Characterization of Carboxylic Acid Reductase from Mycobacterium phlei Immobilized onto Seplite LX120
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Equipment
2.2. Preparation of Purified Recombinant MpCAR
2.3. Protein Content Determination
2.4. Enzyme Activity Assay of MpCAR
2.5. Immobilization of Purified MpCAR
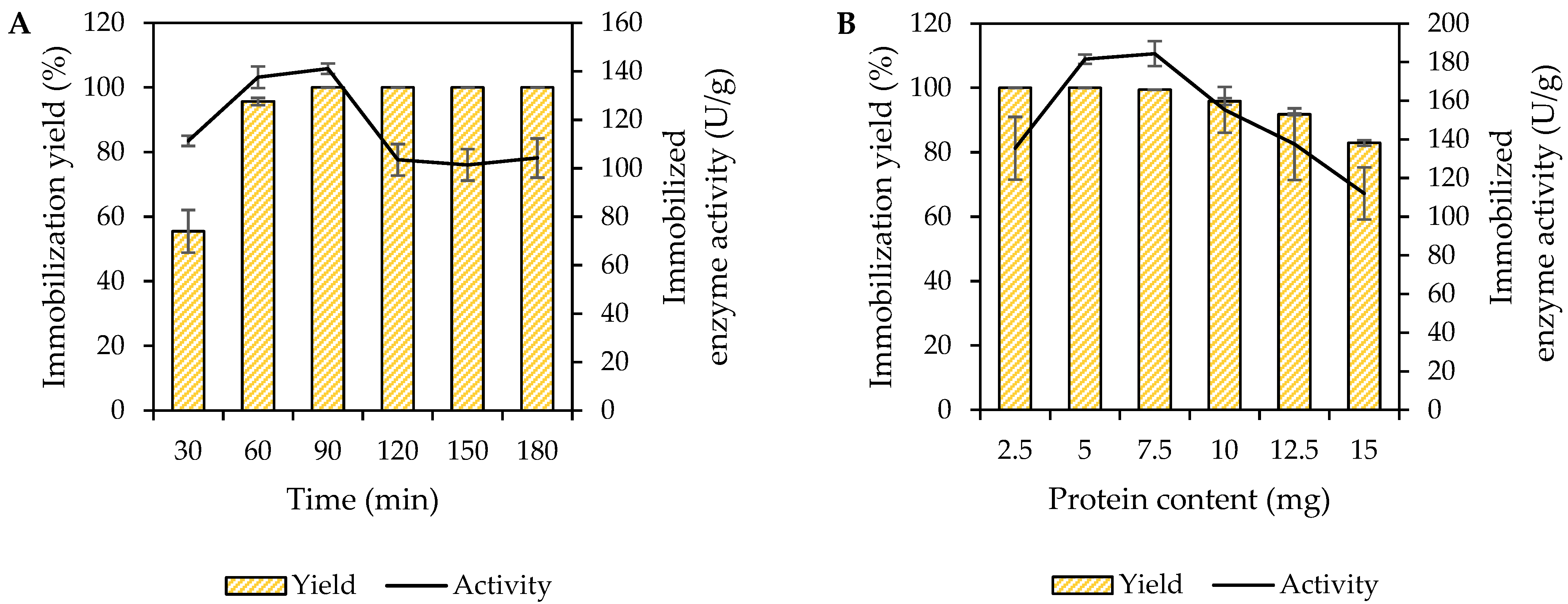
2.5.1. Effect of Immobilization Time of MpCAR
2.5.2. Effect of Immobilization Protein Load of MpCAR
2.6. Characterization of the Immobilized and Free MpCAR
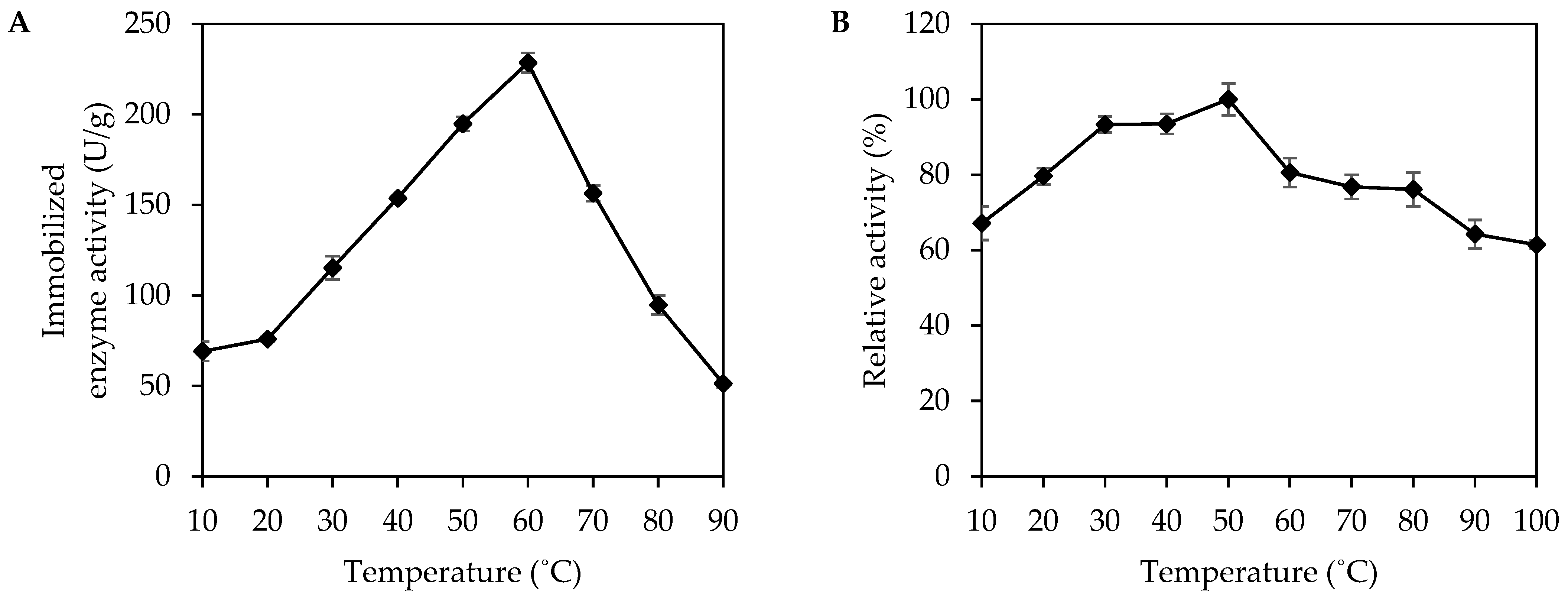
2.6.1. Effect of Temperature
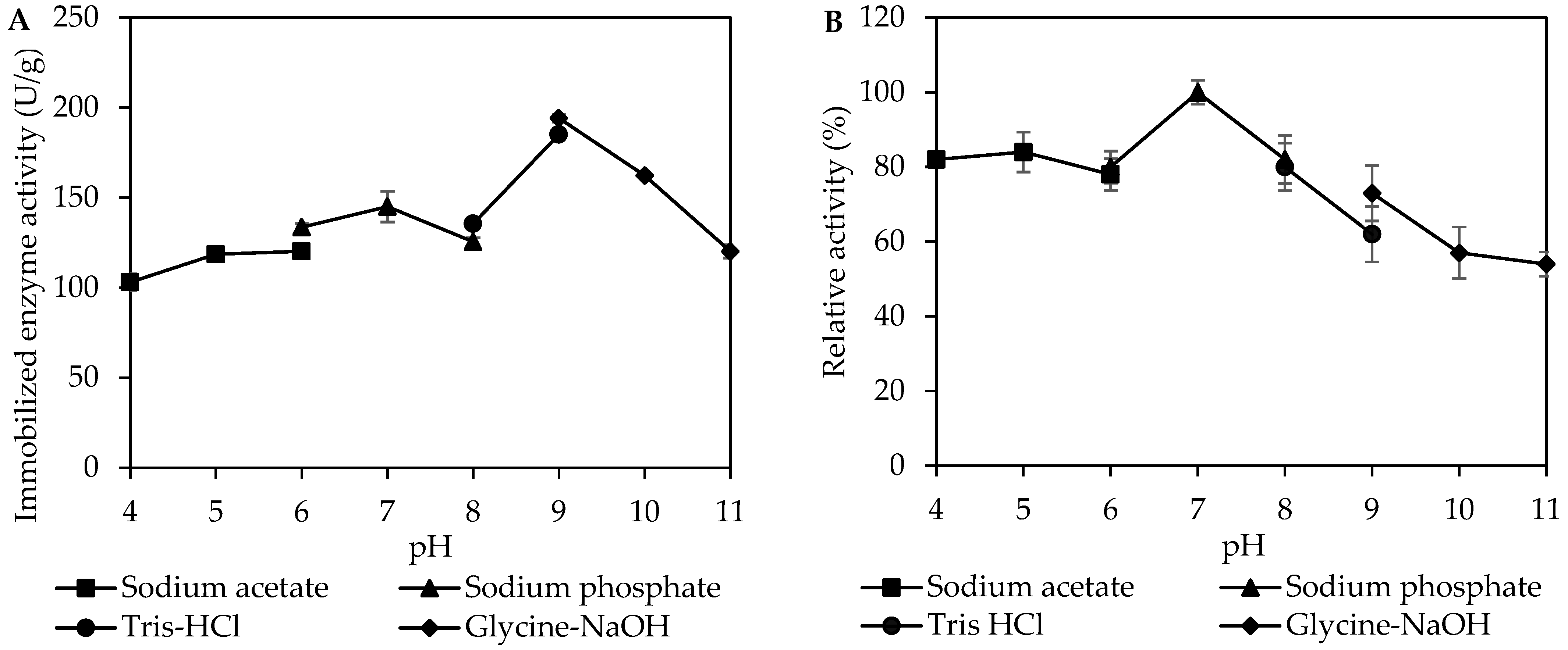
2.6.2. Effect of pH
2.6.3. Effect of Organic Solvents
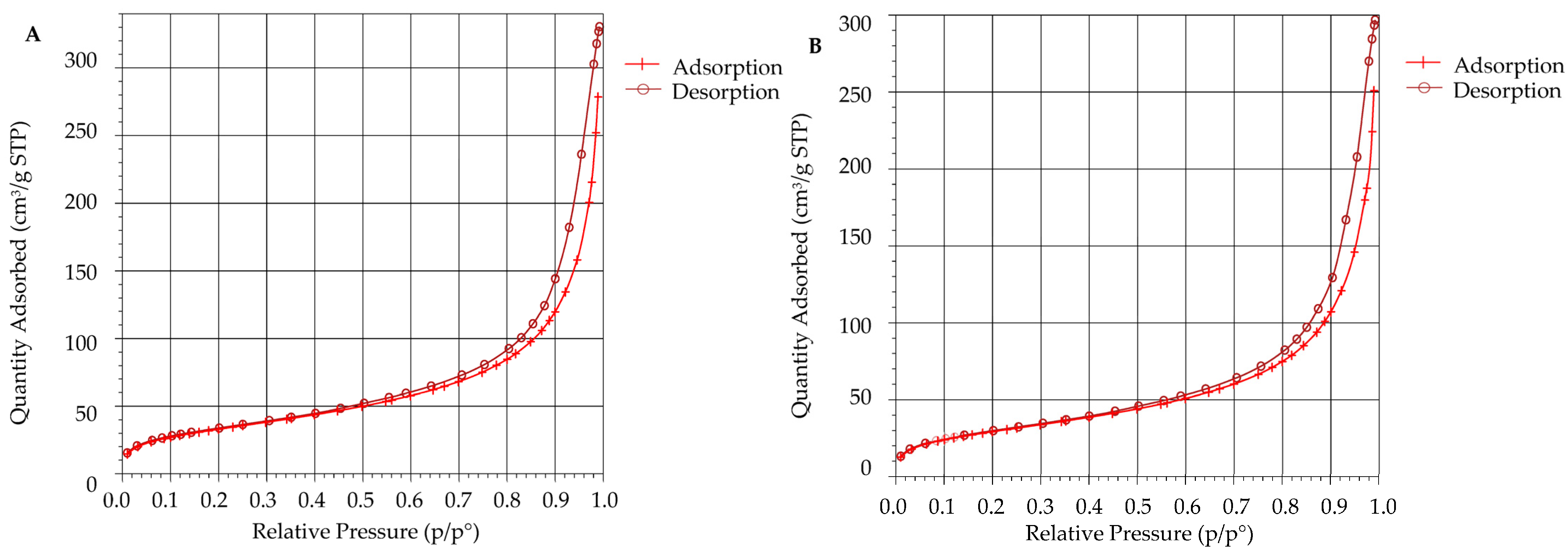
2.6.4. Scanning Electron Microscopy (SEM) and Brunauer–Emmett–Teller (BET) Analysis
2.6.5. Fourier-Transform Infrared (FTIR) Spectroscopy
2.6.6. Storage Stability and Reusability Study
2.7. Bioconversion Analysis of Immobilized MpCAR Using High-Performance Liquid Chromatography (HPLC)
3. Results and Discussion
3.1. Immobilization of MpCAR
3.2. Effect of Temperature on Activity and Stability of Immobilized MpCAR
3.3. Effect of pH on Activity and Stability of Immobilized MpCAR
3.4. Effect of Organic Solvents on the Stability of Immobilized MpCAR
3.5. Morphology Analysis Using SEM
3.6. Surface Area Analysis Using BET
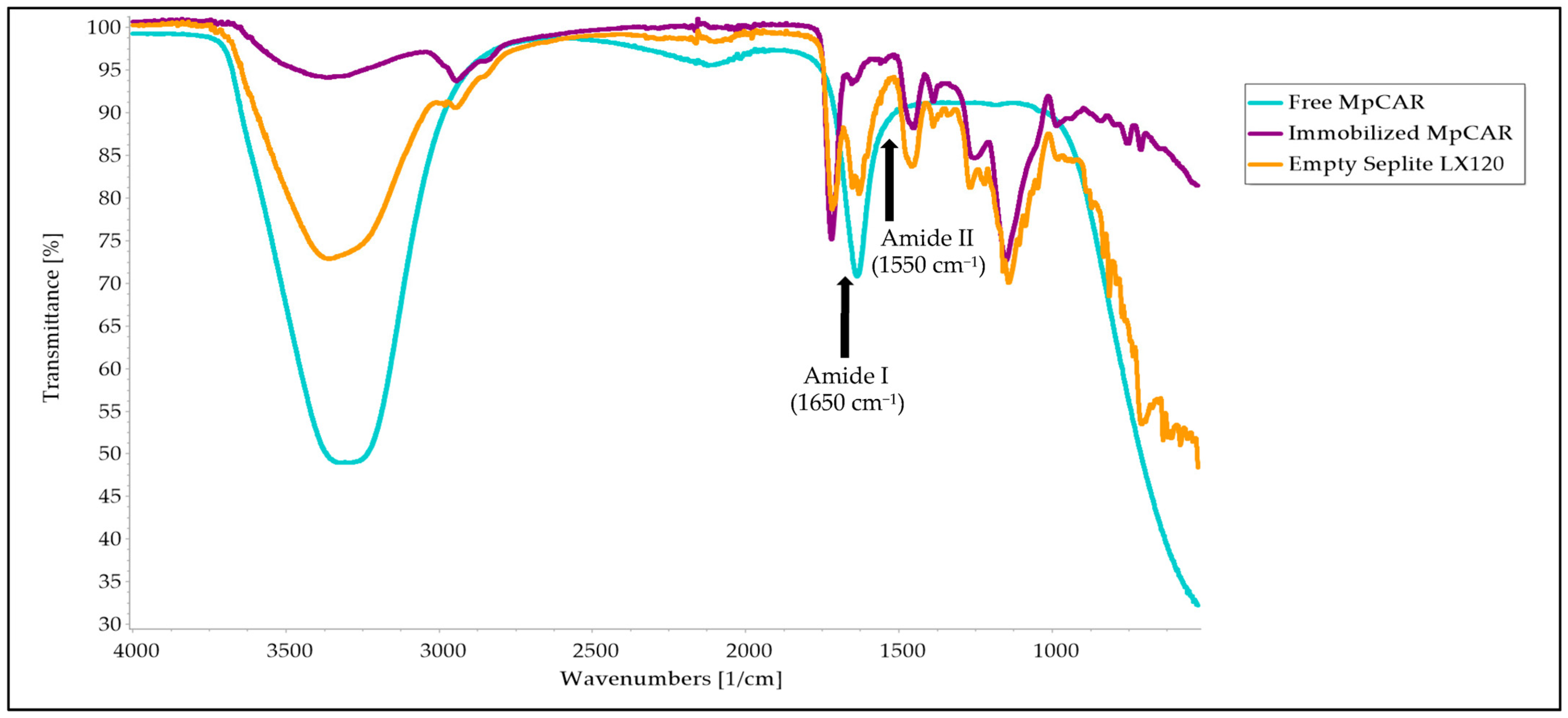
3.7. Structural Analysis Using FTIR
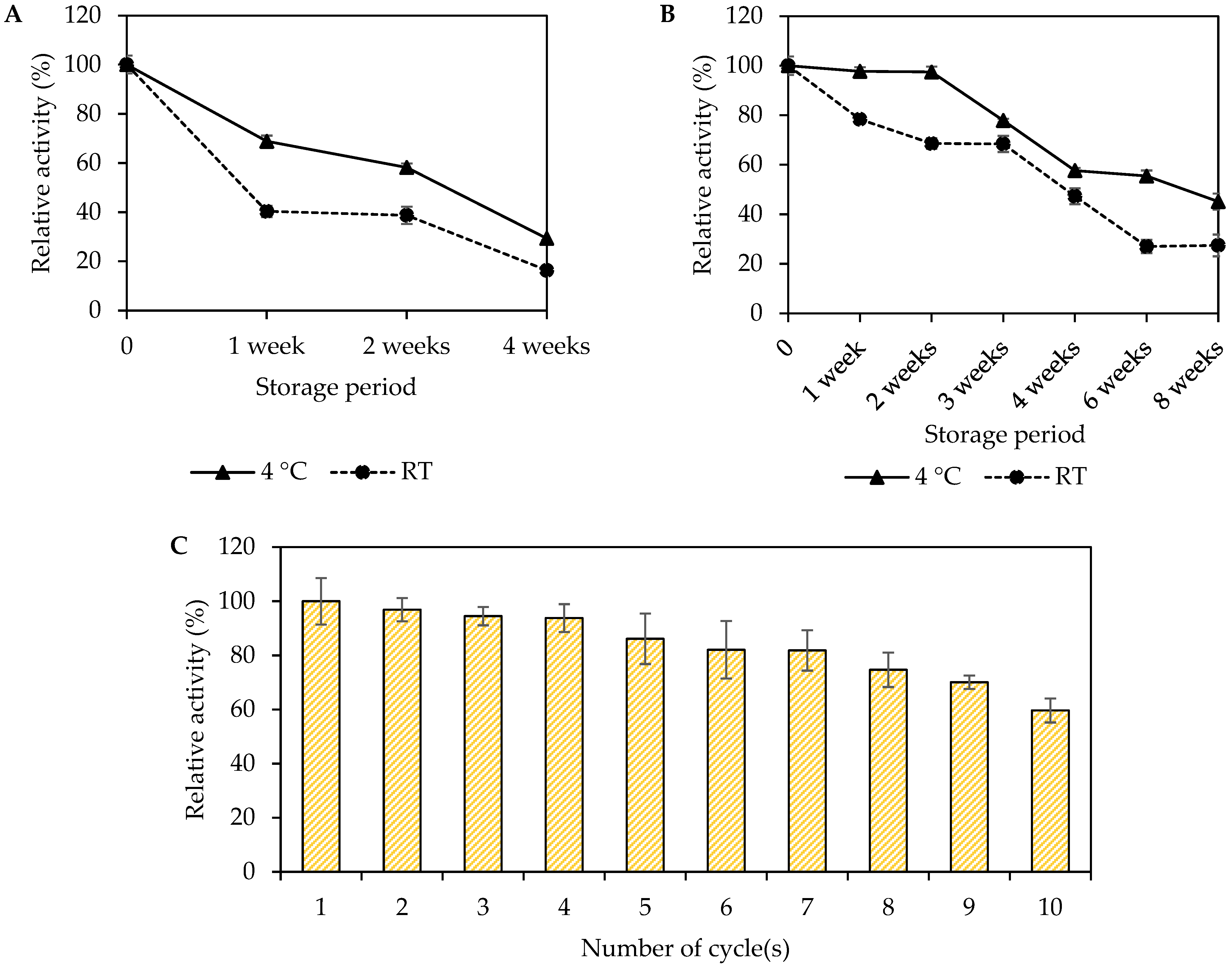
3.8. Storage Stability and Reusability
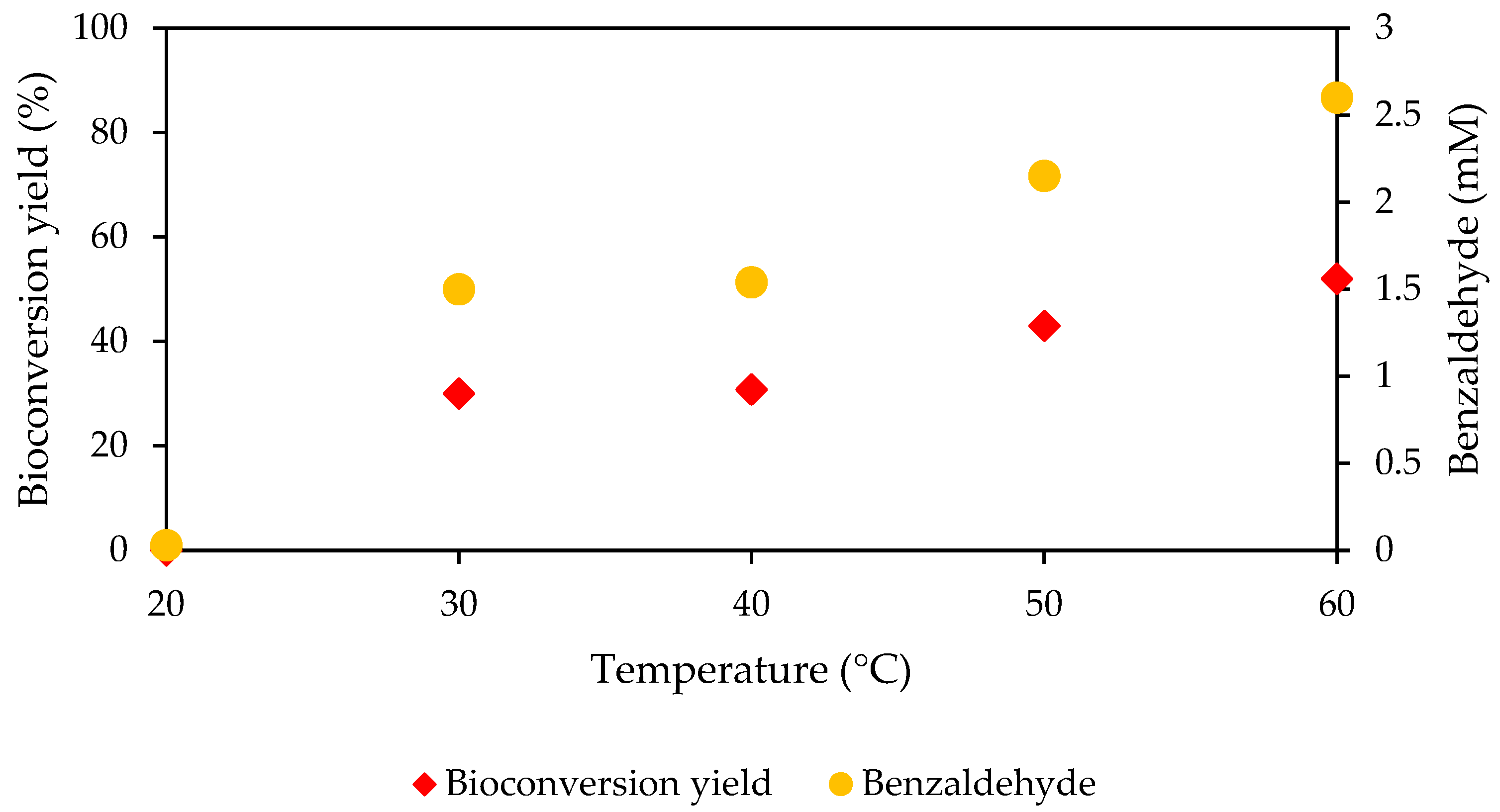
3.9. Bioconversion Using Immobilized MpCAR
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Napora-Wijata, K.; Strohmeier, G.A.; Winkler, M. Biocatalytic reduction of carboxylic acids. Biotechnol. J. 2014, 9, 822–843. [Google Scholar] [CrossRef]
- Li, T.; Rosazza, J.P.N. Purification, characterization, and properties of an aryl aldehyde oxidoreductase from Nocardia sp. strain NRRL 5646. J. Bacteriol. 1997, 179, 3482–3487. [Google Scholar] [CrossRef]
