Cloning, Expression, and Characterization of a Highly Stable Heparinase I from Bacteroides xylanisolvens
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cloning of BxHep-I Gene from B. xylanisolvens
2.3. Expression and Purification of BxHep-I
2.4. Enzyme Assay
2.5. Optimization of BxHep-I Expression
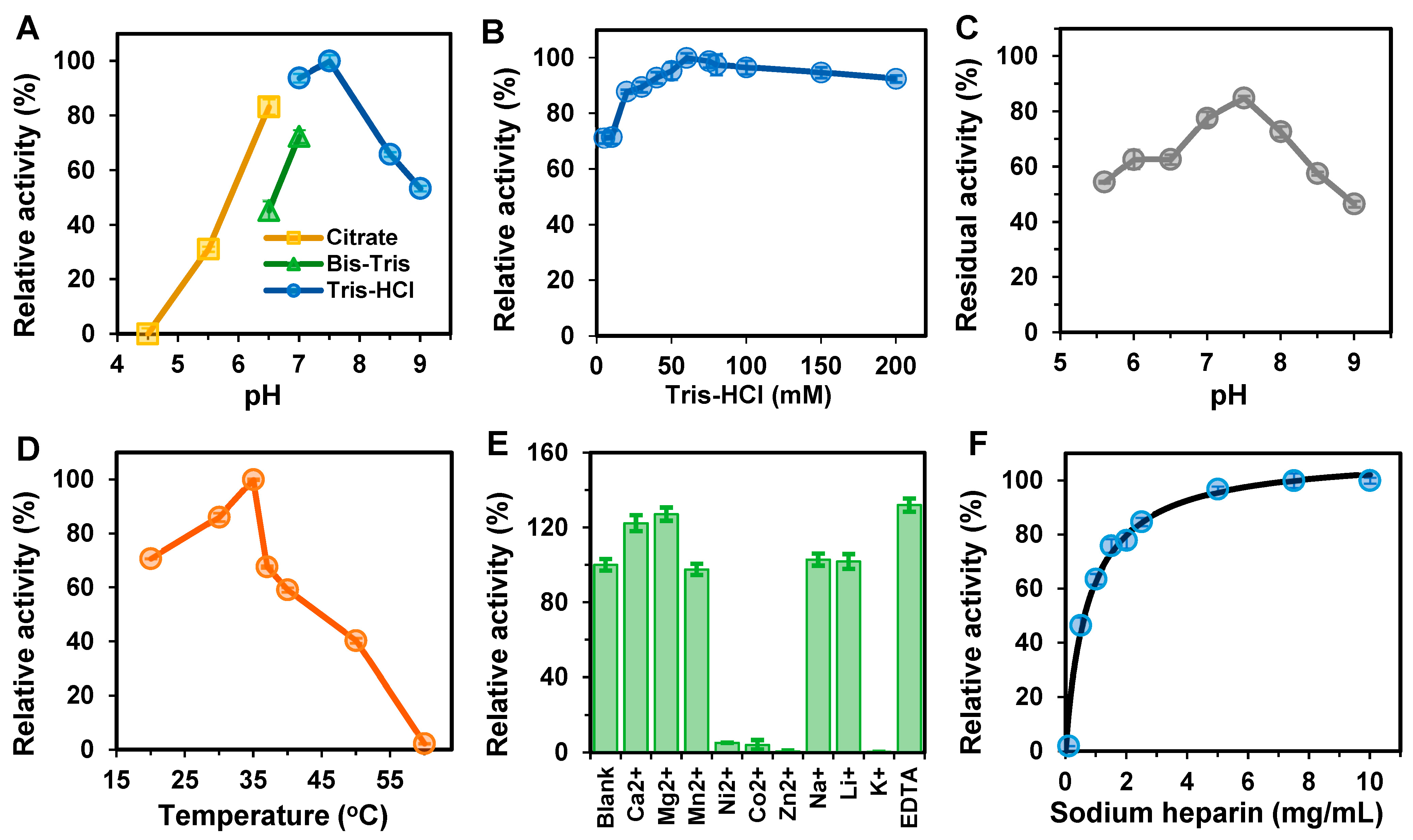
2.6. Biochemical Characterization of BxHep-I
2.6.1. Effects of Temperature on the Activity and Stability of BxHep-I
2.6.2. Effect of pH on the Activity and Stability of BxHep-I
2.6.3. Effects of Metal Ions and EDTA on the Activity of BxHep-I
2.7. Enzyme Kinetics
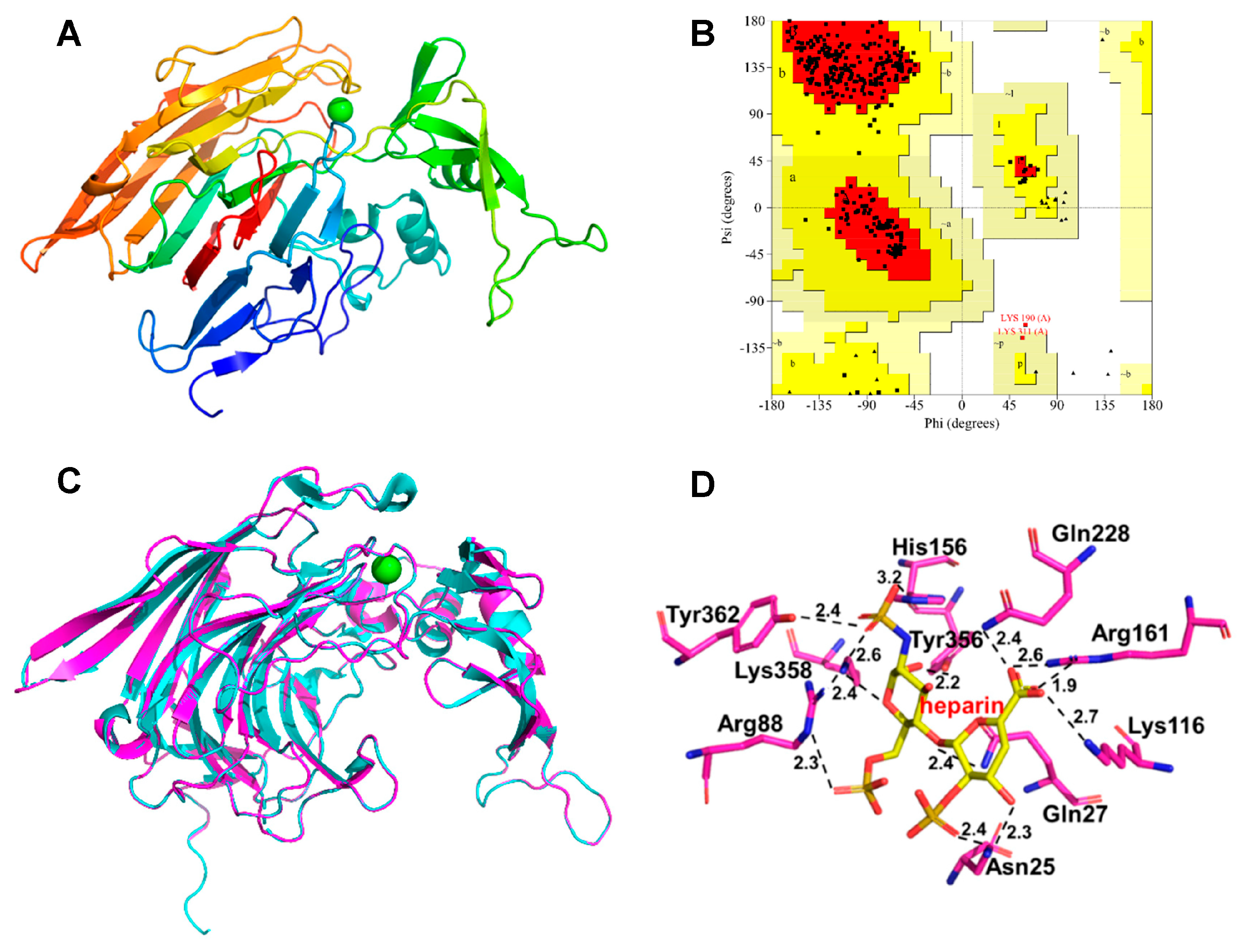
2.8. Homology Modeling and Substrate Docking of BxHep-I
3. Results and Discussion
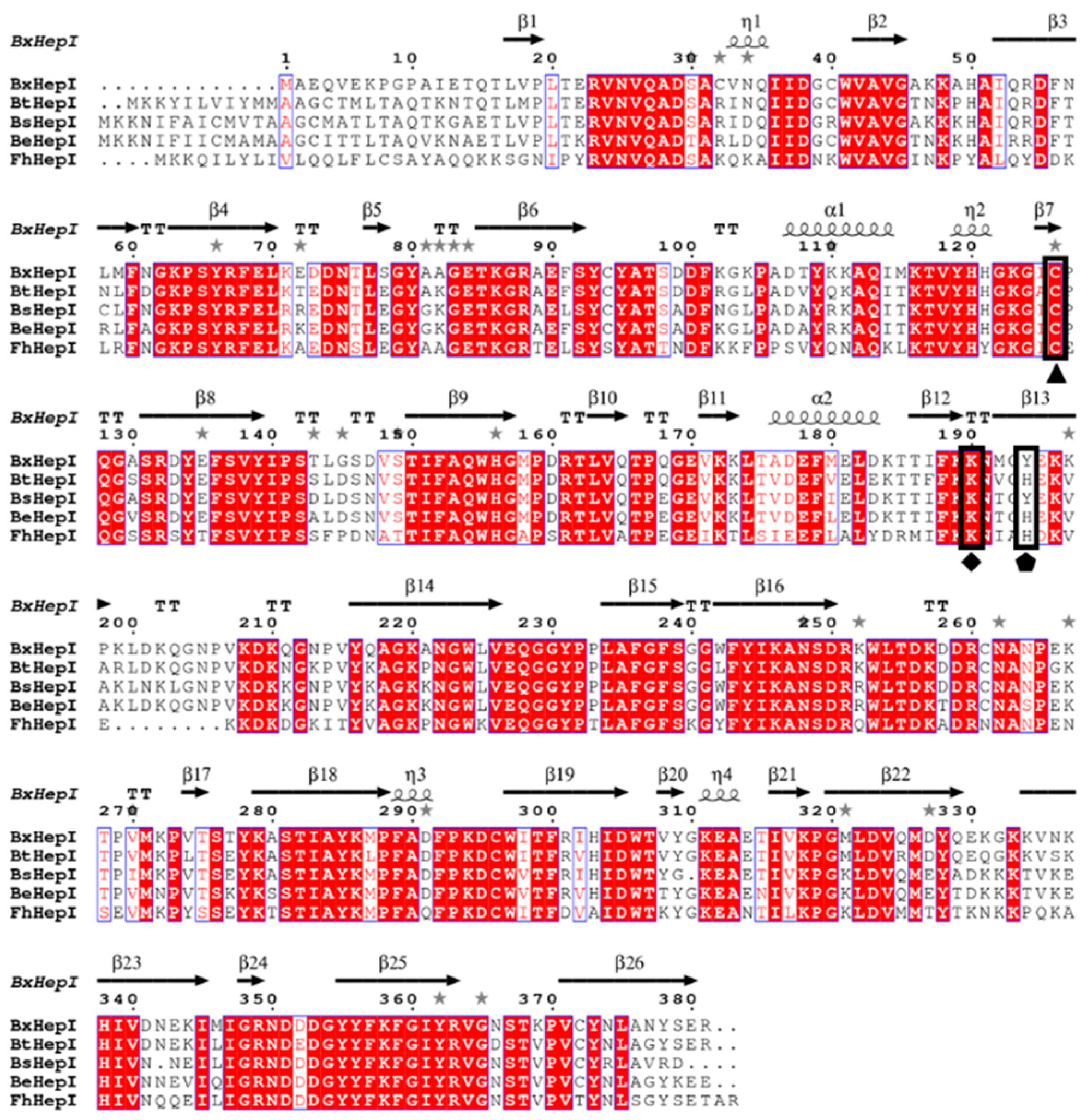
3.1. Sequence Analysis of BxHep-I
3.2. Cloning, Expression, and Purification of BxHep-I
3.3. Biochemical Characterization of BxHep-I
3.4. Thermostability Analysis of BxHep-I
| Organisms | Heparinase I | Specific Activity (U/mg) | Kinetic Parameters | t1/2 (min) | Residual Activity (%) * | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Km (mg/mL) | Vmax (U/mg) | 30 °C | 37 °C | |||||
| F. heparinum | Native | 90.33 | 17.8 | 219.48 | - | - | <30 | [45] |
| F. heparinum | Recombinant MBP-fused | 149 | - | - | 10 | - | 11.3 | [26] |
| F. heparinum | Recombinant MBP-fused mutant | 56.31 | - | - | 130 | - | 85.1 | [46] |
| B. stercoris | Recombinant | 46.02 | 0.023 | 51.81 | - | 30 | - | [19] |
| B. eggerthii | Recombinant | 480 | 3.6 | 647.93 | 350 | 60 | 89.1 | [17] |
| B. cellulosilyticus | Recombinant | 738.8 | 0.17 | 750.58 | 300 | 59 | 58.6 | [18] |
| B. xylanisolvens | Recombinant | 57.6 | 0.79 | 124.58 | 597 | 158 | 73.1 | This work |
3.5. Homology Modeling and Substrate Docking
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, V.C.; Linhardt, R.J.; Bernstein, H.; Cooney, C.L.; Langer, R. Purification and characterization of heparinase from Flavobacterium heparinum. J. Biol. Chem. 1985, 260, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Lohse, D.; Linhardt, R. Purification and characterization of heparin lyases from Flavobacterium heparinum. J. Biol. Chem. 1992, 267, 24347–24355. [Google Scholar] [CrossRef]
