Study on the Biodegradation Process of D-Mannose Glycopolymers in Liquid Media and Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biodegradation Using a Pure Culture of P. mirabilis
2.2. Biodegradation in Soil
2.3. SEM/EDX Analysis
2.4. Mathematical Modeling
3. Results and Discussions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, W.; Song, P.; Zhang, H.; Duan, X.; Wei, Y.; Wang, H.; Wang, S. Microplastic materials in the environment: Problem and strategical solutions. Prog. Mater. Sci. 2023, 123, 101035. [Google Scholar] [CrossRef]
- Tan, Q.; Yang, L.; Wei, F.; Chen, Y.; Li, J. Comparative life cycle assessment of polyethylene agricultural mulching film and alternative options including different end-of-life routes. Renew. Sustain. Energy Rev. 2023, 178, 113239. [Google Scholar] [CrossRef]
- Adegoke, K.A.; Adu, F.A.; Oyebamiji, A.K.; Bamisaye, A.; Adigun, R.A.; Olasoji, S.O.; Ogunjinmi, O.E. Microplastics toxicity, detection and removal from water/wastewater. Marine Pollut. Bull. 2023, 187, 114546. [Google Scholar] [CrossRef]
- Lebreton, L.; Slat, B.; Ferrari, F.; Sainte-Rose, B.; Aitken, J.; Marthouse, R.; Hajbane, S.; Cunsolo, S.; Schwarz, A.; Levivier, A.; et al. Evidence that the Great Pacific Garbage Patch is rapidly accumulating plastic. Sci. Rep. 2018, 8, 4666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czarnecka, E.; Walczak, M.; Kumar, G.; Piechota, G.; Nowaczyk, J. Degradation of biodegradable diapers as an element circular economy in waste containing various plastics. J. Clean. Prod. 2022, 377, 134426. [Google Scholar] [CrossRef]