- Kunjapur, A.M.; Prather, K.L.J. Microbial engineering for aldehyde synthesis. Appl. Environ. Microbiol. 2015, 81, 1892–1901. [Google Scholar] [CrossRef]
- Kato, N.; Joung, E.H.; Yang, H.C.; Masuda, M.; Shimao, M.; Yanase, H. Purification and characterization of aromatic acid reductase from Nocardia asteroides JCM 3016. Agric. Biol. Chem. 1991, 55, 757–762. [Google Scholar] [CrossRef]
- Gahloth, D.; Aleku, G.A.; Leys, D. Carboxylic acid reductase: Structure and mechanism. J. Biotechnol. 2020, 307, 107–113. [Google Scholar] [CrossRef]
- He, A.; Li, T.; Daniels, L.; Fotheringham, I.; Rosazza, J.P.N. Nocardia sp. carboxylic acid reductase: Cloning, expression, and characterization of a new aldehyde oxidoreductase family. Appl. Environ. Microbiol. 2004, 70, 1874–1881. [Google Scholar] [CrossRef]
- Venkitasubramanian, P.; Daniels, L.; Rosazza, J.P.N. Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J. Biol. Chem. 2007, 282, 478–485. [Google Scholar] [CrossRef]
- Qu, G.; Guo, J.; Yang, D.; Sun, Z. Biocatalysis of carboxylic acid reductases: Phylogenesis, catalytic mechanism and potential applications. Green Chem. 2018, 20, 777–792. [Google Scholar] [CrossRef]
- Atsumi, S.; Wu, T.-Y.; Eckl, E.-M.; Hawkins, S.D.; Buelter, T.; Liao, J.C. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2010, 85, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Kramer, L.; Hankore, E.D.; Liu, Y.; Liu, K.; Jimenez, E.; Guo, J.; Niu, W. Characterization of carboxylic acid reductases for biocatalytic synthesis of industrial chemicals. ChemBioChem 2018, 19, 1452–1460. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, M.J.; Kunjapur, A.M.; Wenck, S.J.; Prather, K.L.J. Retro-biosynthetic screening of a modular pathway design achieves selective route for microbial synthesis of 4-methyl-pentanol. Nat. Commun. 2014, 5, 5031. [Google Scholar] [CrossRef] [PubMed]
- Napora-Wijata, K.; Robins, K.; Osorio-Lozada, A.; Winkler, M. Whole-cell carboxylate reduction for the synthesis of 3-hydroxytyrosol. ChemCatChem 2014, 6, 1089–1095. [Google Scholar] [CrossRef]