- Ernst, S.; Langer, R.; Cooney, C.L.; Sasisekharan, R. Enzymatic degradation of glycosaminogIycans. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 387–444. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, C.K.M.; Banga, J.; Mishra, V. Microbial heparin/heparan sulphate lyases: Potential and applications. Appl. Microbiol. Biotechnol. 2012, 94, 307–321. [Google Scholar] [CrossRef]
- Balasubramaniam, K.; Sharma, K.; Rani, A.; Rajulapati, V.; Goyal, A. Deciphering the mode of action, structural and biochemical analysis of heparinase II/III (PsPL12a) a new member of family 12 polysaccharide lyase from Pseudopedobacter saltans. Ann. Microbiol. 2018, 68, 409–418. [Google Scholar] [CrossRef]
- Pervin, A.; Gallo, C.; Jandik, K.A.; Han, X.-J.; Linhardt, R.J. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology 1995, 5, 83–95. [Google Scholar] [CrossRef]
- Loganathan, D.; Wang, H.M.; Mallis, L.M.; Linhardt, R.J. Structural variation in the antithrombin III binding site region and its occurrence in heparin from different sources. Biochemistry 1990, 29, 4362–4368. [Google Scholar] [CrossRef]
- Linhardt, R.J.; Rice, K.G.; Kim, Y.S.; Lohse, D.L.; Wang, H.M.; Loganathan, D. Mapping and quantification of the major oligosaccharide components of heparin. Biochem. J. 1988, 254, 781–787. [Google Scholar] [CrossRef]
- Bhushan, I.; Alabbas, A.; Sistla, J.C.; Saraswat, R.; Desai, U.R.; Gupta, R.B. Heparin depolymerization by immobilized heparinase: A review. Int. J. Biol. Macromol. 2017, 99, 721–730. [Google Scholar] [CrossRef]
- Korir, A.K.; Larive, C.K. Advances in the separation, sensitive detection, and characterization of heparin and heparan sulfate. Anal. Bioanal. Chem. 2009, 393, 155–169. [Google Scholar] [CrossRef]
- Wang, Z.; Chi, L. Recent advances in mass spectrometry analysis of low molecular weight heparins. Chin. Chem. Lett. 2018, 29, 11–18. [Google Scholar] [CrossRef]
- Forte, K.; Abshire, T. The use of hepzyme in removing heparin from blood samples drawn from central venous access devices. J. Pediatr. Oncol. Nurs. 2000, 17, 179–181. [Google Scholar] [CrossRef]
