Polyethylene Glycol-Mediated Directional Conjugation of Biological Molecules for Enhanced Immunoassays at the Point-of-Care
Abstract
1. Introduction
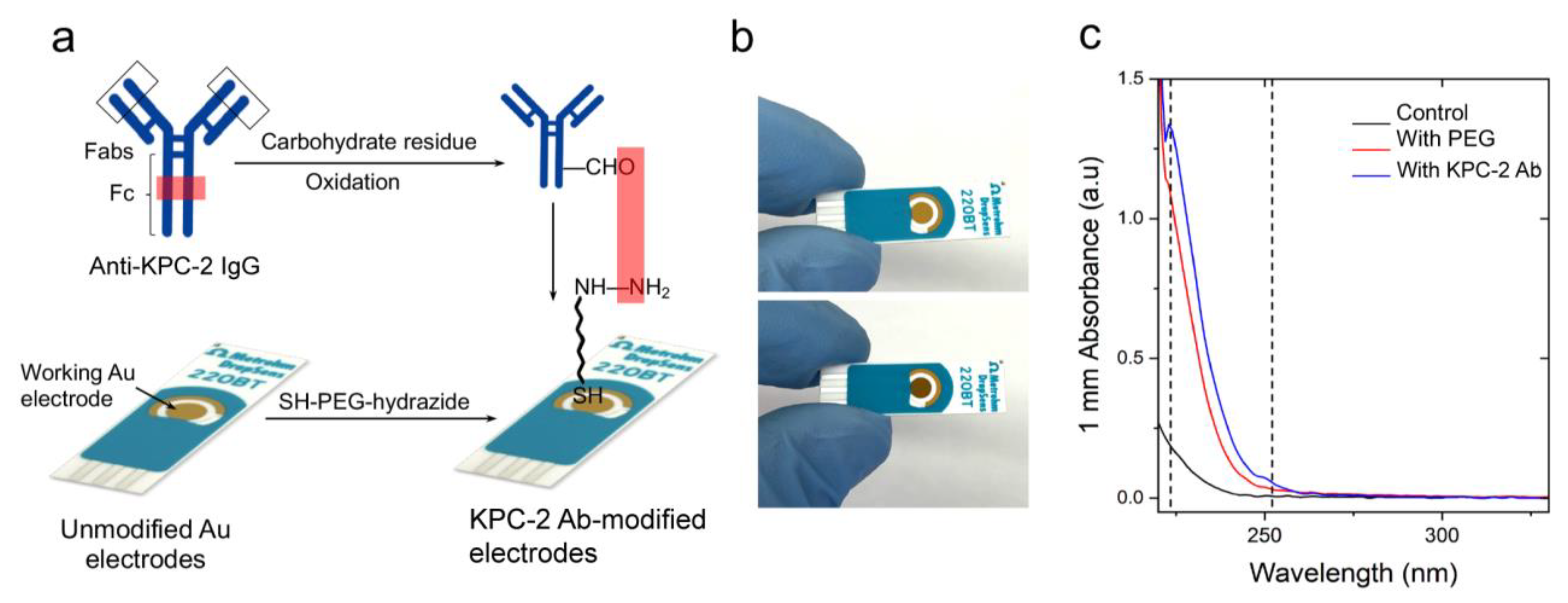
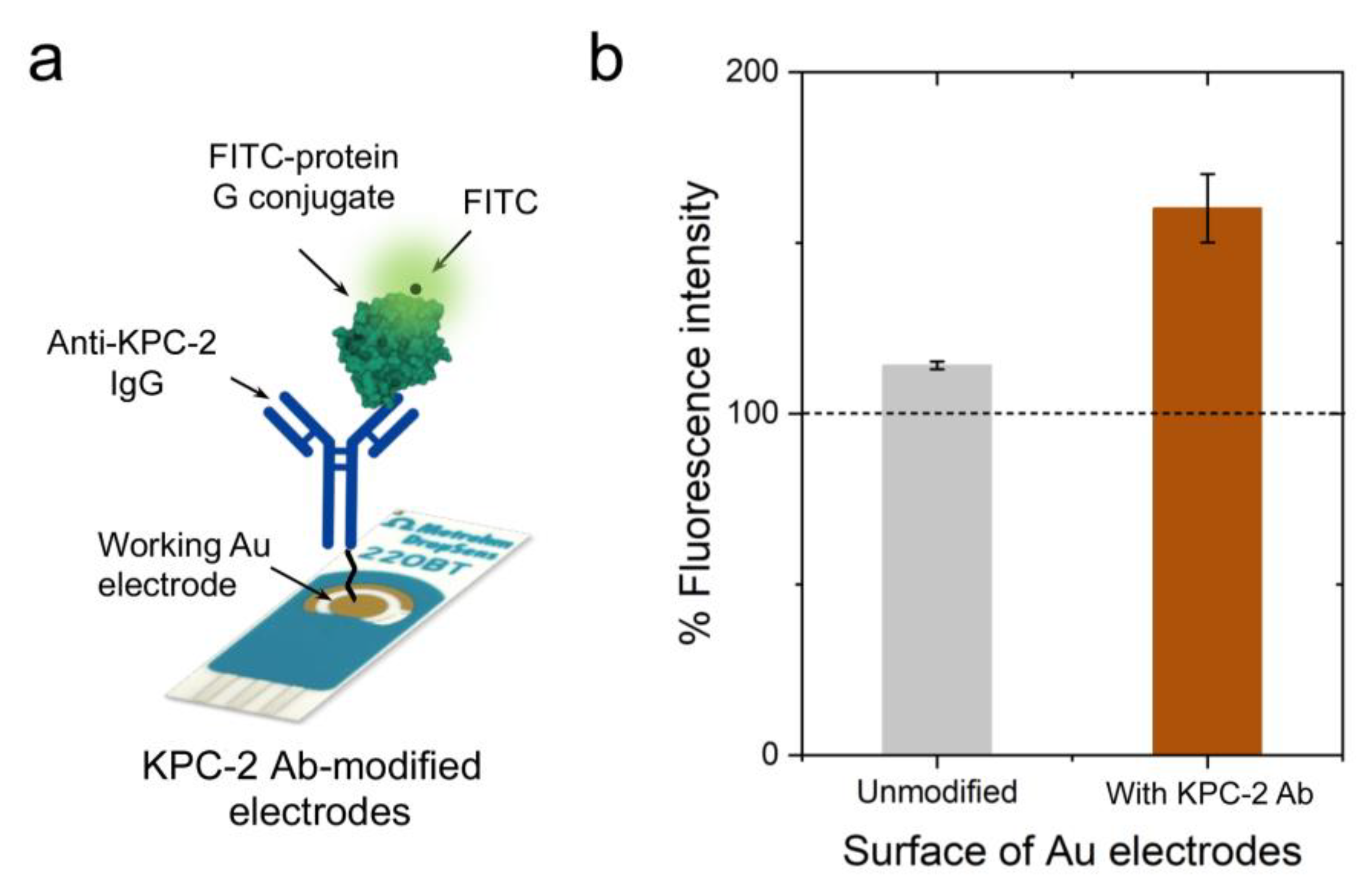
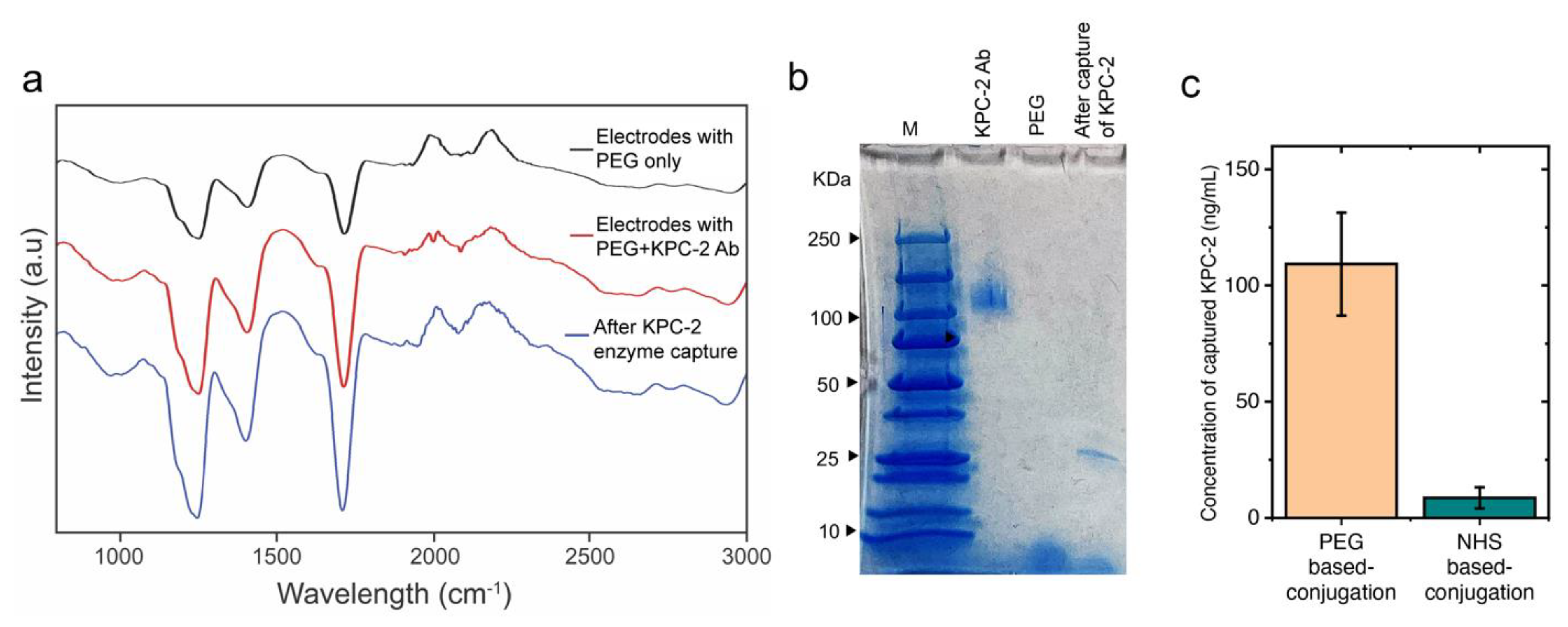
2. Results and Discussion
3. Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yager, P.; Domingo, G.J.; Gerdes, J. Point-of-care diagnostics for global health. Annu. Rev. Biomed. Eng. 2008, 10, 107–144. [Google Scholar] [CrossRef] [PubMed]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno) assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Lequin, R.M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [PubMed]
- Junutula, J.R.; Flagella, K.M.; Graham, R.A.; Parsons, K.L.; Ha, E.; Raab, H.; Bhakta, S.; Nguyen, T.; Dugger, D.L.; Li, G. Engineered Thio-Trastuzumab-DM1 Conjugate with an Improved Therapeutic Index to Target Human Epidermal Growth Factor Receptor 2–Positive Breast CancerEngineered Trastuzumab-DM1 THIOMAB Drug Conjugate. Clin. Cancer Res. 2010, 16, 4769–4778. [Google Scholar] [CrossRef]
- Strop, P.; Liu, S.-H.; Dorywalska, M.; Delaria, K.; Dushin, R.G.; Tran, T.-T.; Ho, W.-H.; Farias, S.; Casas, M.G.; Abdiche, Y. Location matters: Site of conjugation modulates stability and pharmacokinetics of antibody drug conjugates. Chem. Biol. 2013, 20, 161–167. [Google Scholar] [CrossRef]
- Sadiki, A.; Vaidya, S.R.; Abdollahi, M.; Bhardwaj, G.; Dolan, M.E.; Turna, H.; Arora, V.; Sanjeev, A.; Robinson, T.D.; Koid, A. Site-specific conjugation of native antibody. Antib. Ther. 2020, 3, 271–284. [Google Scholar] [CrossRef]
- Ismail, N.F.; Lim, T.S. Site-specific scFv labelling with invertase via Sortase A mechanism as a platform for antibody-antigen detection using the personal glucose meter. Sci. Rep. 2016, 6, 19338. [Google Scholar] [CrossRef]
- Draz, M.S.; Kochehbyoki, K.M.; Vasan, A.; Battalapalli, D.; Sreeram, A.; Kanakasabapathy, M.K.; Kallakuri, S.; Tsibris, A.; Kuritzkes, D.R.; Shafiee, H. DNA engineered micromotors powered by metal nanoparticles for motion based cellphone diagnostics. Nat. Commun. 2018, 9, 4282. [Google Scholar] [CrossRef]
