Candidate Acetic Acid Bacteria Strains for Levan Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains
2.2. Media and Culture Conditions
2.3. Levan Production
2.4. Levan Extraction and Estimation
2.5. NMR Spectroscopy
2.6. Determination of Sucrose, Sugar Monomer Residues, and Gluconic Acid
2.7. Statistical Methods
3. Results
3.1. Effect of Sucrose Concentration on Levan Yield
3.2. Gluconic Acid Formation and pH Reduction
3.3. Carbon Sources Utilization and Consumption during Levan Production
3.4. NMR Analysis for Levan Characterization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arvidson, S.A.; Rinehart, B.T.; Gadala-Maria, F. Concentration regimes of solutions of levan polysaccharide from Bacillus sp. Carbohydr. Polym. 2006, 65, 144–149. [Google Scholar] [CrossRef]
- Han, Y.W.; Clarke, M.A. Production and characterization of microbial levan. J. Agric. Food Chem. 1990, 38, 393–396. [Google Scholar] [CrossRef]
- Jakob, F.; Pfaff, A.; Novoa-carballal, R.; Rübsam, H.; Becker, T.; Vogel, R.F. Structural analysis of fructans produced by acetic acid bacteria reveals a relation to hydrocolloid function. Carbohydr. Polym. 2013, 92, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Peng, J.; Ni, D.; Zhang, W.; Wu, H.; Mu, W. Preparation, characterization and application of levan/montmorillonite biocomposite and levan/BSA nanoparticle. Carbohydr. Polym. 2020, 234, 115921. [Google Scholar] [CrossRef]
- Banguela, A.; Hernández, L. Fructans: From natural sources to transgenic plants. Biotecnol. Apl. 2006, 23, 202–210. [Google Scholar]
- Toksoy, E.; Hernández, L.; Combie, J.; Öner, E.T.; Hernández, L.; Combie, J. Review of levan polysaccharide: From a century of past experiences to future prospects. Biotechnol. Adv. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Moscovici, M. Present and future medical applications of microbial exopolysaccharides. Front. Microbiol. 2015, 6, 1012. [Google Scholar] [CrossRef]
- Srikanth, R.; Reddy, C.H.S.S.S.; Siddartha, G.; Ramaiah, M.J.; Uppuluri, K.B. Review on production, characterization and applications of microbial levan. Carbohydr. Polym. 2015, 120, 102–114. [Google Scholar] [CrossRef]
- Rairakhwada, D.; Pal, A.; Bhathena, Z.; Sahu, N.; Jha, A.; Mukherjee, S. Dietary microbial levan enhances cellular non-specific immunity and survival of common carp (Cyprinus carpio) juveniles. Fish Shellfish. Immunol. 2007, 22, 477–486. [Google Scholar] [CrossRef]
- Nasir, D.Q.; Wahyuningrum, D.; Hertadi, R. Screening and characterization of levan secreted by halophilic bacterium of Halomonas and Chromohalobacter genuses originated from bledug kuwu mud crater. Procedia Chem. 2015, 16, 272–278. [Google Scholar] [CrossRef]
- Stasiak-Różańska, L.; Kupiec, M. Industrial applications of wild and genetically-modified strains of acetic acid bacteria. Postępy Mikrobiol.—Adv. Microbiol. 2019, 57, 398–402. [Google Scholar] [CrossRef]
- La China, S.; Zanichelli, G.; De Vero, L.; Gullo, M. Oxidative fermentations and exopolysaccharides production by acetic acid bacteria: A mini review. Biotechnol. Lett. 2018, 40, 1289–1302. [Google Scholar] [CrossRef] [PubMed]