- Higashi, K.; Hosoyama, S.; Ohno, A.; Masuko, S.; Yang, B.; Sterner, E.; Wang, Z.; Linhardt, R.J.; Toida, T. Photochemical preparation of a novel low molecular weight heparin. Carbohydr. Polym. 2012, 87, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Szajek, A.Y.; Chess, E.; Johansen, K.; Gratzl, G.; Gray, E.; Keire, D.; Linhardt, R.J.; Liu, J.; Morris, T.; Mulloy, B.; et al. The US regulatory and pharmacopeia response to the global heparin contamination crisis. Nat. Biotechnol. 2016, 34, 625–630. [Google Scholar] [CrossRef]
- Mackman, N.; Bergmeier, W.; Stouffer, G.A.; Weitz, J.I. Therapeutic strategies for thrombosis: New targets and approaches. Nat. Rev. Drug Discov. 2020, 19, 333–352. [Google Scholar] [CrossRef]
- Ye, F.; Kuang, Y.; Chen, S.; Zhang, C.; Chen, Y.; Xing, X.-H. Characteristics of low molecular weight heparin production by an ultrafiltration membrane bioreactor using maltose binding protein fused heparinase I. Biochem. Eng. J. 2009, 46, 193–198. [Google Scholar] [CrossRef]
- Gao, L.-W.; Zhu, H.-T.; Liu, C.-Y.; Lv, Z.-X.; Fan, X.-M.; Zhang, Y.-W. A highly active heparinase I from Bacteroides cellulosilyticus: Cloning, high level expression, and molecular characterization. PLoS ONE 2020, 15, e0240920. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-Y.; Su, W.-B.; Guo, L.-B.; Zhang, Y.-W. Cloning, expression, and characterization of a novel heparinase I from Bacteroides eggerthii. Prep. Biochem. Biotechnol. 2020, 50, 477–485. [Google Scholar] [CrossRef]
- Hyun, Y.-J.; Jung, I.-H.; Kim, D.-H. Expression of heparinase I of Bacteroides stercoris HJ-15 and its degradation tendency toward heparin-like glycosaminoglycans. Carbohydr. Res. 2012, 359, 37–43. [Google Scholar] [CrossRef]
- Zhou, H.-P.; Wang, D.-R.; Xu, C.-L.; Zhang, Y.-W. Combination of engineering the substrate and Ca2+ binding domains of heparinase I to improve the catalytic activity. Prep. Biochm. Biotechnol. 2023, in press. [Google Scholar] [CrossRef]