- Draz, M.S.; Fang, B.A.; Li, L.; Chen, Z.; Wang, Y.; Xu, Y.; Yang, J.; Killeen, K.; Chen, F.F. Hybrid nanocluster plasmonic resonator for immunological detection of hepatitis B virus. ACS Nano 2012, 6, 7634–7643. [Google Scholar] [CrossRef]
- Draz, M.S.; Vasan, A.; Muthupandian, A.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Sreeram, A.; Krishnakumar, S.; Yogesh, V.; Lin, W.; Yu, X.G. Virus detection using nanoparticles and deep neural network–enabled smartphone system. Sci. Adv. 2020, 6, eabd5354. [Google Scholar] [CrossRef]
- Draz, M.S.; Moazeni, M.; Venkataramani, M.; Lakshminarayanan, H.; Saygili, E.; Lakshminaraasimulu, N.K.; Kochehbyoki, K.M.; Kanakasabapathy, M.K.; Shabahang, S.; Vasan, A. Hybrid paper–plastic microchip for flexible and high-performance point-of-care diagnostics. Adv. Funct. Mater. 2018, 28, 1707161. [Google Scholar] [CrossRef]
- Draz, M.S.; Venkataramani, M.; Lakshminarayanan, H.; Saygili, E.; Moazeni, M.; Vasan, A.; Li, Y.; Sun, X.; Hua, S.; Xu, G.Y. Nanoparticle-enhanced electrical detection of Zika virus on paper microchips. Nanoscale 2018, 10, 11841–11849. [Google Scholar] [CrossRef] [PubMed]
- Draz, M.S.; Tang, Y.; Zhang, P. Bio-nanoparticles: Nanoscale probes for nanoscale pathogens. In 21st Century Nanoscience—A Handbook; CRC Press: Boca Raton, FL, USA, 2020; pp. 20-21–20-23. [Google Scholar]
- Jevševar, S.; Kunstelj, M.; Porekar, V.G. PEGylation of therapeutic proteins. Biotechnol. J. Healthc. Nutr. Technol. 2010, 5, 113–128. [Google Scholar]
- Roberts, M.; Bentley, M.; Harris, J. Chemistry for peptide and protein PEGylation. Adv. Drug Deliv. Rev. 2002, 54, 459–476. [Google Scholar] [CrossRef] [PubMed]
- Veronese, F.M.; Pasut, G. PEGylation, successful approach to drug delivery. Drug Discov. Today 2005, 10, 1451–1458. [Google Scholar] [CrossRef]
- Harris, J.M.; Chess, R.B. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug Discov. 2003, 2, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Caliceti, P.; Veronese, F.M. Pharmacokinetic and biodistribution properties of poly (ethylene glycol)–protein conjugates. Adv. Drug Deliv. Rev. 2003, 55, 1261–1277. [Google Scholar] [CrossRef]