- La China, S.; De Vero, L.; Anguluri, K.; Brugnoli, M.; Mamlouk, D.; Gullo, M. Kombucha tea as a reservoir of cellulose producing bacteria: Assessing diversity among Komagataeibacter isolates. Appl. Sci. 2021, 11, 1595. [Google Scholar] [CrossRef]
- La China, S.; Bezzecchi, A.; Moya, F.; Petroni, G.; Di Gregorio, S.; Gullo, M. Genome sequencing and phylogenetic analysis of K1G4: A new Komagataeibacter strain producing bacterial cellulose from different carbon sources. Biotechnol. Lett. 2020, 42, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Antolak, H.; Piechota, D.; Kucharska, A. Kombucha Tea—A double power of bioactive compounds from tea and symbiotic culture of bacteria and yeasts (SCOBY). Antioxidants 2021, 10, 1541. [Google Scholar] [CrossRef]
- Gojgic-Cvijovic, G.D.; Jakovljevic, D.M.; Loncarevic, B.D.; Todorovic, N.M.; Pergal, M.V.; Ciric, J.; Loos, K.; Beskoski, V.P.; Vrvic, M.M. Production of levan by Bacillus licheniformis NS032 in sugar beet molasses-based medium. Int. J. Biol. Macromol. 2019, 121, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Hestrin, S.; Feingold, D.S.; Avigad, G. The mechanism of polysaccharide production from sucrose. 3. Donor–acceptor specificity of levansucrase from Aerobacter levanicum. Biochem. J. 1956, 64, 340–351. [Google Scholar] [CrossRef]
- Bekers, M.; Upite, D.; Kaminska, E.; Laukevics, J.; Grube, M.; Vigants, A.; Linde, R. Stability of levan produced by Zymomonas mobilis. Process Biochem. 2005, 40, 1535–1539. [Google Scholar] [CrossRef]
- Caputi, L.; Nepogodiev, S.A.; Malnoy, M.; Rejzek, M.; Field, R.A.; Benini, S. Biomolecular characterization of the levansucrase of Erwinia amylovora, a promising biocatalyst for the synthesis of fructooligosaccharides. J. Agric. Food Chem. 2013, 61, 12265–12273. [Google Scholar] [CrossRef]
- Hernandez, L.; Arrieta, J.; Menendez, C.; Vazquez, R.; Coego, A.; Suarez, V.; Selman, G.; Petit-Glatron, M.F.; Chambert, R. Isolation and enzymic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem. J. 1995, 309, 113–118. [Google Scholar] [CrossRef]
- Visnapuu, T.; Mardo, K.; Alamäe, T. Levansucrases of a Pseudomonas syringae pathovar as catalysts for the synthesis of potentially prebiotic oligo- and polysaccharides. N. Biotechnol. 2015, 32, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Donot, F.; Fontana, A.; Baccou, J.C.; Schorr-Galindo, S. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 2012, 87, 951–962. [Google Scholar] [CrossRef]
- González-Garcinuño, Á.; Tabernero, A.; Sánchez-Álvarez, J.M.; Galán, M.A.; Martin del Valle, E.M. Effect of bacteria type and sucrose concentration on levan yield and its molecular weight. Microb. Cell Fact. 2017, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.L.; Boiardi, J.L. Levans production by Gluconacetobacter diazotrophicus. Electron. J. Biotechnol. 2013, 16, 0717–3458. [Google Scholar] [CrossRef]
- Tajima, K.; Uenishi, N.; Fujiwara, M.; Erata, T.; Munekata, M.; Takai, M. The production of a new water-soluble polysaccharide by Acetobacter xylinum NCI 1005 and its structural analysis by NMR spectroscopy. Carbohydr. Res. 1997, 305, 117–122. [Google Scholar] [CrossRef]