- Dunn, R.A.; Welden, N.A. Management of Environmental Plastic Pollution: A Comparison of Existing Strategies and Emerging Solutions from Nature. Water Air Soil Pollut. 2023, 234, 201. [Google Scholar] [CrossRef]
- Andrady, A.; Barnes, P.; Bornman, J.; Gouin, T.; Madronich, S.; White, C.; Zepp, R.; Jansen, M. Oxidation and fragmentation of plastics in a changing environment; from UV-radiation to biological degradation. Sci. Total Environ. 2022, 851, 158022. [Google Scholar] [CrossRef]
- Battista, F.; Frison, N.; Bolzonella, D. Can bioplastics be treated in conventional anaerobic digesters for food waste treatment? Environ. Technol. Innov. 2021, 22, 101393. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, Y.; Liang, C.; Song, J.; Yu, S.; Liao, G.; Zou, P.; Tang, K.H.D.; Wu, C. Airborne microplastics: Occurrence, sources, fate, risks and mitigation. Sci. Total Environ. 2023, 858, 159943. [Google Scholar] [CrossRef]
- An, R.; Liu, C.; Wang, J.; Jia, P. Recent Advances in Degradation of Polymer Plastics by Insects Inhabiting Microorganisms. Polymers 2023, 15, 1307. [Google Scholar] [CrossRef]
- Cholewinski, A.; Dadzie, E.; Sherlock, C.; Anderson, W.A.; Charles, T.C.; Habib, K.; Young, S.B.; Zhao, B. A critical review of microplastic degradation and material flow analysis towards a circular economy. Environ. Pollut. 2022, 315, 120334. [Google Scholar] [CrossRef] [PubMed]
- Orlando, M.; Molla, G.; Castellani, P.; Pirillo, V.; Torretta, V.; Ferronato, N. Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives. Int. J. Mol. Sci. 2023, 24, 3877. [Google Scholar] [CrossRef]