- Venkitasubramanian, P.; Daniels, L.; Das, S.; Lamm, A.S.; Rosazza, J.P.N. Aldehyde oxidoreductase as a biocatalyst: Reductions of vanillic acid. Enzyme Microb. Technol. 2008, 42, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech. 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. Developments in support materials for immobilization of oxidoreductases: A comprehensive review. Adv. Colloid Interface Sci. 2018, 258, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.Y.; Li, F.L.; Zhang, Y.W.; Gupta, R.K.; Patel, S.K.S.; Lee, J.K. Recent strategies for the immobilization of therapeutic enzymes. Polymers 2022, 14, 1409. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.S.; Choi, H.; Lee, J.K. Multimetal-based inorganic-protein hybrid system for enzyme immobilization. ACS Sustain. Chem. Eng. 2019, 7, 13633–13638. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Anwar, M.Z.; Kumar, A.; Otari, S.V.; Pagolu, R.T.; Kim, S.Y.; Kim, I.W.; Lee, J.K. Fe2O3 yolk-shell particle-based laccase biosensor for efficient detection of 2,6-dimethoxyphenol. Biochem. Eng. J. 2018, 132, 1–8. [Google Scholar] [CrossRef]
- Lyu, X.; Gonzalez, R.; Horton, A.; Li, T. Immobilization of enzymes by polymeric materials. Catalysts 2021, 11, 1211. [Google Scholar] [CrossRef]
- Jia, Y.; Chen, Y.; Luo, J.; Hu, Y. Immobilization of laccase onto meso-MIL-53 (Al) via physical adsorption for the catalytic conversion of triclosan. Ecotoxicol. Environ. Saf. 2019, 184, 109670. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.M.; Chen, J.; Shi, Y.P. Tyrosinase immobilization on aminated magnetic nanoparticles by physical adsorption combined with covalent crosslinking with improved catalytic activity, reusability and storage stability. Anal. Chim. Acta 2018, 1006, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Mostafa, F.A.; Ouis, M.A. Enhancement stability and catalytic activity of immobilized α-amylase using bioactive phospho-silicate glass as a novel inorganic support. Int. J. Biol. Macromol. 