- Jakob, F. Novel Fructans from Acetic Acid Bacteria; Technische Universität München: München, Germany, 2014. [Google Scholar]
- Semjonovs, P.; Shakirova, L.; Treimane, R.; Shvirksts, K.; Auzina, L.; Cleenwerck, I.; Zikmanis, P. Production of extracellular fructans by Gluconobacter nephelii P1464. Lett. Appl. Microbiol. 2016, 62, 145–152. [Google Scholar] [CrossRef]
- Jakob, F.; Quintero, Y.; Musacchio, A.; Estrada-de los Santos, P.; Hernández, L.; Vogel, R.F. Acetic acid bacteria encode two levansucrase types of different ecological relationship. Environ. Microbiol. 2019, 21, 4151–4165. [Google Scholar] [CrossRef]
- Hermann, M.; Petermeier, H.; Vogel, R.F. Development of novel sourdoughs with in situ formed exopolysaccharides from acetic acid bacteria. Eur. Food Res. Technol. 2015, 241, 185–197. [Google Scholar] [CrossRef]
- Yukphan, P.; Malimas, T.; Potacharoen, W.; Tanasupawat, S.; Tanticharoen, M.; Yamada, Y. Neoasaia chiangmaiensis gen. nov., sp. nov., a novel osmotolerant acetic acid bacterium in the ALPHA-Proteobacteria. J. Gen. Appl. Microbiol. 2005, 51, 301–311. [Google Scholar] [CrossRef][Green Version]
- Brandt, J.U.; Jakob, F.; Geissler, A.J.; Behr, J.; Vogel, R.F. Multiple genome sequences of heteropolysaccharide-forming acetic acid bacteria. Genome Announc. 2017, 5, 3–5. [Google Scholar] [CrossRef]
- Brandt, J.U.; Jakob, F.; Behr, J.; Geissler, A.J.; Vogel, R.F. Dissection of exopolysaccharide biosynthesis in Kozakia baliensis. Microb. Cell Fact. 2016, 15, 170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Calazans, G.M.T.; Lima, R.C.; de França, F.P.; Lopes, C.E.; Lima, C.; Franc, F.P. De Molecular weight and antitumour activity of Zymomonas mobilis levans. Int. J. Biol. Macromol. 2000, 27, 245–247. [Google Scholar] [CrossRef]
- Esawy, M.A.; Abdel-Fattah, A.M.; Ali, M.M.; Helmy, W.A.; Salama, B.M.; Taie, H.A.A.; Hashem, A.M.; Awad, G.E.A. Levansucrase optimization using solid state fermentation and levan biological activities studies. Carbohydr. Polym. 2013, 96, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Ua-Arak, T.; Jakob, F.; Vogel, R.F. Characterization of growth and exopolysaccharide production of selected acetic acid bacteria in buckwheat sourdoughs. Int. J. Food Microbiol. 2016, 239, 103–112. [Google Scholar] [CrossRef]
- Jakob, F.; Steger, S.; Vogel, R.F. Influence of novel fructans produced by selected acetic acid bacteria on the volume and texture of wheat breads. Eur. Food Res. Technol. 2012, 234, 493–499. [Google Scholar] [CrossRef]
- Brandão, P.R.; Crespo, M.T.B.; Nascimento, F.X. Phylogenomic and comparative analyses support the reclassification of several Komagataeibacter species as novel members of the Novacetimonas gen. nov. and bring new insights into the evolution of cellulose synthase genes. Int. J. Syst. Evol. Microbiol. 2022, 72, 005252. [Google Scholar] [CrossRef]
- Kim, D.H.; Chon, J.W.; Kim, H.; Seo, K.H. Development of a novel selective medium for the isolation and enumeration of acetic acid bacteria from various foods. Food Control 2019, 106, 106717. [Google Scholar] [CrossRef]
- Hestrin, S.; Schramm, M. Synthesis of cellulose by Acetobacter xylinum. 2. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem. J. 1954, 58, 345–352. [Google Scholar] [CrossRef]