- Yu, C.; Dongsu, B.; Tao, Z.; Xintong, J.; Ming, C.; Siqi, W.; Zheng, S.; Yalei, Z. Anaerobic co-digestion of three commercial bio-plastic bags with food waste: Effects on methane production and microbial community structure. Sci. Total Environ. 2023, 859, 159967. [Google Scholar] [CrossRef] [PubMed]
- Swetha, T.A.; Ananthi, V.; Bora, A.; Sengottuvelan, N.; Ponnuchamy, K.; Muthusamy, G.; Arun, A. A review on biodegradable polylactic acid (PLA) production from fermentative food waste-Its applications and degradation. Int. J. Biol. Macromol. 2023, 234, 123703. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, L.; Wang, D.; Wang, L.; Zhang, Y.; Feng, S. Study on Rapid Detection Method for Degradation Performance of Polyolefin-Based Degradable Plastics. Polymers 2023, 15, 183. [Google Scholar] [CrossRef]
- Lai, J.; Huang, H.; Lin, M.; Xu, Y.; Li, X.; Sun, B. Enzyme catalyzes ester bond synthesis and hydrolysis: The key step for sustainable usage of plastics. Front. Microbiol. 2023, 13, 1113705. [Google Scholar] [CrossRef]
- Ghatge, S.; Yang, Y.; Ahn, J.-H.; Hur, H.-G. Biodegradation of polyethylene: A brief review. Appl. Biol. Chem. 2020, 63, 27. [Google Scholar] [CrossRef]
- Slezak, R.; Krzystek, L.; Puchalski, M.; Krucińska, I.; Sitarski, A. Degradation of bio-based film plastics in soil under natural conditions. Sci. Total Environ. 2023, 866, 161401. [Google Scholar] [CrossRef]
- Bhavsar, P.; Bhave, M.; Webb, H.K. Solving the plastic dilemma: The fungal and bacterial biodegradability of polyurethanes. World J. Microbiol. Biotechnol. 2023, 39, 122. [Google Scholar] [CrossRef]
- Machona, O.; Chidzwondo, F.; Mangoyi, R. Tenebrio molitor: Possible source of polystyrene-degrading bacteria. BMC Biotechnol. 2022, 22, 2. [Google Scholar] [CrossRef]