- Milla, P.; Dosio, F.; Cattel, L. PEGylation of proteins and liposomes: A powerful and flexible strategy to improve the drug delivery. Curr. Drug Metab. 2012, 13, 105–119. [Google Scholar] [CrossRef]
- Pasut, G.; Veronese, F.M. State of the art in PEGylation: The great versatility achieved after forty years of research. J. Control. Release 2012, 161, 461–472. [Google Scholar] [CrossRef]
- Huang, C.; Wen, T.; Shi, F.-J.; Zeng, X.-Y.; Jiao, Y.-J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega 2020, 5, 12550–12556. [Google Scholar] [CrossRef]
- Khawar, I.A.; Kim, J.H.; Kuh, H.-J. Improving drug delivery to solid tumors: Priming the tumor microenvironment. J. Control. Release 2015, 201, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.S.; de Camargo, A.S. Exploring the use of upconversion nanoparticles in chemical and biological sensors: From surface modifications to point-of-care devices. Nanoscale Adv. 2021, 3, 5135–5165. [Google Scholar] [CrossRef] [PubMed]
- Gopal, A.; Yan, L.; Kashif, S.; Munshi, T.; Roy, V.A.; Voelcker, N.H.; Chen, X. Biosensors and Point-of-Care Devices for Bacterial Detection: Rapid Diagnostics Informing Antibiotic Therapy. Adv. Healthc. Mater. 2022, 11, 2101546. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Jaiswal, A. Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst 2018, 143, 1970–1996. [Google Scholar] [CrossRef]
- Nath, P.; Kabir, A.; Khoubafarin Doust, S.; Kreais, Z.J.; Ray, A. Detection of bacterial and viral pathogens using photonic point-of-care devices. Diagnostics 2020, 10, 841. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Qi, M.; Hutchinson, M.R.; Yang, G.; Goldys, E.M. Recent advances in cytokine detection by immunosensing. Biosens. Bioelectron. 2016, 79, 810–821. [Google Scholar] [CrossRef]
- Kinstler, O.; Molineux, G.; Treuheit, M.; Ladd, D.; Gegg, C. Mono-N-terminal poly (ethylene glycol)–protein conjugates. Adv. Drug Deliv. Rev. 2002, 54, 477–485. [Google Scholar] [CrossRef]
