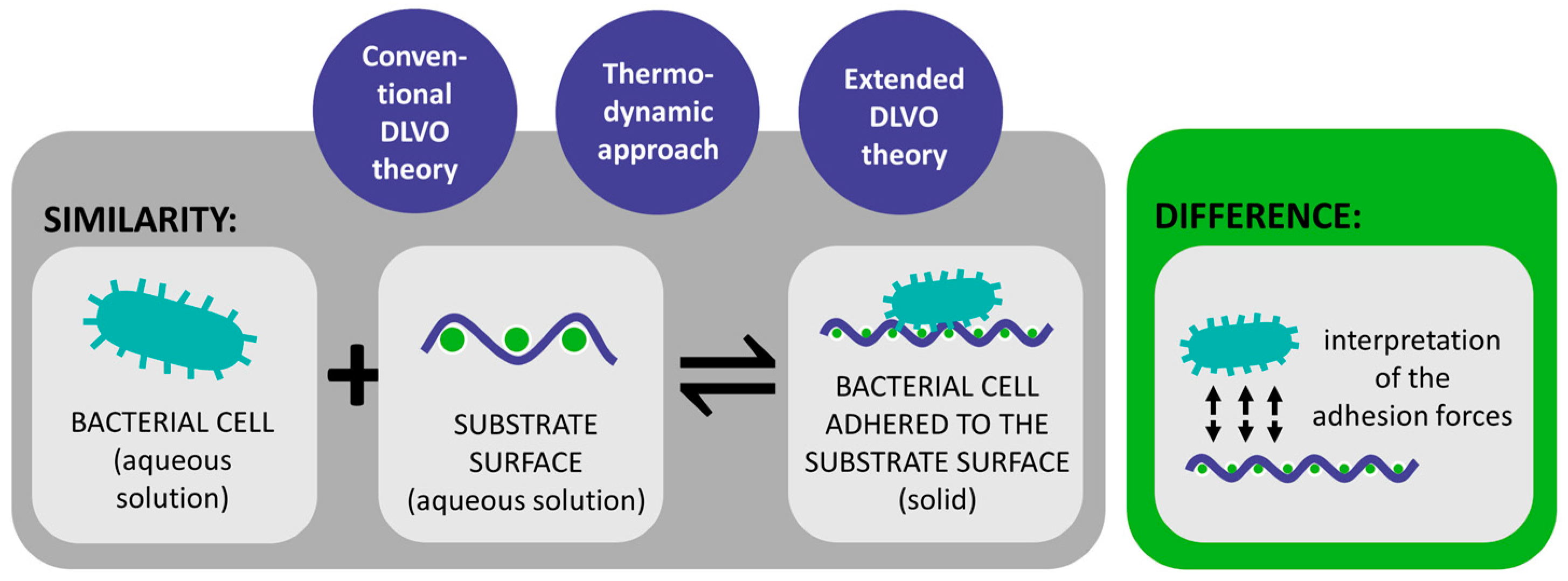


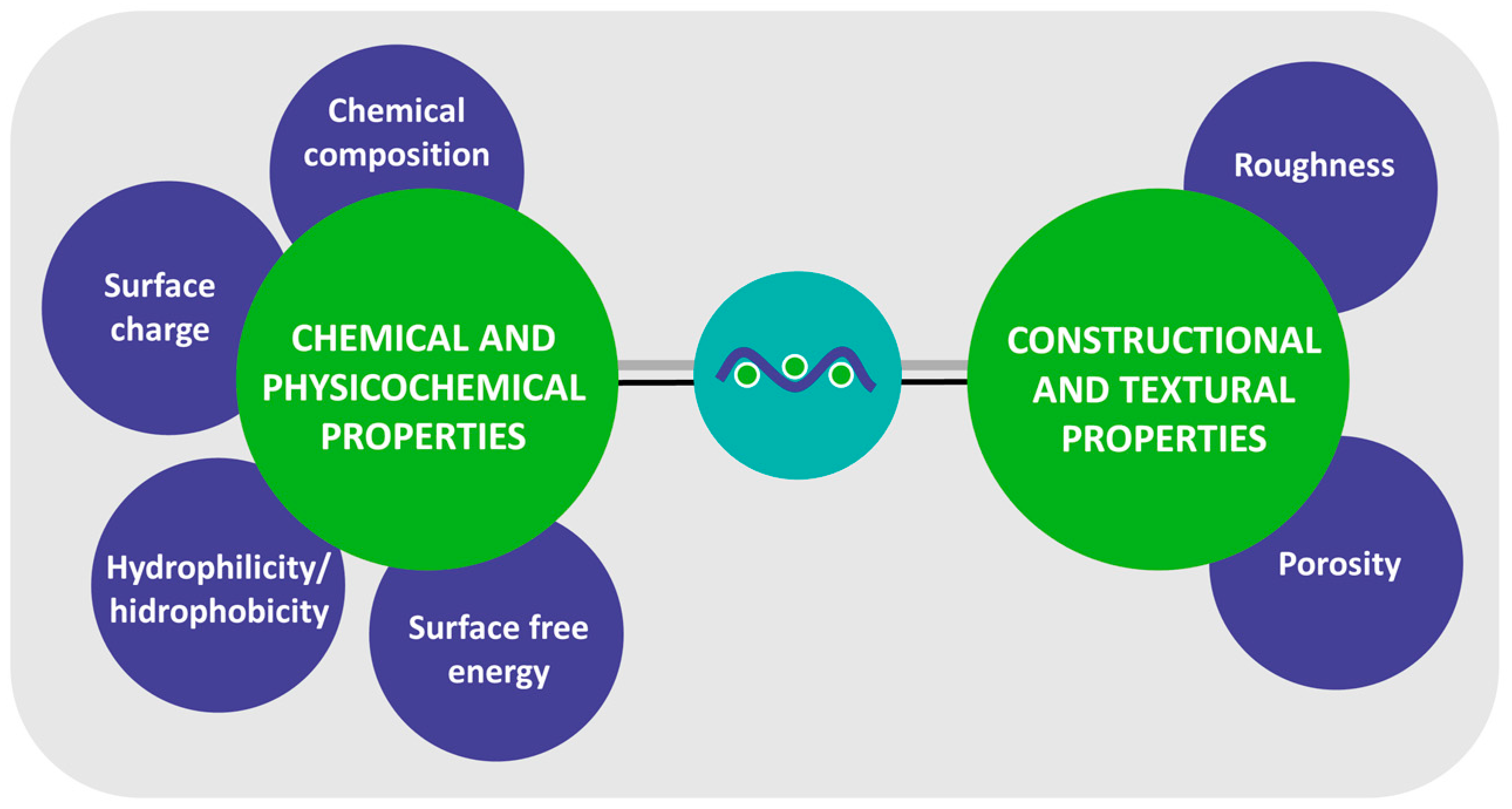
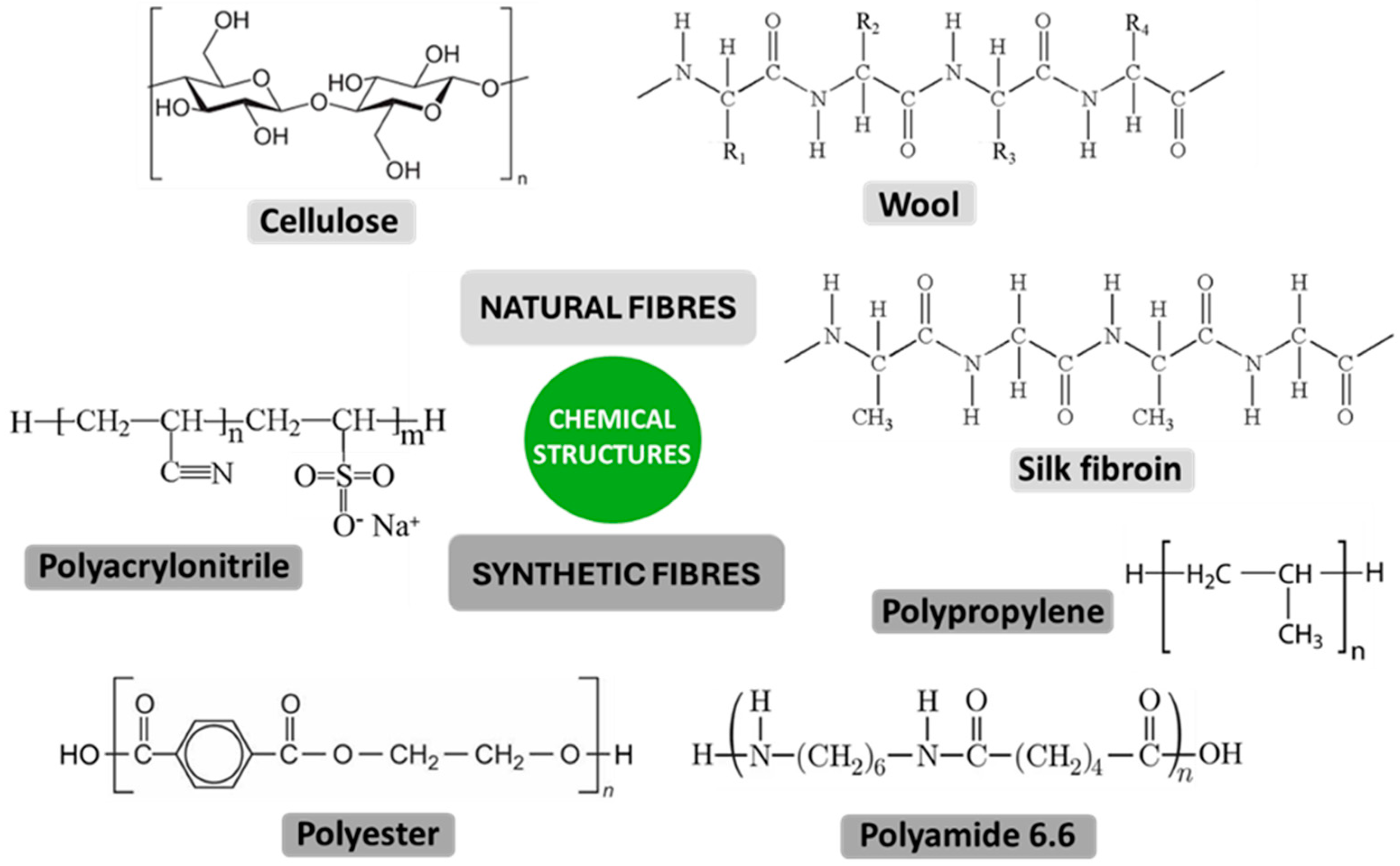

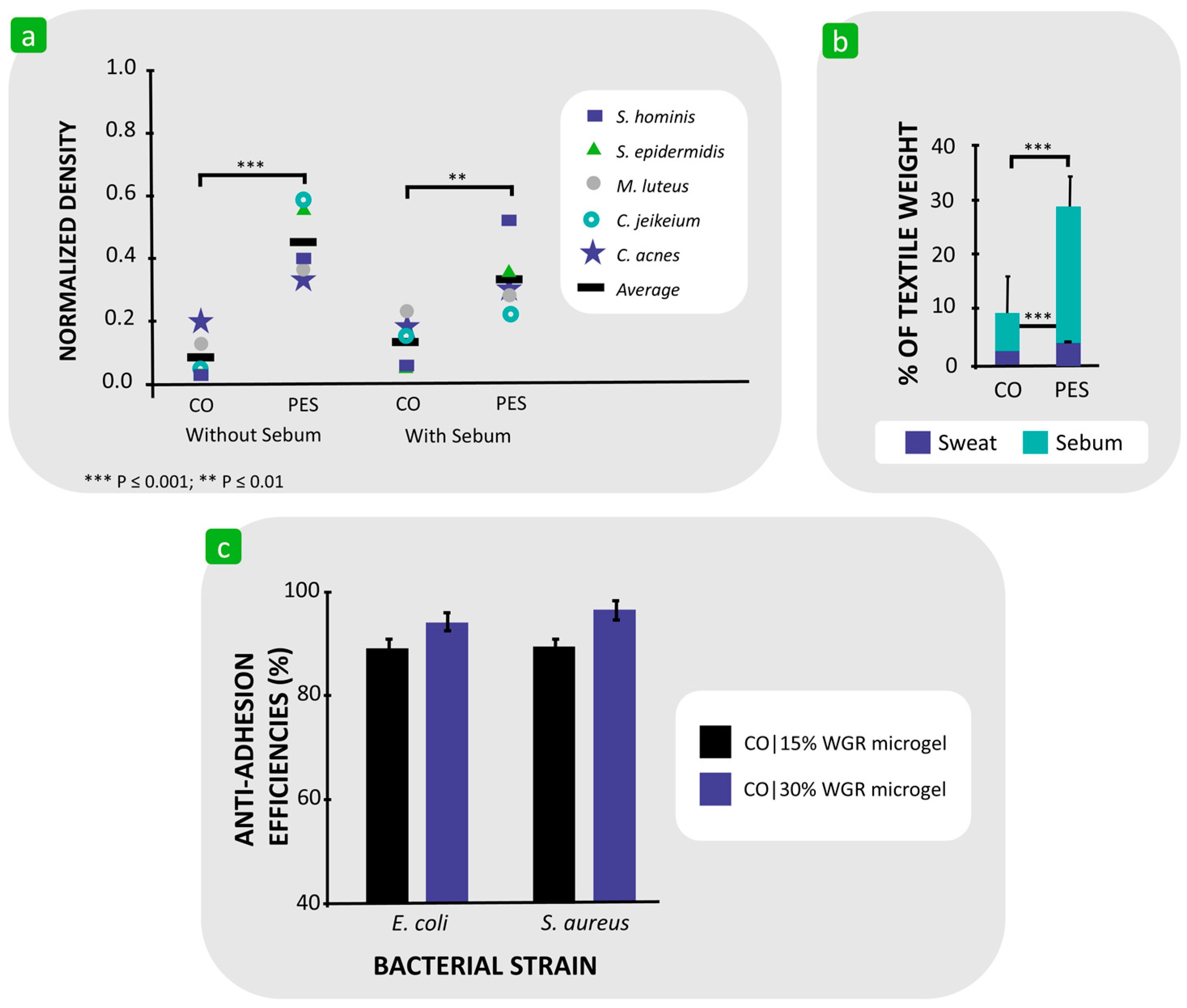

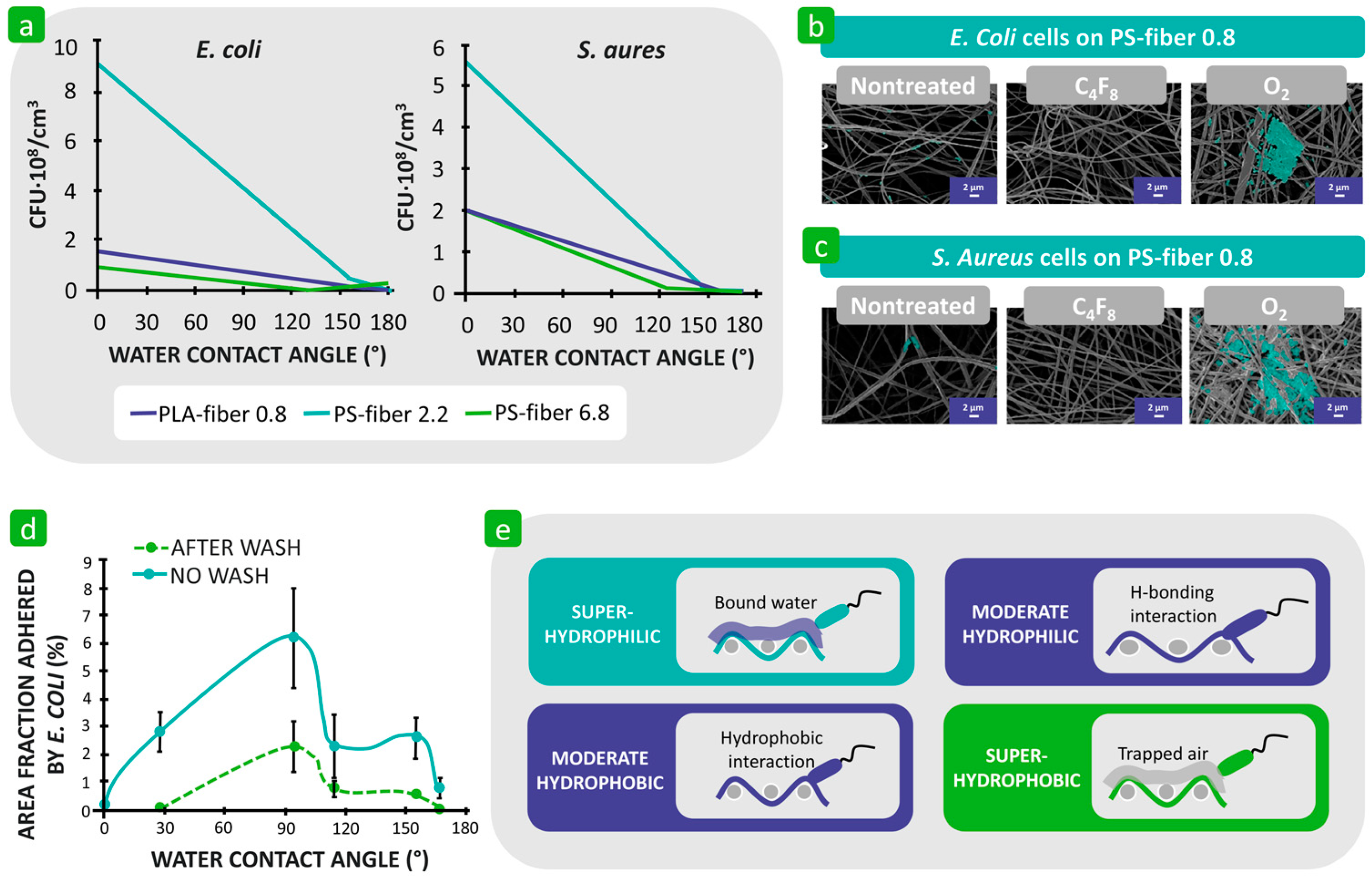
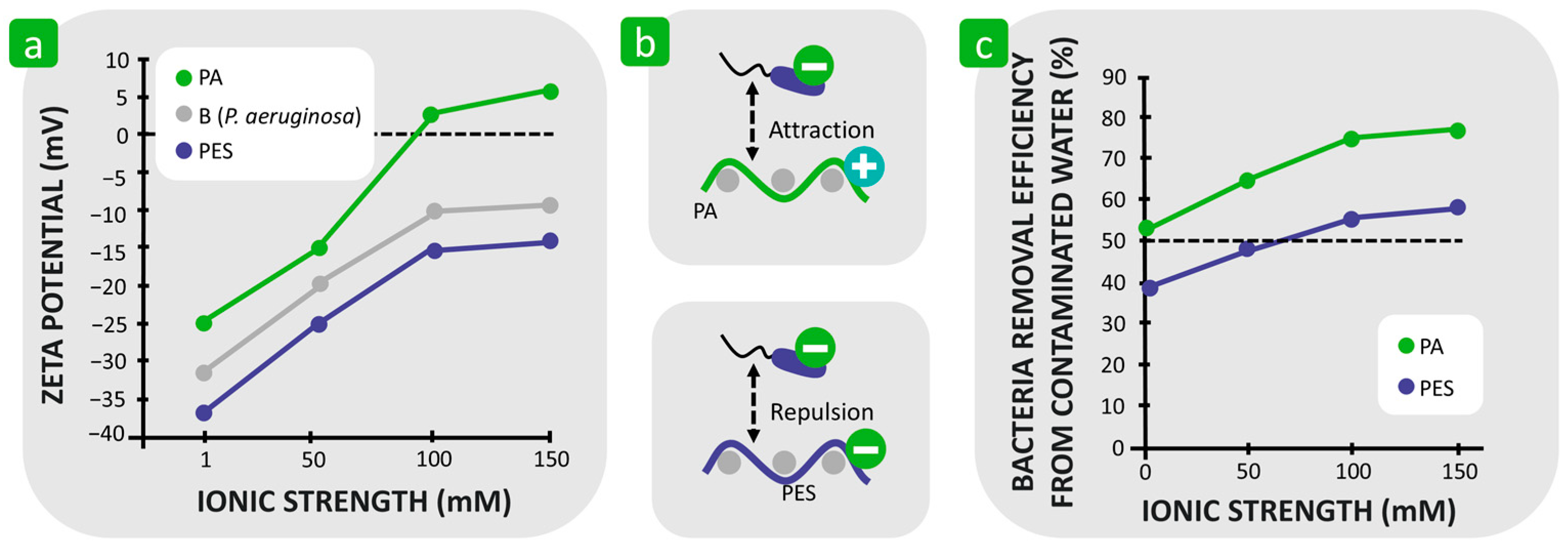
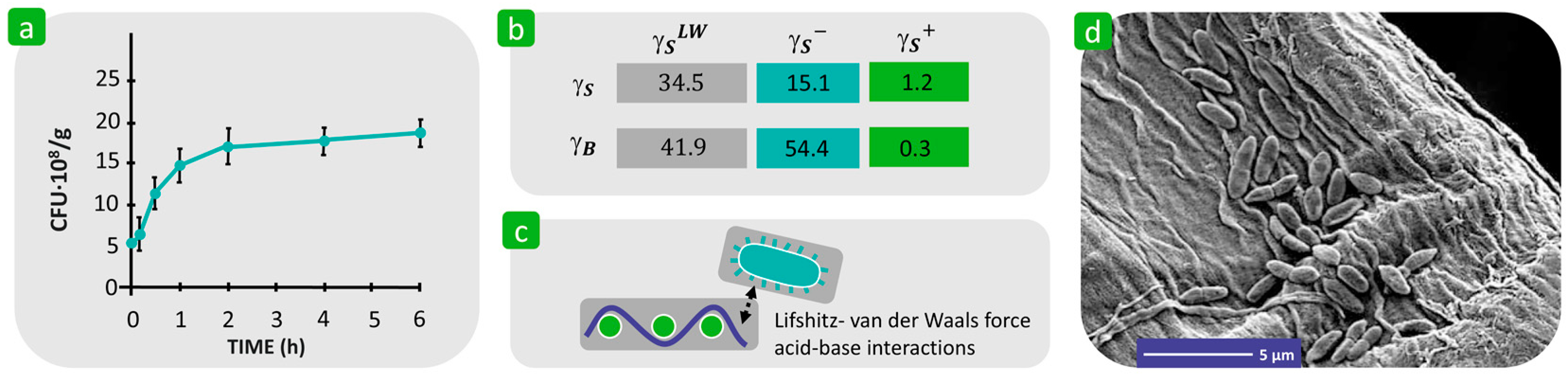
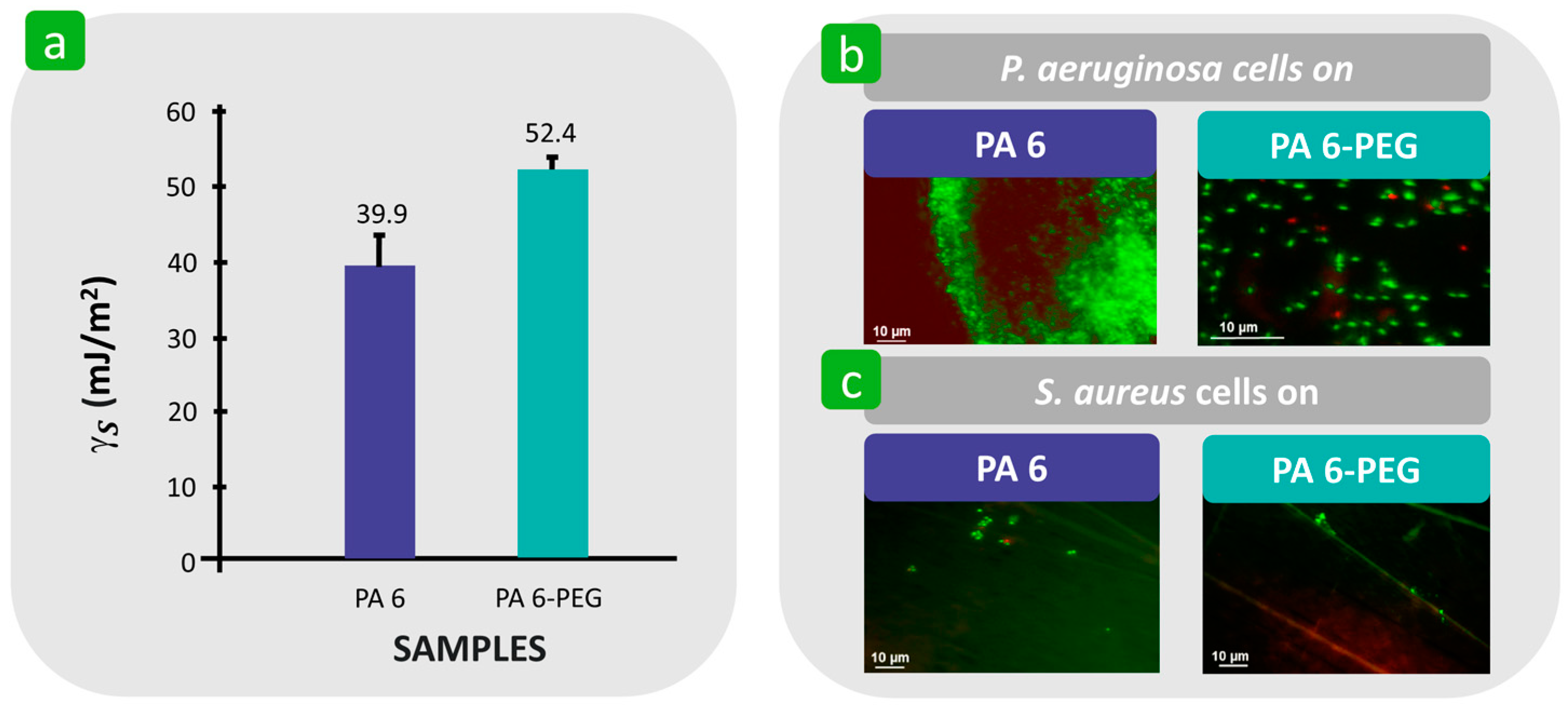




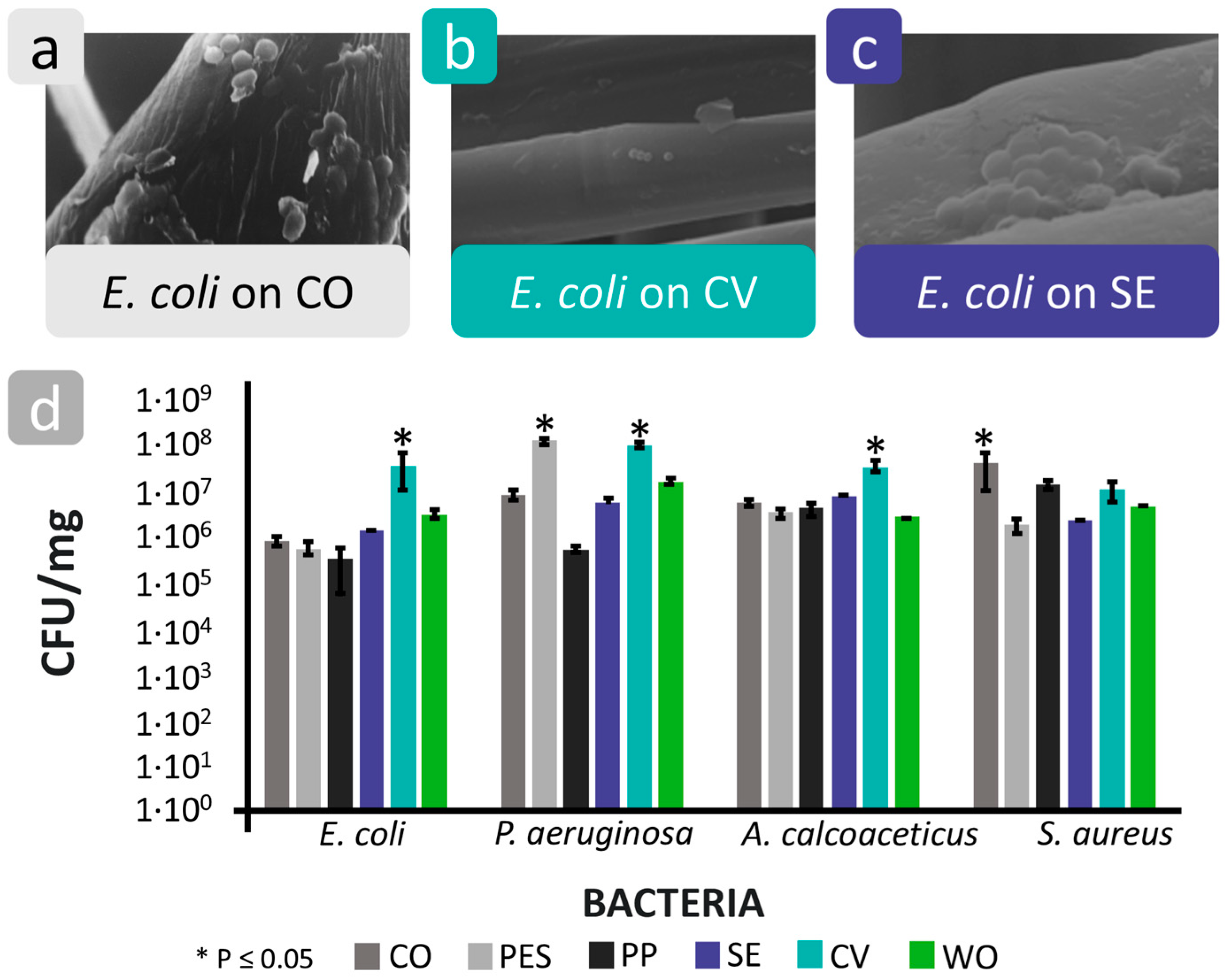
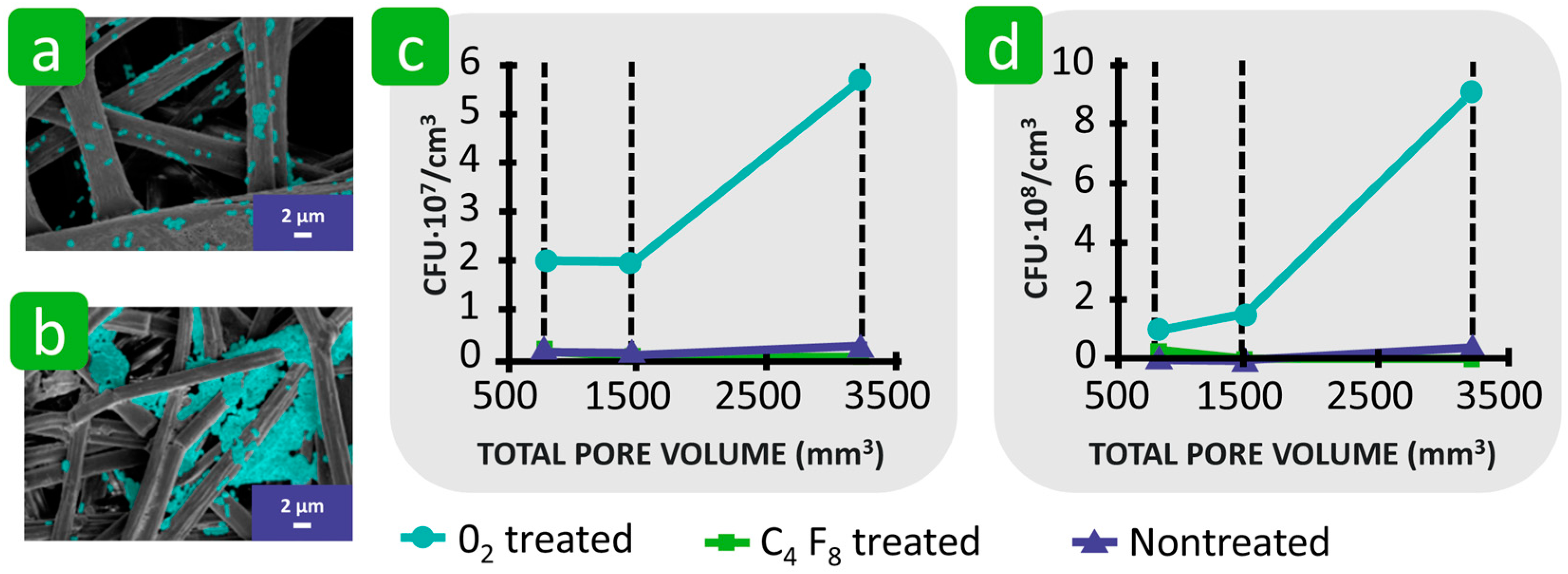
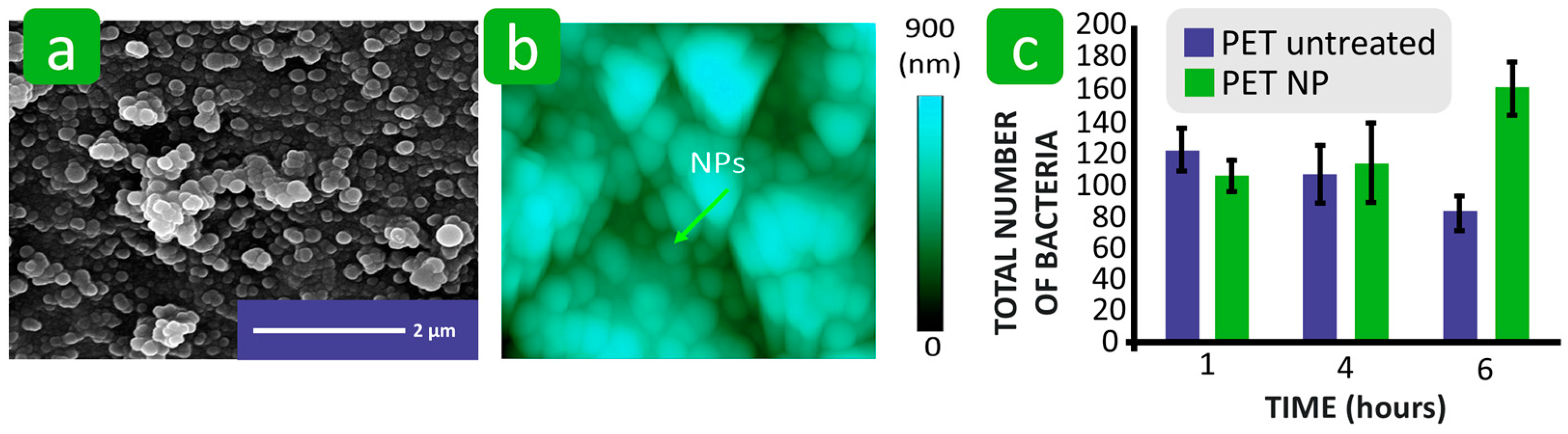