- Von Vacano, B.; Mangold, H.; Vandermeulen, G.W.M.; Battagliarin, G.; Hofmann, M.; Bean, J.; Künkel, A. Sustainable Design of Structural and Functional Polymers for a Circular Economy. Angew. Chem. Int. Ed. 2023, 62, e202210823. [Google Scholar] [CrossRef]
- Steiner, T.; Zhang, Y.; Möller, J.N.; Agarwal, S.; Löder, M.G.J.; Greiner, A.; Laforsch, C.; Freitag, R. Municipal biowaste treatment plants contribute to the contamination of the environment with residues of biodegradable plastics with putative higher persistence potential. Sci. Rep. 2022, 12, 9021. [Google Scholar] [CrossRef]
- Li, H.; Zhou, M.; Mohammed, A.E.A.Y.; Chen, L.; Zhou, C. From fruit and vegetable waste to degradable bioplastic films and advanced materials: A review. Sustain. Chem. Pharm. 2022, 30, 100859. [Google Scholar] [CrossRef]
- Al-Khairy, D.; Fu, W.; Alzahmi, A.S.; Twizere, J.-C.; Amin, S.A.; Salehi-Ashtiani, K.; Mystikou, A. Closing the Gap between Bio-Based and Petroleum-Based Plastic through Bioengineering. Microorganisms 2022, 10, 2320. [Google Scholar] [CrossRef]
- Easton, Z.H.W.; Essink, M.A.J.; Comas, L.R.; Wurm, F.R.; Gojzewski, H. Acceleration of Biodegradation Using Polymer Blends and Composites. Macromol. Chem. Phys. 2023, 224, 2200421. [Google Scholar] [CrossRef]
- Grossule, V.; Zanatta, S.; Modesti, M.; Lavagnolo, M.C. Treatment of food waste contaminated by bioplastics using BSF larvae: Impact and fate of starch-based bioplastic films. J. Environ. Manag. 2023, 330, 117229. [Google Scholar] [CrossRef]
- Aleksanyan, K.V. Polysaccharides for Biodegradable Packaging Materials: Past, Present, and Future (Brief Review). Polymers 2023, 15, 451. [Google Scholar] [CrossRef]