- Ananthalakshmy, V.K.; Gunasekaran, P. Optimization of levan production by Zymomonas mobilis. Brazilian Arch. Biol. Technol. 1999, 42, 291–298. [Google Scholar] [CrossRef]
- Shih, I.-L.; Wang, T.-C.; Chou, S.-Z.; Lee, G.-D. Sequential production of two biopolymers-levan and poly-ε-lysine by microbial fermentation. Bioresour. Technol. 2011, 102, 3966–3969. [Google Scholar] [CrossRef]
- Ua-Arak, T.; Jakob, F.; Vogel, R.F. Influence of levan-producing acetic acid bacteria on buckwheat-sourdough breads. Food Microbiol. 2017, 65, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Katina, K.; Henry, N.; Juvonen, R.; Flander, L.; Johansson, L.; Virkki, L.; Tenkanen, M.; Laitila, A. In situ production and analysis of Weissella confusa dextran in wheat sourdough. Food Microbiol. 2009, 26, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Mondal, H. Value of r2 in Statistical Analysis by Pearson Correlation Coefficient. J. Clin. Diagn. Res. 2017, 11, CL01. [Google Scholar] [CrossRef] [PubMed]
- Matulová, M.; Husárová, S.; Capek, P.; Sancelme, M.; Delort, A.-M. NMR structural study of fructans produced by Bacillus sp. 3B6, bacterium isolated in cloud water. Carbohydr. Res. 2011, 346, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Aramsangtienchai, P.; Kongmon, T.; Pechroj, S.; Srisook, K. Enhanced production and immunomodulatory activity of levan from the acetic acid bacterium, Tanticharoenia sakaeratensis. Int. J. Biol. Macromol. 2020, 163, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Veerapandian, B.; Shanmugam, S.R.; Varadhan, S.; Sarwareddy, K.K.; Mani, K.P.; Ponnusami, V. Levan production from sucrose using chicken feather peptone as a low cost supplemental nutrient source. Carbohydr. Polym. 2020, 227, 115361. [Google Scholar] [CrossRef]
- Khan, R.A.; Singh, A.K.; Agrawal, P.K. Sitosterol sucroside from the suckers of Mentha arvensis. Phytochemistry 1997, 45, 1295–1296. [Google Scholar] [CrossRef]
- Lopez, M.G.; Mancilla-Margalli, N.A.; Mendoza-Diaz, G. Molecular structures of fructans from Agave tequilana Weber var. azul. J. Agric. Food Chem. 2003, 51, 7835–7840. [Google Scholar] [CrossRef]
- Joaquim, E.O.; Hayashi, A.H.; Torres, L.M.B.; Figueiredo-ribeiro, R.C.L.; Shiomi, N.; De Sousa, F.S.; Lago, J.H.G. Chemical structure and localization of levan, the predominant fructan type in underground systems of Gomphrena marginata (Amaranthaceae ). Front. Plant Sci. 2018, 9, 1745. [Google Scholar] [CrossRef]
- Hövels, M.; Kosciow, K.; Kniewel, J.; Jakob, F.; Deppenmeier, U. High yield production of levan-type fructans by Gluconobacter japonicus LMG Int. J. Biol. Macromol. 2020, 164, 295–303. [Google Scholar] [CrossRef]
- Ryngajłło, M.; Kubiak, K.; Jędrzejczak-Krzepkowska, M.; Jacek, P.; Bieleck, S. Comparative genomics of the Komagataeibacter strains—Efficient bionanocellulose producers. Microbiologyopen 2019, 8, e00731. [Google Scholar] [CrossRef] [PubMed]
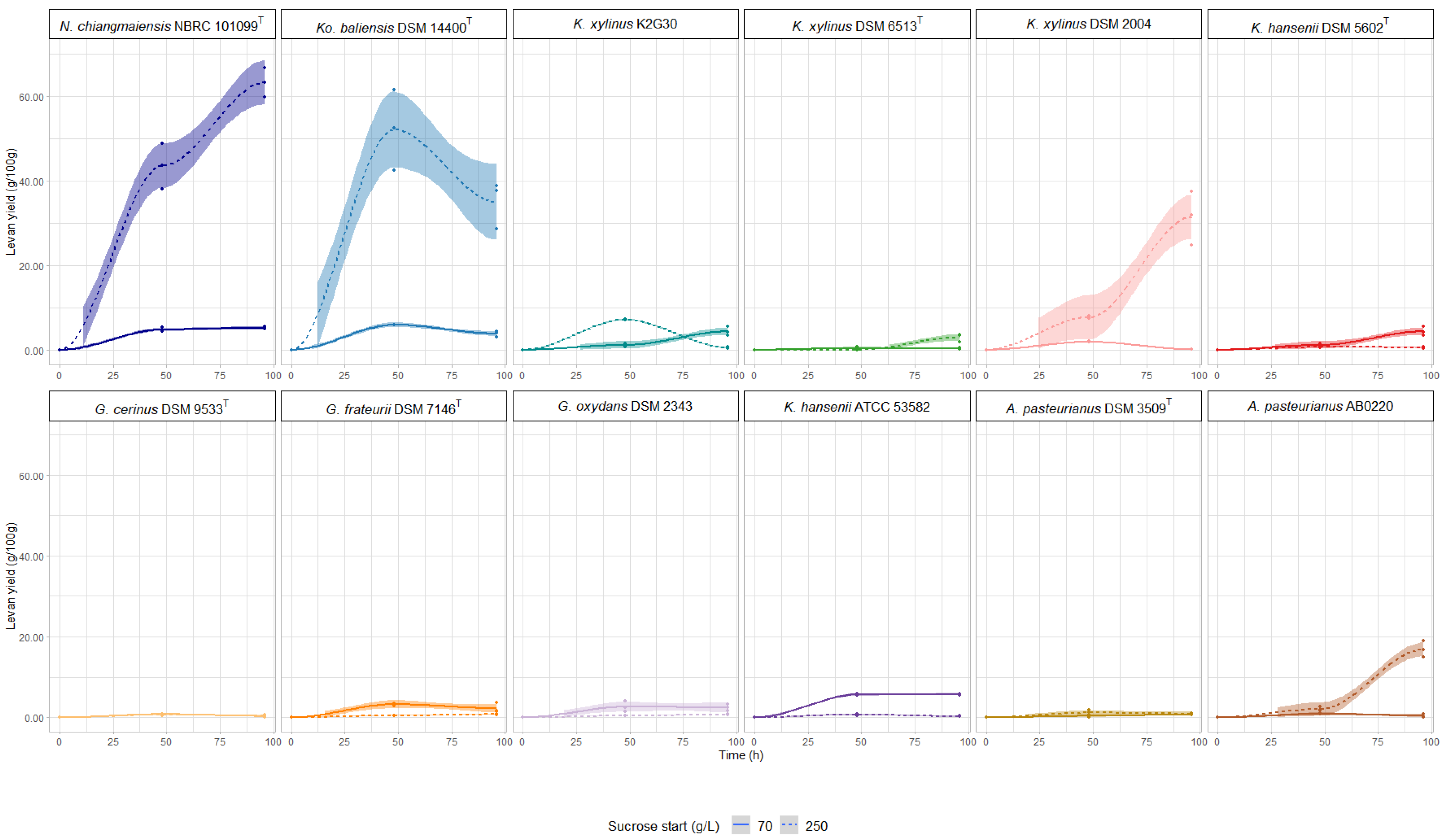
 —48 h of incubation;
—48 h of incubation;  —96 h of incubation;
—96 h of incubation;  —color refers to 250 g/L sucrose);
—color refers to 250 g/L sucrose);  —color refers to 70 g/L sucrose.