- Banga, J.; Tripathi, C. Response surface methodology for optimization of medium components in submerged culture of Aspergillus flavus for enhanced heparinase production. Lett. Appl. Microbiol. 2009, 49, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xing, X.-H.; Ye, F.; Kuang, Y.; Luo, M. Production of MBP–HepA fusion protein in recombinant Escherichia coli by optimization of culture medium. Biochem. Eng. J. 2007, 34, 114–121. [Google Scholar] [CrossRef]
- Ernst, S.; Venkataraman, G.; Winkler, S.; Godavarti, R.; Langer, R.; Cooney, C.L.; Sasisekharan, R. Expression in Escherichia coli, purification and characterization of heparinase I from Flavobacterium heparinum. Biochem. J. 1996, 315, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xing, X.-H.; Lou, K. Construction of recombinant Escherichia coli for over-production of soluble heparinase I by fusion to maltose-binding protein. Biochem. Eng. J. 2005, 23, 155–159. [Google Scholar] [CrossRef]
- Yu, P.; Yang, J.; Gu, H. Expression of HpaI in Pichia pastoris and optimization of conditions for the heparinase I production. Carbohydr. Polym. 2014, 106, 223–229. [Google Scholar] [CrossRef]
- Chen, S.; Ye, F.; Chen, Y.; Chen, Y.; Zhao, H.; Yatsunami, R.; Nakamura, S.; Arisaka, F.; Xing, X.-H. Biochemical analysis and kinetic modeling of the thermal inactivation of MBP-fused heparinase I: Implications for a comprehensive thermostabilization strategy. Biotechnol. Bioeng. 2011, 108, 1841–1851. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved Prediction of Signal Peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Godavarti, R.; Sasisekharan, R. Heparinase I from Flavobacterium heparinum. J. Biol. Chem. 1998, 273, 248–255. [Google Scholar] [CrossRef]
- Sasisekharan, R.; Leckband, D.; Godavarti, R.; Venkataraman, G.; Cooney, C.L.; Langer, R. Heparinase I from Flavobacterium heparinum: The role of the cysteine residue in catalysis as probed by chemical modification and site-directed mutagenesis. Biochemistry 1995, 34, 14441–14448. [Google Scholar] [CrossRef]
- Córdula, C.R.; Lima, M.A.; Shinjo, S.K.; Gesteira, T.F.; Pol-Fachin, L.; Coulson-Thomas, V.J.; Verli, H.; Yates, E.A.; Rudd, T.R.; Pinhal, M.A.S.; et al. On the catalytic mechanism of polysaccharide lyases: Evidence of His and Tyr involvement in heparin lysis by heparinase I and the role of Ca2+. Mol. Biosyst. 2014, 10, 54–64. [Google Scholar] [CrossRef]