- Bulatović, V.O.; Grgić, D.K.; Mandić, V.; Miloloža, M.; Dybal, J.; Gajdosova, V.; Slouf, M. Biodegradation of LDPE_TPS blends under controlled composting conditions. Polym. Bull. 2022, 80, 3331–3357. [Google Scholar] [CrossRef]
- Perez-Puyana, V.; Jiménez-Rosado, M.; Escribano, D.; Romero, A.; Martínez, I. Influence of the Aliphatic Chain Length on the Crosslinking Properties of Aldehydes on Sustainable Bioplastics Obtained from Pea Protein. J. Polym. Environ. 2022, 30, 5163–5172. [Google Scholar] [CrossRef]
- Hogan, K.J.; Mikos, A.G. Biodegradable thermoresponsive polymers: Applications in drug delivery and tissue engineering. Polymer 2020, 211, 123063. [Google Scholar] [CrossRef]
- Chen, X.; Wu, B.; Perera, H.A.; Yan, M. Synthesis of Glycopolymer Micelles for Antibiotic Delivery. Molecules 2023, 28, 4031. [Google Scholar] [CrossRef]
- Mei, R.; Heng, X.; Liu, X.; Chen, G. Glycopolymers for Antibacterial and Antiviral Applications. Molecules 2023, 28, 985. [Google Scholar] [CrossRef]
- Zashikhina, N.; Levit, M.; Dobrodumov, A.; Gladnev, S.; Lavrentieva, A.; Tennikova, T.; Korzhikova-Vlakh, E. Biocompatible Nanoparticles Based on Amphiphilic Random Polypeptides and Glycopolymers as Drug Delivery Systems. Polymers 2022, 14, 1677. [Google Scholar] [CrossRef]
- Pană, A.-M.; Rusnac, L.-M.; Bandur, G.; Silion, M.; Deleanu, C.; Bălan, M. Novel D-glucose and D-mannose based oligomers: Synthesis and characterization. E-Polymers 2011, 11, 29–42. [Google Scholar] [CrossRef] [Green Version]
- Pană, A.-M.; Gherman, V.; Sfîrloagă, P.; Bandur, G.; Ştefan, L.-M.; Popa, M.; Rusnac, L.-M. Thermal stability and biodegradation of novel D-mannose based glycopolymers. Polym. Test. 2012, 31, 384–392. [Google Scholar] [CrossRef]