- Junutula, J.R.; Raab, H.; Clark, S.; Bhakta, S.; Leipold, D.D.; Weir, S.; Chen, Y.; Simpson, M.; Tsai, S.P.; Dennis, M.S. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008, 26, 925–932. [Google Scholar] [CrossRef]
- Casi, G.; Neri, D. Antibody–drug conjugates: Basic concepts, examples and future perspectives. J. Control. Release 2012, 161, 422–428. [Google Scholar] [CrossRef]
- Swierczewska, M.; Lee, K.C.; Lee, S. What is the future of PEGylated therapies? Expert Opin. Emerg. Drugs 2015, 20, 531–536. [Google Scholar] [CrossRef]
- Luppa, P.B.; Sokoll, L.J.; Chan, D.W. Immunosensors—Principles and applications to clinical chemistry. Clin. Chim. Acta 2001, 314, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Corstjens, P.; Malamud, D. Point-of-care diagnostics for infectious diseases. In Salivary Diagnostics; Wiley-Blackwell: Hoboken, NJ, USA, 2008. [Google Scholar]
- Chin, C.D.; Linder, V.; Sia, S.K. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 2012, 12, 2118–2134. [Google Scholar] [CrossRef] [PubMed]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly (ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. 2010, 49, 6288–6308. [Google Scholar] [CrossRef] [PubMed]
- Veronese, F.M.; Mero, A. The impact of PEGylation on biological therapies. BioDrugs 2008, 22, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Jefferis, R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 2009, 8, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. [Google Scholar] [CrossRef]
- Metters, J.P.; Kadara, R.O.; Banks, C.E. New directions in screen printed electroanalytical sensors: An overview of recent developments. Analyst 2011, 136, 1067–1076. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Kalia, J.; Raines, R.T. Reactivity of intein thioesters: Appending a functional group to a protein. ChemBioChem 2006, 7, 1375–1383. [Google Scholar] [CrossRef]
- Jeon, O.; Song, S.J.; Kang, S.-W.; Putnam, A.J.; Kim, B.-S. Enhancement of ectopic bone formation by bone morphogenetic protein-2 released from a heparin-conjugated poly (L-lactic-co-glycolic acid) scaffold. Biomaterials 2007, 28, 2763–2771. [Google Scholar] [CrossRef]