2018, 112, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Liu, Z.; Zeng, G.; Liu, Y.; Yang, X.; Zhou, C.; Chen, M.; Liu, Y.; Jiang, Y.; Yan, M. Immobilization of laccase on hollow mesoporous carbon nanospheres: Noteworthy immobilization, excellent stability and efficacious for antibiotic contaminants removal. J. Hazard. Mater. 2019, 362, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Kuribayashi, L.M.; Ribeiro, V.P.D.R.; de Santana, R.C.; Ribeiro, E.J.; dos Santos, M.G.; Falleiros, L.N.S.S.; Guidini, C.Z. Immobilization of β-galactosidase from Bacillus licheniformis for application in the dairy industry. Appl. Microbiol. Biotechnol. 2021, 105, 3601–3610. [Google Scholar] [CrossRef]
- Guidini, C.Z.; Fischer, J.; Santana, L.N.S.; Cardoso, V.L.; Ribeiro, E.J. Immobilization of Aspergillus oryzae β-galactosidase in ion exchange resins by combined ionic-binding method and cross-linking. Biochem. Eng. J. 2010, 52, 137–143. [Google Scholar] [CrossRef]
- Finnigan, W.; Thomas, A.; Cromar, H.; Gough, B.; Snajdrova, R.; Adams, J.P.; Littlechild, J.A.; Harmer, N.J. Characterization of carboxylic acid reductases as enzymes in the toolbox for synthetic chemistry. ChemCatChem 2017, 9, 1005–1017. [Google Scholar] [CrossRef]
- Thomas, A.; Cutlan, R.; Finnigan, W.; van der Mark, G.; Harmer, N. Highly thermostable carboxylic acid reductases generated by ancestral sequence reconstruction. Commun. Biol. 2019, 2, 429. [Google Scholar] [CrossRef]
- Finnigan, W.; Cutlan, R.; Snajdrova, R.; Adams, J.P.; Littlechild, J.A.; Harmer, N.J. Engineering a seven enzyme biotransformation using mathematical modelling and characterized enzyme parts. ChemCatChem 2019, 11, 3474–3489. [Google Scholar] [CrossRef]
- Thompson, M.P.; Derrington, S.R.; Heath, R.S.; Porter, J.L.; Mangas-Sanchez, J.; Devine, P.N.; Truppo, M.D.; Turner, N.J. A generic platform for the immobilisation of engineered biocatalysts. Tetrahedron 2019, 75, 327–334. [Google Scholar] [CrossRef]