- Pană, A.-M.; Ştefan, L.-M.; Bandur, G.; Sfîrloagă, P.; Gherman, V.; Silion, M.; Popa, M.; Rusnac, L.-M. Novel Glycopolymers Based on d-Mannose and Methacrylates: Synthesis, Thermal Stability and Biodegradability Testing. J. Polym. Environ. 2013, 21, 981–994. [Google Scholar] [CrossRef]
- Pană, A.M.; Rusnac, L.M.; Bandur, G.; Deleanu, C.; Bălan, M.; Silion, M. Synthesis and characterization of new glycopolymers based on monosaccharides and maleic anhydride. II. Mannose derivatives. Mat. Plast. 2010, 47, 299–305. [Google Scholar]
- Stanila, A.-L.; Simota, C.C.; Dumitru, M. Contributions to the Knowledge of Sandy Soils from Oltenia Plain. Rev. Chim. 2020, 71, 192–200. [Google Scholar] [CrossRef]
- Calina, J.; Calina, A. Evolution of the Mollic Reddish Preluvisol in a Romanian Riverine Region and the assessment of its agro-productive properties in forms and agro-touristic households. Environ. Eng. Manag. J. 2019, 18, 2729–2738. [Google Scholar] [CrossRef]
- Kiani, D.; Santus, W.; Kiernan, K.A.; Behnsen, J. Proteus mirabilis Employs a Contact-Dependent Killing System against Competing Enterobacteriaceae. Msphere 2021, 6, e0032121. [Google Scholar] [CrossRef]
- Gmiter, D.; Kaca, W. Into the understanding the multicellular lifestyle of Proteus mirabilis on solid surfaces. Front. Cell. Infect. Microbiol. 2022, 12, 864305. [Google Scholar] [CrossRef]
- Stickler, D.J. 7—Proteus mirabilis biofilm formation and catheter design. In Woodhead Publishing Series in Biomaterials, Biomaterials and Tissue Engineering in Urology; Denstedt, J., Atale, A., Eds.; Woodhead Publishing: Sawston, UK, 2009; pp. 157–190. [Google Scholar]