—color refers to 70 g/L sucrose.
 —48 h of incubation;
—48 h of incubation;  —96 h of incubation;
—96 h of incubation;  —color refers to 250 g/L sucrose);
—color refers to 250 g/L sucrose);  —color refers to 70 g/L sucrose.
—color refers to 70 g/L sucrose.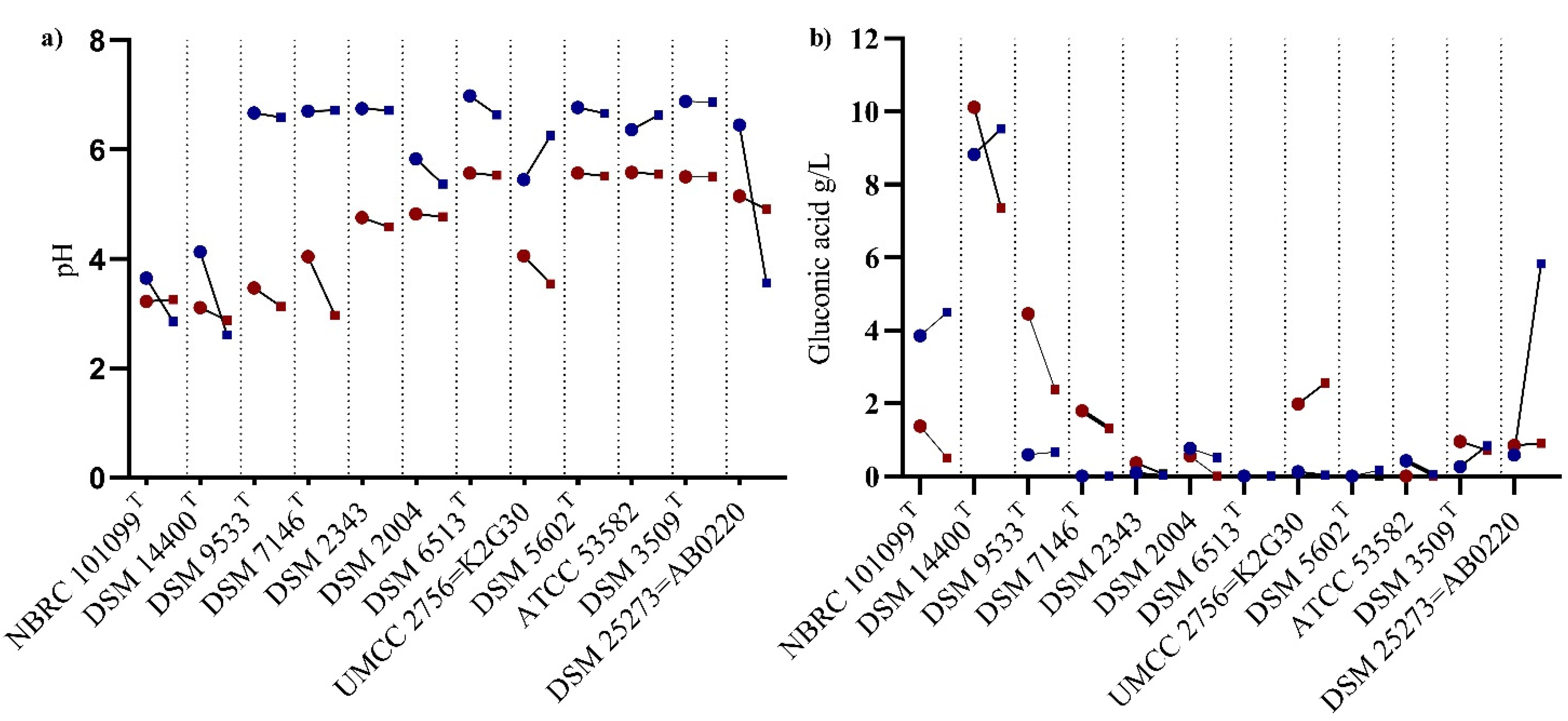


| Strain | Collection ID | * Sucrose (w/v) | Reference |
|---|---|---|---|
| N. chiangmaiensis | NBRC 101099 T | 80/50-100 | [29,36] |
| Ko. baliensis | DSM 14400 T | 80/50–100 | [29,36] |
| G. cerinus | DSM 9533 T | 80 | [36] |
| G. frateurii | DSM 7146 T | 80 | [36] |
| G. oxydans | DSM 2343 | 80 | [36] |
| K. xylinus | DSM 2004 | - | - |
| K. xylinus | DSM 6513 T | - | - |
| K. xylinus | UMCC 2756 = K2G30 | - | - |
| ** K. hansenii | DSM 5602 T | 80 | [36] |
| ** K. hansenii | ATCC 53582 | - | - |
| A. pasteurianus | DSM 3509 T | - | - |
| A. pasteurianus | UMCC 1754 = AB0220 | - | - |
| Component (% w/v) | Medium | ||
|---|---|---|---|
| GYC | HS | HS-Sucrose (HS-S) | |
| Glucose | 10 | 2 | - |
| Sucrose | - | - | 7/25 |
| Yeast extract | 1 | 0.5 | 0.5 |
| Poly-peptone | - | 0.5 | 0.5 |
| Na2HPO4 | - | 0.73 | 0.73 |
| Citric acid | - | 0.115 | 0.115 |
| MgSO4 | - | 0.05 | 0.05 |
| CaCO3 | 2 | - | - |
| Agar | 1.5 | - | - |
| Strain | Chemical Shifts (ppm) Carbon Atoms | |||||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | |
| Levan control flour (Megazyme) | 59.89 | 104.21 | 76.26 | 75.16 | 80.26 | 63.33 |