- Makrides, S.C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996, 60, 512–538. [Google Scholar] [CrossRef] [PubMed]
- Schrödel, A.; de Marco, A. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 2005, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.G.; Baneyx, F. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing heat-shock proteins. J. Biol. Chem. 1996, 271, 11141–11147. [Google Scholar] [CrossRef]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, X.; McKeehan, W.L. High yield, purity and activity of soluble recombinant Bacteroides thetaiotaomicron GST-heparinase I from Escherichia coli. Arch. Biochem. Biophys. 2007, 460, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Wu, Y. Expression of the heparinase gene from Flavobacterium heparinum in Escherichia coli and its enzymatic properties. Carbohydr. Polym. 2012, 90, 348–352. [Google Scholar] [CrossRef]
- Watanabe, M.; Tsuda, H.; Yamada, S.; Shibata, Y.; Nakamura, T.; Sugahara, K. Characterization of heparinase from an oral bacterium Prevotella heparinolytica. J. Biochem. 1998, 123, 283–288. [Google Scholar] [CrossRef]
- Shriver, Z.; Liu, D.; Hu, Y.; Sasisekharan, R. Biochemical investigations and mapping of the calcium-binding sites of heparinase I from Flavobacterium heparinum. J. Biol. Chem. 1999, 274, 4082–4088. [Google Scholar] [CrossRef]
- Liu, D.; Shriver, Z.; Godavarti, R.; Venkataraman, G.; Sasisekharan, R. The calcium-binding sites of heparinase I from Flavobacterium heparinum are essential for enzymatic activity. J. Biol. Chem. 1999, 274, 4089–4095. [Google Scholar] [CrossRef]
- Kuang, Y.; Xing, X.-H.; Chen, Y.; Ye, F.; Chen, Y.; Yan, Y.; Liu, Z.; Bi, R. Production of heparin oligosaccharides by fusion protein of MBP–heparinase I and the enzyme thermostability. J. Mol. Catal. B Enzym. 2006, 43, 90–95. [Google Scholar] [CrossRef]