| Mechanism | Characteristics of the Phase | Characteristics of the Bacterial Cells Involved | Forces/Interactions Involved |
|---|---|---|---|
| Reversible adhesion | Immediate attraction of planktonic bacteria to a substrate surface [54,58,60,61,66]. | Passive and/or active movement, but Brownian motion is still present [54,58,60,61,66]. | Attractive (van der Walls) and repulsive (electrostatic) physicochemical forces [60,61]. |
| Irreversible adhesion | Bacteria adhere firmly to the substrate surface [58,60]. The bacterial cells can also adhere to each other and form aggregates on the substrate [60]. | Bacteria no longer show Brownian movement [58,60]. Production of exopolysaccharides that form a complex with the substrate surface [60]. | Molecular and cellular interactions and the production of specific adhesin molecules [58,60]. |
| Theory | Strengths | Limitations | Applications |
|---|---|---|---|
| DLVO theory | Bacterial adhesion is assumed to be balanced by two main forces between the bacterial cells and the substrate surface [18,29,72,73,74,75]. | Bacteria are more complex than inert spherical colloid particles [29,90,91]. Does not consider various factors, which also influence bacterial adhesion [29,90,91]. | Only applicable when a new cell-substrate interface is formed [75]. Great importance in colloid and surface chemistry [71]. |