- Love, J.C.; Estroff, L.A.; Kriebel, J.K.; Nuzzo, R.G.; Whitesides, G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005, 105, 1103–1170. [Google Scholar] [CrossRef] [PubMed]
- Gubala, V.; Harris, L.F.; Ricco, A.J.; Tan, M.X.; Williams, D.E. Point of care diagnostics: Status and future. Anal. Chem. 2012, 84, 487–515. [Google Scholar] [CrossRef] [PubMed]
- Tadepalli, S.; Kuang, Z.; Jiang, Q.; Liu, K.-K.; Fisher, M.A.; Morrissey, J.J.; Kharasch, E.D.; Slocik, J.M.; Naik, R.R.; Singamaneni, S. Peptide functionalized gold nanorods for the sensitive detection of a cardiac biomarker using plasmonic paper devices. Sci. Rep. 2015, 5, 16206. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-M.; Jin, X.; Zhao, G.-C. Amplified QCM biosensor for type IV collagenase based on collagenase-cleavage of gold nanoparticles functionalized peptide. Biosens. Bioelectron. 2018, 106, 111–116. [Google Scholar] [CrossRef]
- Draz, M.S.; Lakshminaraasimulu, N.K.; Krishnakumar, S.; Battalapalli, D.; Vasan, A.; Kanakasabapathy, M.K.; Sreeram, A.; Kallakuri, S.; Thirumalaraju, P.; Li, Y. Motion-based immunological detection of Zika virus using Pt-nanomotors and a cellphone. ACS Nano 2018, 12, 5709–5718. [Google Scholar] [CrossRef]
- Bhardwaj, S.K.; Bhardwaj, N.; Mohanta, G.C.; Kumar, P.; Sharma, A.L.; Kim, K.-H.; Deep, A. Immunosensing of atrazine with antibody-functionalized Cu-MOF conducting thin films. ACS Appl. Mater. Interfaces 2015, 7, 26124–26130. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Ezhov, A.A.; Vigovskiy, M.A.; Grigorieva, O.A.; Dyachkova, U.D.; Belogurova, N.G.; Kudryashova, E.V. Application Prospects of FTIR Spectroscopy and CLSM to Monitor the Drugs Interaction with Bacteria Cells Localized in Macrophages for Diagnosis and Treatment Control of Respiratory Diseases. Diagnostics 2023, 13, 698. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Mouwen, D.; López, M.; Prieto, M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J. Microbiol. Methods 2011, 84, 369–378. [Google Scholar] [CrossRef]