- Maphatsoe, M.M.; Hashem, C.; Ling, J.G.; Horvat, M.; Rumbold, K.; Bakar, F.D.A.; Winkler, M. Characterization and immobilization of Pycnoporus cinnabarinus carboxylic acid reductase, PcCAR2. J. Biotechnol. 2022, 345, 47–54. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 254, 248–254. [Google Scholar] [CrossRef]
- Akhtara, M.K.; Turner, N.J.; Jones, P.R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. USA 2013, 110, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Horvat, M.; Winkler, M. In vivo reduction of medium- to long-chain fatty acids by carboxylic acid reductase (CAR) enzymes: Limitations and solutions. ChemCatChem 2020, 12, 5076–5090. [Google Scholar] [CrossRef]
- Costa, V.M.; de Souza, M.C.M.; Fechine, P.B.A.; Macedo, A.C.; Gonçalves, L.R.B. Nanobiocatalytic systems based on lipase-Fe3O4 and conventional systems for isoniazid synthesis: A comparative study. Braz. J. Chem. Eng. 2016, 33, 661–673. [Google Scholar] [CrossRef]
- Liu, G.; Sun, L.; Wu, X.; Zhang, W.; Feng, J.; Cui, Y.; Lu, Z.; Shen, J.; Liu, Z.; Yuan, S. Immobilization of puerarin glycosidase from Microbacterium oxydans CGMCC 1788 increases puerarin transformation efficiency. Braz. J. Chem. Eng. 2014, 31, 325–333. [Google Scholar] [CrossRef]
- Keerti; Gupta, A.; Kumar, V.; Dubey, A.; Verma, A.K. Kinetic characterization and effect of immobilized thermostable β-glucosidase in alginate gel beads on sugarcane juice. ISRN Biochem. 2014, 2014, 178498. [Google Scholar] [CrossRef]
- Wu, E.; Li, Y.; Huang, Q.; Yang, Z.; Wei, A.; Hu, Q. Laccase immobilization on amino-functionalized magnetic metal organic framework for phenolic compound removal. Chemosphere 2019, 233, 327–335. [Google Scholar] [CrossRef]
- Bayramoǧlu, G.; Yalçin, E.; Arica, M.Y. Immobilization of urease via adsorption onto L-histidine-Ni(II) complexed poly(HEMA-MAH) microspheres: Preparation and characterization. Process Biochem. 2005, 40, 3505–3513. [Google Scholar] [CrossRef]