- Babaeva, A.A.; Grigorieva, E.V. Household Waste Recycling Technologies. IOP Conf. Series Earth Environ. Sci. 2021, 720, 012005. [Google Scholar] [CrossRef]
- Li, J.; Song, G.; Cai, M.; Bian, J.; Mohammed, B.S. Green environment and circular economy: A state-of-the-art analysis. Sustain. Energy Technol. Assess. 2022, 52, 102106. [Google Scholar] [CrossRef]
- Tores, N.; Santos, G. The (mathematical) modeling process in biosciences. Front. Genet. 2015, 6, 354. [Google Scholar]
- Asuero, A.G.; Sayago, A.; González, A.G. The Correlation Coefficient: An Overview. Crit. Rev. Anal. Chem. 2006, 36, 41–59. [Google Scholar] [CrossRef]
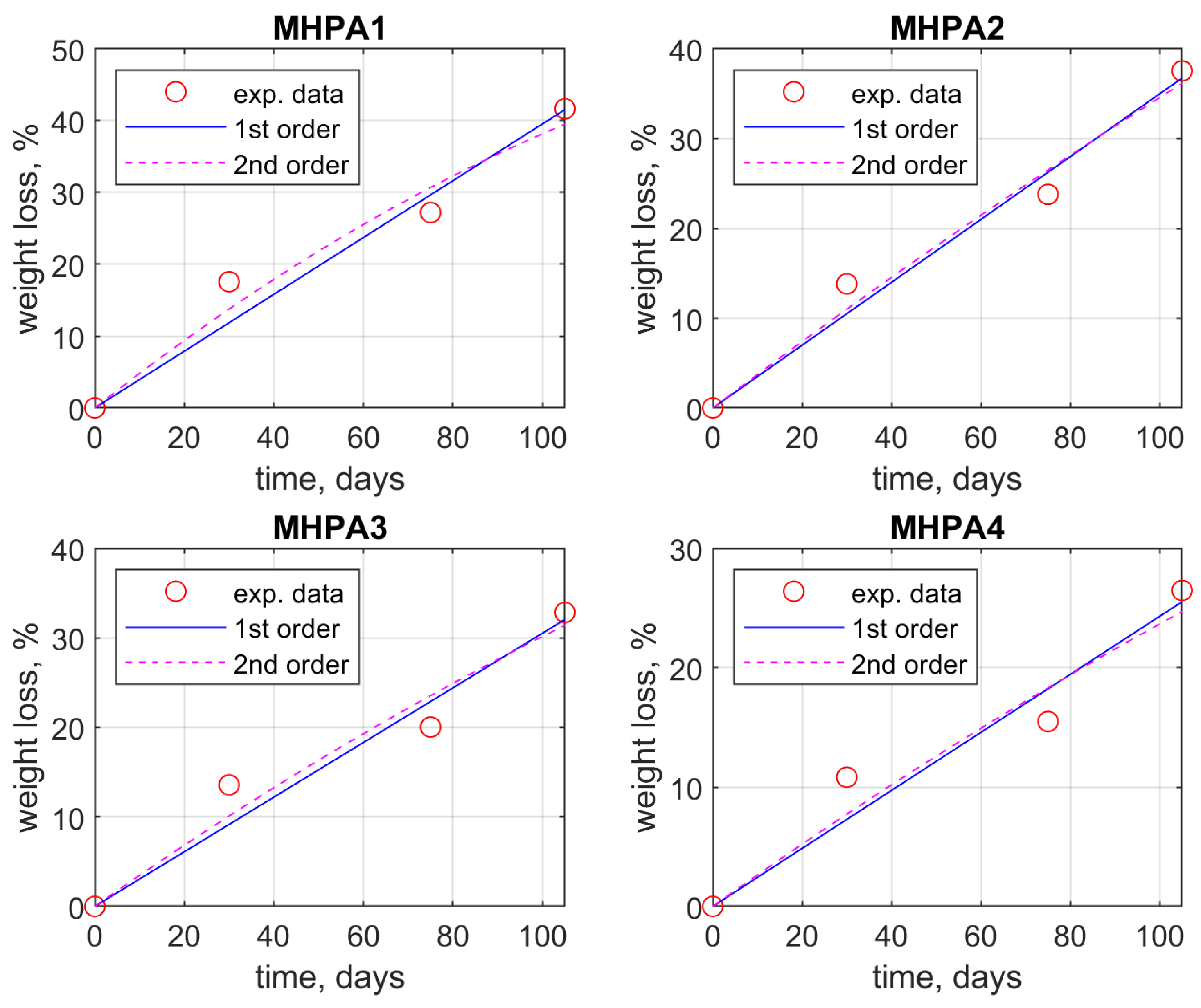
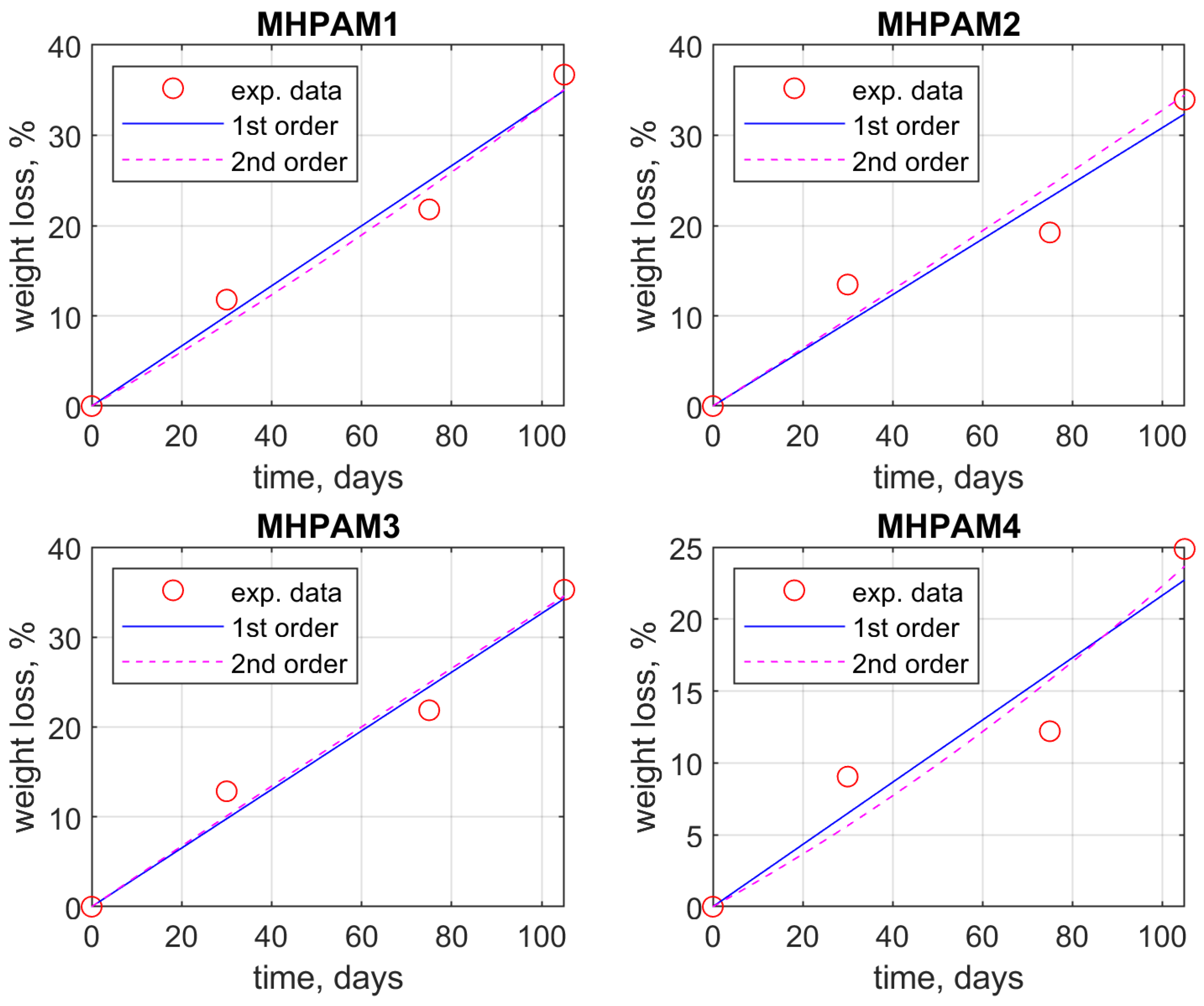
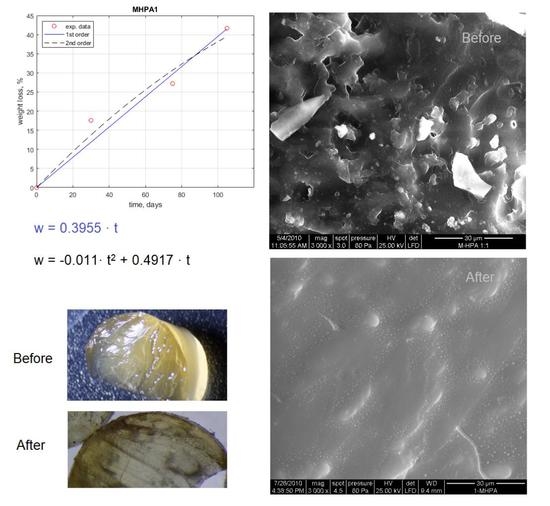
| Sample | Weight Loss, % | ||||
|---|---|---|---|---|---|
| Time, Days | 0 | 30 | 75 | 105 | |
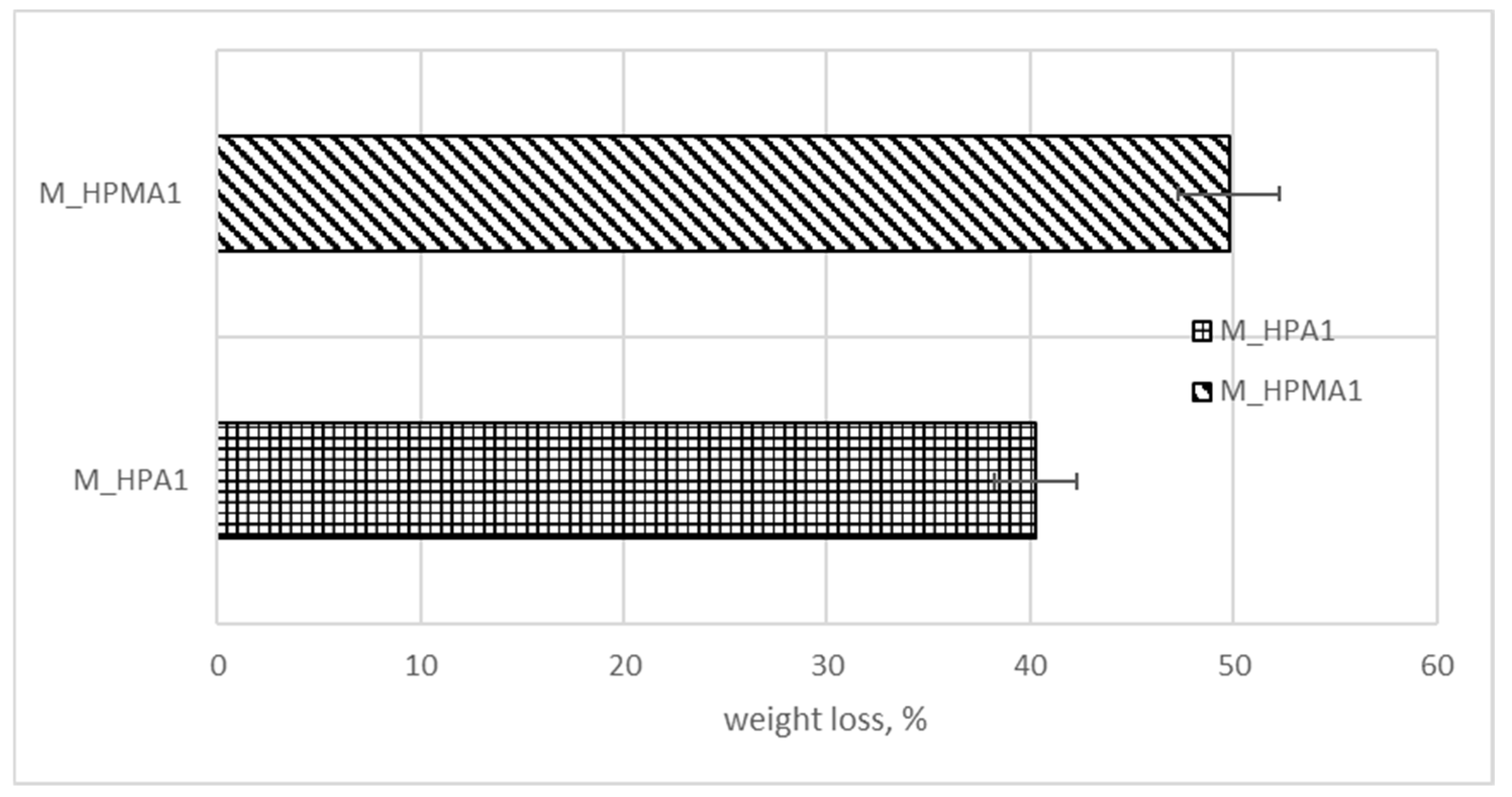
| M_HPA1 | 0 | 17.57 | 27.2 | 41.65 | |
| M_HPA2 | 0 | 13.82 | 23.79 | 37.54 | |
| M_HPA3 | 0 | 13.58 | 20.04 | 32.84 | |
| M_HPA4 | 0 | 10.82 | 15.5 | 26.47 | |
| M_HPMA1 | 0 | 11.79 | 21.79 | 36.72 | |
| M_HPMA2 | 0 | 13.46 | 19.23 | 33.95 | |
| M_HPMA3 | 0 | 12.85 | 21.87 | 35.26 | |
| M_HPMA4 | 0 | 9.05 | 12.21 | 24.87 | |
| Sample | Mathematical Model | Root-Mean Standard Deviation (RMSD) | Correlation Coefficient (R2) |
|---|---|---|---|
| M_HPA1 | w = 0.3955 · t w = −0.011 · t2 + 0.4917 · t | 3.5882 3.9334 | 0.9578 0.9662 |
| M_HPA2 | w = 0.3499 · t w = −0.0003 · t2 + 0.3757 · t | 2.4288 2.9278 | 0.9765 0.9772 |
| M_HPA3 | w = 0.3053 · t w = −0.0005 · t2 + 0.3516 · t | 3.0727 3.6435 | 0.9494 0.9526 |