- Zhou, F.; Fu, T.; Huang, Q.; Kuai, H.; Mo, L.; Liu, H.; Wang, Q.; Peng, Y.; Han, D.; Zhao, Z. Hypoxia-activated PEGylated conditional aptamer/antibody for cancer imaging with improved specificity. J. Am. Chem. Soc. 2019, 141, 18421–18427. [Google Scholar] [CrossRef]
- Hu, Y.; Cortez-Jugo, C.; Ju, Y.; Zheng, T.; Zhou, J.; Lin, Z.; De Rose, R.; Hagemeyer, C.E.; Alt, K.; Caruso, F. Poly (ethylene glycol) Cross-Linked Antibody Nanoparticles for Tunable Biointeractions. Chem. Mater. 2023, 35, 4965–4978. [Google Scholar] [CrossRef]
- Ho, J.-A.A.; Chang, H.-C.; Shih, N.-Y.; Wu, L.-C.; Chang, Y.-F.; Chen, C.-C.; Chou, C. Diagnostic detection of human lung cancer-associated antigen using a gold nanoparticle-based electrochemical immunosensor. Anal. Chem. 2010, 82, 5944–5950. [Google Scholar] [CrossRef] [PubMed]
- Rusling, J.F.; Kumar, C.V.; Gutkind, J.S.; Patel, V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst 2010, 135, 2496–2511. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battalapalli, D.; Chakraborty, P.; Jain, D.; Obaro, S.K.; Gurkan, U.A.; Bonomo, R.A.; Draz, M.S. Polyethylene Glycol-Mediated Directional Conjugation of Biological Molecules for Enhanced Immunoassays at the Point-of-Care. Polymers 2023, 15, 3316. https://doi.org/10.3390/polym15153316
Battalapalli D, Chakraborty P, Jain D, Obaro SK, Gurkan UA, Bonomo RA, Draz MS. Polyethylene Glycol-Mediated Directional Conjugation of Biological Molecules for Enhanced Immunoassays at the Point-of-Care. Polymers. 2023; 15(15):3316. https://doi.org/10.3390/polym15153316
Chicago/Turabian StyleBattalapalli, Dheerendranath, Purbali Chakraborty, Disha Jain, Stephen K. Obaro, Umut A. Gurkan, Robert A. Bonomo, and Mohamed S. Draz. 2023. "Polyethylene Glycol-Mediated Directional Conjugation of Biological Molecules for Enhanced Immunoassays at the Point-of-Care" Polymers 15, no. 15: 3316. https://doi.org/10.3390/polym15153316
APA StyleBattalapalli, D., Chakraborty, P., Jain, D., Obaro, S. K., Gurkan, U. A., Bonomo, R. A., & Draz, M. S. (2023). Polyethylene Glycol-Mediated Directional Conjugation of Biological Molecules for Enhanced Immunoassays at the Point-of-Care. Polymers, 15(15), 3316. https://doi.org/10.3390/polym15153316