| N. chiangmaiensis NBRC 101099T | 59.89 | 104.12 | 76.29 | 75.16 | 80.26 | 63.36 |
| Ko. baliensis DSM 14400 T | 59.91 | 104.16 | 76.32 | 75.18 | 80.21 | 63.33 |
| K. hansenii ATCC 53582 | 59.89 | 104.12 | 76.26 | 75.16 | 80.23 | 63.26 |
| A. pasteurianus UMCC 1754 = AB0220 | 60.0 | 104.23 | 76.38 | 75.18 | 80.44 | 63.35 |
| G. cerinus DSM 9533 T | 92.16 | 71.10 | 72.51 | 69.22 | 72.38 | 60.15 |
| 61.39 | 103.81 | 76.35 | 74.01 | 81.42 | 62.38 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anguluri, K.; La China, S.; Brugnoli, M.; De Vero, L.; Pulvirenti, A.; Cassanelli, S.; Gullo, M. Candidate Acetic Acid Bacteria Strains for Levan Production. Polymers 2022, 14, 2000. https://doi.org/10.3390/polym14102000
Anguluri K, La China S, Brugnoli M, De Vero L, Pulvirenti A, Cassanelli S, Gullo M. Candidate Acetic Acid Bacteria Strains for Levan Production. Polymers. 2022; 14(10):2000. https://doi.org/10.3390/polym14102000
Chicago/Turabian StyleAnguluri, Kavitha, Salvatore La China, Marcello Brugnoli, Luciana De Vero, Andrea Pulvirenti, Stefano Cassanelli, and Maria Gullo. 2022. "Candidate Acetic Acid Bacteria Strains for Levan Production" Polymers 14, no. 10: 2000. https://doi.org/10.3390/polym14102000
APA StyleAnguluri, K., La China, S., Brugnoli, M., De Vero, L., Pulvirenti, A., Cassanelli, S., & Gullo, M. (2022). Candidate Acetic Acid Bacteria Strains for Levan Production. Polymers, 14(10), 2000. https://doi.org/10.3390/polym14102000