| M_HPA4 | w = 0.2431 · t w = −0.0003 · t2 + 0.2667 · t | 2.6330 3.1889 | 0.9424 0.9437 |
| Sample | Mathematical Model | Root-Mean Standard Deviation (RMSD) | Correlation Coefficient (R2) |
|---|---|---|---|
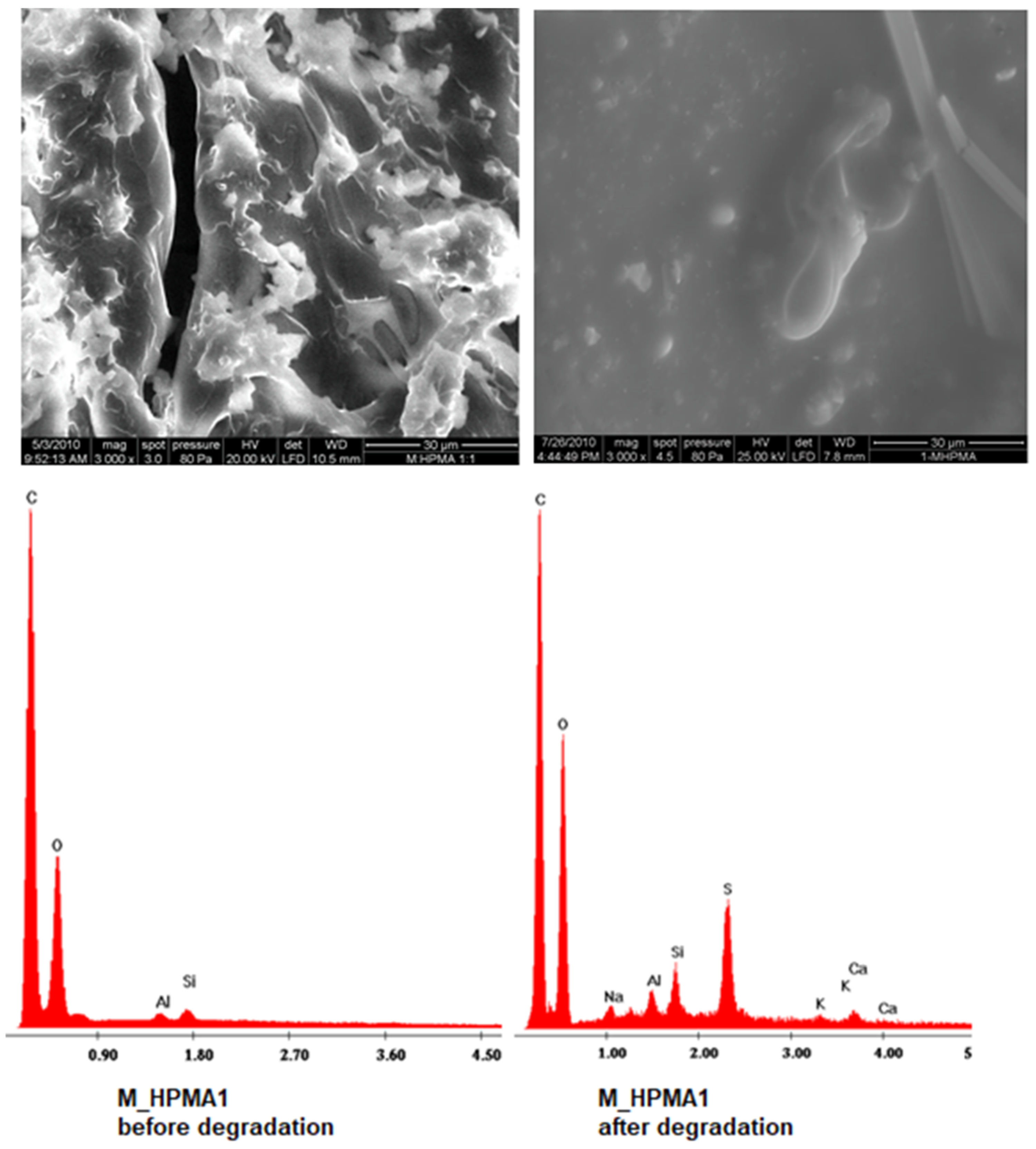
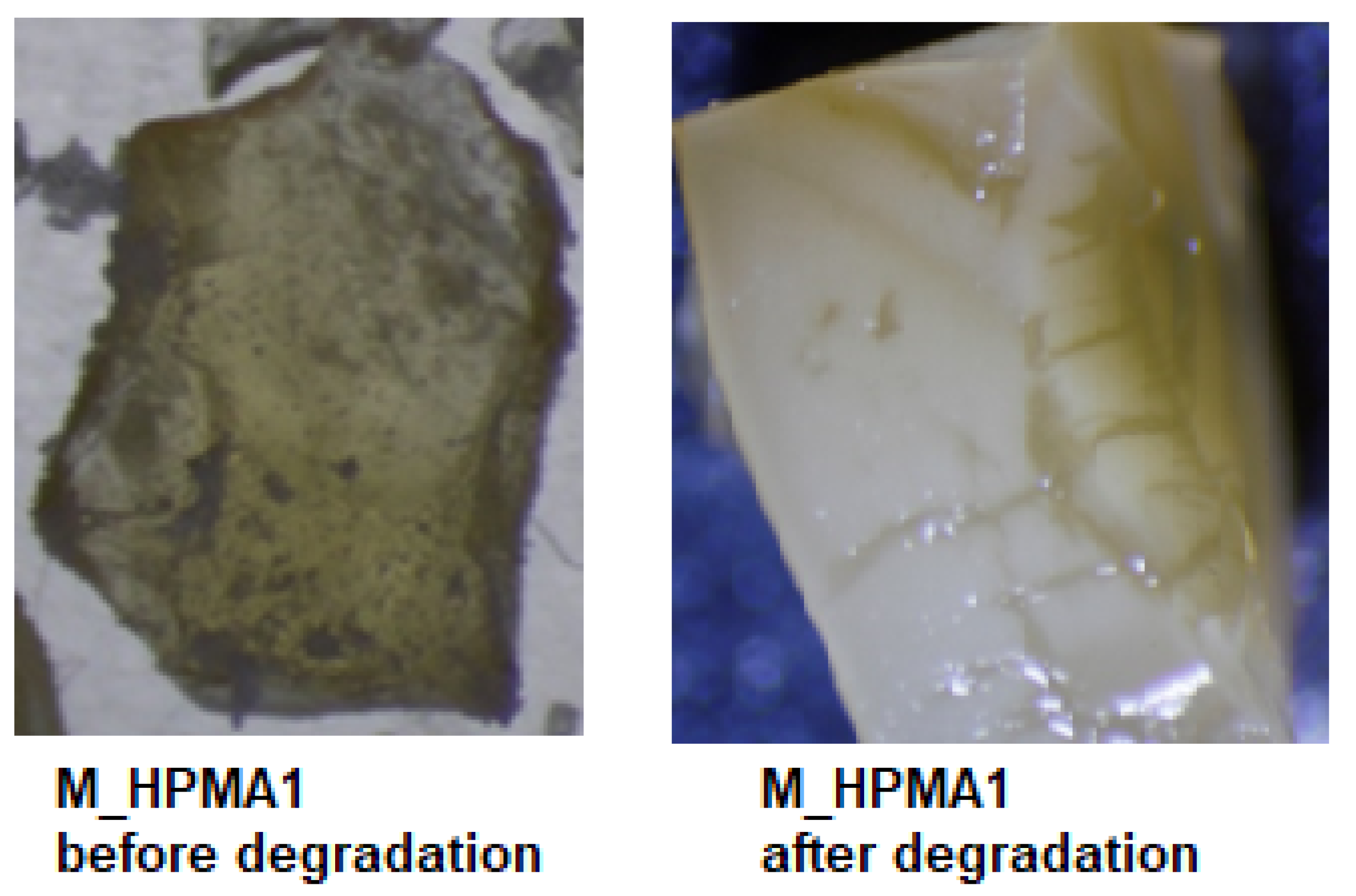
| M_HPMA1 | w = 0.3330 · t w = 0.0004 · t2 + 0.2920 · t | 2.3427 2.7453 | 0.9773 0.9793 |
| M_HPMA2 | w = 0.3083 · t w = 0.0001 · t2 + 0.3177 · t | 3.4339 4.2013 | 0.9404 0.9405 |
| M_HPMA3 | w = 0.3264 · t w = −0.0001 · t2 + 0.3397 · t | 2.3902 2.9148 | 0.9741 0.9743 |
| M_HPMA4 | w = 0.2164 · t w = 0.0005 · t2 + 0.1729 · t | 3.0176 3.5881 | 0.9140 0.9189 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pană, A.-M.; Ordodi, V.; Gherman, V.; Sfîrloagă, P.; Dumitrel, G.-A. Study on the Biodegradation Process of D-Mannose Glycopolymers in Liquid Media and Soil. Polymers 2023, 15, 3194. https://doi.org/10.3390/polym15153194
Pană A-M, Ordodi V, Gherman V, Sfîrloagă P, Dumitrel G-A. Study on the Biodegradation Process of D-Mannose Glycopolymers in Liquid Media and Soil. Polymers. 2023; 15(15):3194. https://doi.org/10.3390/polym15153194
Chicago/Turabian StylePană, Ana-Maria, Valentin Ordodi, Vasile Gherman, Paula Sfîrloagă, and Gabriela-Alina Dumitrel. 2023. "Study on the Biodegradation Process of D-Mannose Glycopolymers in Liquid Media and Soil" Polymers 15, no. 15: 3194. https://doi.org/10.3390/polym15153194
APA StylePană, A.-M., Ordodi, V., Gherman, V., Sfîrloagă, P., & Dumitrel, G.-A. (2023). Study on the Biodegradation Process of D-Mannose Glycopolymers in Liquid Media and Soil. Polymers, 15(15), 3194. https://doi.org/10.3390/polym15153194