- Erginer, R.; Toppare, L.; Alkan, S.; Bakir, U. Immobilization of invertase in functionalized copolymer matrices. React. Funct. Polym. 2000, 45, 227–233. [Google Scholar] [CrossRef]
- Hung, T.C.; Giridhar, R.; Chiou, S.H.; Wu, W.T. Binary immobilization of Candida rugosa lipase on chitosan. J. Mol. Catal. B Enzym. 2003, 26, 69–78. [Google Scholar] [CrossRef]
- Miao, Q.; Zhang, C.; Zhou, S.; Meng, L.; Huang, L.; Ni, Y.; Chen, L. Immobilization and characterization of pectinase onto the cationic polystyrene resin. ACS Omega 2021, 6, 31683–31688. [Google Scholar] [CrossRef]
- Cao, M.; Li, Z.; Wang, J.; Ge, W.; Yue, T.; Li, R.; Colvin, V.L.; Yu, W.W. Food related applications of magnetic iron oxide nanoparticles: Enzyme immobilization, protein purification, and food analysis. Trends Food Sci. Technol. 2012, 27, 47–56. [Google Scholar] [CrossRef]
- Almulaiky, Y.Q.; Al-Harbi, S.A. A novel peroxidase from Arabian balsam (Commiphora gileadensis) stems: Its purification, characterization and immobilization on a carboxymethylcellulose/Fe3O4 magnetic hybrid material. Int. J. Biol. Macromol. 2019, 133, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef]
- Wang, S.; Meng, X.; Zhou, H.; Liu, Y.; Secundo, F.; Liu, Y. Enzyme stability and activity in non-aqueous reaction systems: A mini review. Catalysts 2016, 6, 32. [Google Scholar] [CrossRef]
- Trodler, P.; Pleiss, J. Modeling structure and flexibility of Candida antarctica lipase B in organic solvents. BMC Struct. Biol. 2008, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Fasoli, E.; Ferrer, A.; Barletta, G.L. Hydrogen/deuterium exchange study of subtilisin carlsberg during prolonged exposure to organic solvents. Biotechnol. Bioeng. 2009, 102, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Chu, J.; Yue, J.; Qin, S.; Li, Y.; Wu, B.; He, B. Biocatalysis for rare ginsenoside Rh2 production in high level with co-immobilized UDP-glycosyltransferase Bs-YjiC mutant and sucrose synthase AtSuSy. Catalysts 2021, 11, 132. [Google Scholar] [CrossRef]