- Kumar, S.; Tsai, C.-J.; Nussinov, R. Factors enhancing protein thermostability. Protein Eng. Des. Sel. 2000, 13, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-H.; Garron, M.-L.; Kim, H.-Y.; Kim, W.-S.; Zhang, Z.; Ryu, K.-S.; Shaya, D.; Xiao, Z.; Cheong, C.; Kim, Y.S.; et al. Structural snapshots of heparin depolymerization by heparin lyase I. J. Biol. Chem. 2009, 284, 34019–34027. [Google Scholar] [CrossRef]
- Sasisekharan, R.; Venkataraman, G.; Godavarti, R.; Ernst, S.; Cooney, C.L.; Langer, R. Heparinase I from Flavobacterium heparinum: Mapping and characterization of the heparin binding domain. J. Biol. Chem. 1996, 271, 3124–3131. [Google Scholar] [CrossRef]
- Ma, X.; Wang, Z.; Li, S.; Shen, Q.; Yuan, Q. Effect of CaCl2 as activity stabilizer on purification of heparinase I from Flavobacterium heparinum. J. Chromatogr. B 2006, 843, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, Z.; Wu, J.; Chen, Y.; Ye, F.; Zhang, C.; Yatsunami, R.; Nakamura, S.; Xing, X.-H. Combination of site-directed mutagenesis and calcium ion addition for enhanced production of thermostable MBP-fused heparinase I in recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2907–2916. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, J.-L.; Wei, W.; Wang, D.-R.; Liu, C.-Y.; Zhou, H.-P.; Xu, C.-L.; Zhang, Y.-W. Cloning, Expression, and Characterization of a Highly Stable Heparinase I from Bacteroides xylanisolvens. Polymers 2023, 15, 1776. https://doi.org/10.3390/polym15071776
Pei J-L, Wei W, Wang D-R, Liu C-Y, Zhou H-P, Xu C-L, Zhang Y-W. Cloning, Expression, and Characterization of a Highly Stable Heparinase I from Bacteroides xylanisolvens. Polymers. 2023; 15(7):1776. https://doi.org/10.3390/polym15071776
Chicago/Turabian StylePei, Jia-Lu, Wei Wei, Ding-Ran Wang, Cai-Yun Liu, Hua-Ping Zhou, Chen-Lu Xu, and Ye-Wang Zhang. 2023. "Cloning, Expression, and Characterization of a Highly Stable Heparinase I from Bacteroides xylanisolvens" Polymers 15, no. 7: 1776. https://doi.org/10.3390/polym15071776
APA StylePei, J.-L., Wei, W., Wang, D.-R., Liu, C.-Y., Zhou, H.-P., Xu, C.-L., & Zhang, Y.-W. (2023). Cloning, Expression, and Characterization of a Highly Stable Heparinase I from Bacteroides xylanisolvens. Polymers, 15(7), 1776. https://doi.org/10.3390/polym15071776