| Thermodynamic approach | Explains bacterial adhesion to solid substrates based on the interfacial free energy balance [18,29,69,92]. | The approach requires the estimation of the numerical values of the thermodynamic parameters [75]. Assumes a closed energy system although bacteria are living organisms [75]. | Different thermodynamic approaches can be used to calculate Gibs free energy of the system [94]. |
| Extended DLVO theory | Generally, predicts adhesion and its reversibility more accurately than the DLVO theory and the thermodynamic approach [29,75,89,105]. | The theory was not rigorously tested [75]. | It allows for more accurate predictions and effective solutions in managing bacterial contamination on textile surfaces. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čuk, N.; Simončič, B.; Fink, R.; Tomšič, B. Bacterial Adhesion to Natural and Synthetic Fibre-Forming Polymers: Influence of Material Properties. Polymers 2024, 16, 2409. https://doi.org/10.3390/polym16172409
Čuk N, Simončič B, Fink R, Tomšič B. Bacterial Adhesion to Natural and Synthetic Fibre-Forming Polymers: Influence of Material Properties. Polymers. 2024; 16(17):2409. https://doi.org/10.3390/polym16172409
Chicago/Turabian StyleČuk, Nina, Barbara Simončič, Rok Fink, and Brigita Tomšič. 2024. "Bacterial Adhesion to Natural and Synthetic Fibre-Forming Polymers: Influence of Material Properties" Polymers 16, no. 17: 2409. https://doi.org/10.3390/polym16172409
APA StyleČuk, N., Simončič, B., Fink, R., & Tomšič, B. (2024). Bacterial Adhesion to Natural and Synthetic Fibre-Forming Polymers: Influence of Material Properties. Polymers, 16(17), 2409. https://doi.org/10.3390/polym16172409