- Liao, L.; Meng, Y.; Wang, R.; Jia, B.; Li, P. Coupling and regulation of porous carriers using plasma and amination to improve the catalytic performance of glucose oxidase and catalase. Front. Bioeng. Biotechnol. 2019, 7, 426. [Google Scholar] [CrossRef]
- Shahrin, N.A.; Yong, G.G.; Serri, N.A. Fructose stearate esterify in packed bed reactor using immobilized lipase. IOP Conf. Ser. Mater. Sci. Eng. 2020, 716, 012017. [Google Scholar] [CrossRef]
- Sarma, G.K.; Gupta, S.S.; Bhattacharyya, K.G. Removal of hazardous basic dyes from aqueous solution by adsorption onto kaolinite and acid-treated kaolinite: Kinetics, isotherm and mechanistic study. SN Appl. Sci. 2019, 1, 211. [Google Scholar] [CrossRef]
- Adnan, M.; Li, K.; Xu, L.; Yan, Y. X-shaped ZIF-8 for immobilization Rhizomucor miehei lipase via encapsulation and its application toward biodiesel production. Catalysts 2018, 8, 96. [Google Scholar] [CrossRef]
- Seki, T.; Chiang, K.Y.; Yu, C.C.; Yu, X.; Okuno, M.; Hunger, J.; Nagata, Y.; Bonn, M. The bending mode of water: A powerful probe for hydrogen bond structure of aqueous systems. J. Phys. Chem. Lett. 2020, 11, 8459–8469. [Google Scholar] [CrossRef]
- Kowalczuk, D.; Pitucha, M. Application of FTIR method for the assessment of immobilization of active substances in the matrix of biomedical materials. Materials 2019, 12, 2972. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.E.; Lassalle, V.; Ferreira, M.L. FTIR-ATR characterization of free Rhizomucor meihei lipase (RML), Lipozyme RM im and chitosan-immobilized RML. J. Mol. Catal. B Enzym. 2011, 72, 220–228. [Google Scholar] [CrossRef]
- Delfino, I.; Portaccio, M.; Della Ventura, B.; Mita, D.G.; Lepore, M. Enzyme distribution and secondary structure of sol-gel immobilized glucose oxidase by micro-attenuated total reflection FT-IR spectroscopy. Mater. Sci. Eng. C 2013, 33, 304–310. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization-Aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszynska, D. Immobilization as a strategy for improving enzyme properties-Application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.Z.; Kim, D.J.; Kumar, A.; Patel, S.K.S.; Otari, S.; Mardina, P.; Jeong, J.-H.; Sohn, J.-H.; Kim, J.H.; Park, J.T.; et al. SnO2 hollow nanotubes: A novel and efficient support matrix for enzyme immobilization. Sci. Rep. 2017, 7, 1533. [Google Scholar] [CrossRef]
- Handayani, N.; Loos, K.; Wahyuningrum, D.; Buchari; Zulfikar, M.A. Immobilization of Mucor miehei lipase onto macroporous aminated polyethersulfone membrane for enzymatic reactions. Membranes 2012, 2, 198–213. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Gupta, R.K.; Kim, S.Y.; Kim, I.W.; Kalia, V.C.; Lee, J.K. Rhus vernicifera laccase immobilization on magnetic nanoparticles to improve stability and its potential application in bisphenol A degradation. Indian J. Microbiol. 2021, 61, 45–54. [Google Scholar] [CrossRef]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basri, R.S.; Rahman, R.N.Z.R.A.; Kamarudin, N.H.A.; Latip, W.; Ali, M.S.M. Characterization of Carboxylic Acid Reductase from Mycobacterium phlei Immobilized onto Seplite LX120. Polymers 2022, 14, 4375. https://doi.org/10.3390/polym14204375
Basri RS, Rahman RNZRA, Kamarudin NHA, Latip W, Ali MSM. Characterization of Carboxylic Acid Reductase from Mycobacterium phlei Immobilized onto Seplite LX120. Polymers. 2022; 14(20):4375. https://doi.org/10.3390/polym14204375
Chicago/Turabian StyleBasri, Rose Syuhada, Raja Noor Zaliha Raja Abd. Rahman, Nor Hafizah Ahmad Kamarudin, Wahhida Latip, and Mohd Shukuri Mohamad Ali. 2022. "Characterization of Carboxylic Acid Reductase from Mycobacterium phlei Immobilized onto Seplite LX120" Polymers 14, no. 20: 4375. https://doi.org/10.3390/polym14204375
APA StyleBasri, R. S., Rahman, R. N. Z. R. A., Kamarudin, N. H. A., Latip, W., & Ali, M. S. M. (2022). Characterization of Carboxylic Acid Reductase from Mycobacterium phlei Immobilized onto Seplite LX120. Polymers, 14(20), 4375. https://doi.org/10.3390/polym14204375





