Detection of Streptavidin Based on Terminal Protection and Cationic Conjugated Polymer-Mediated Fluorescence Resonance Energy Transfer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Measurements
2.2. Apparatus
2.3. Method Optimization
2.4. Detection of SA
3. Results
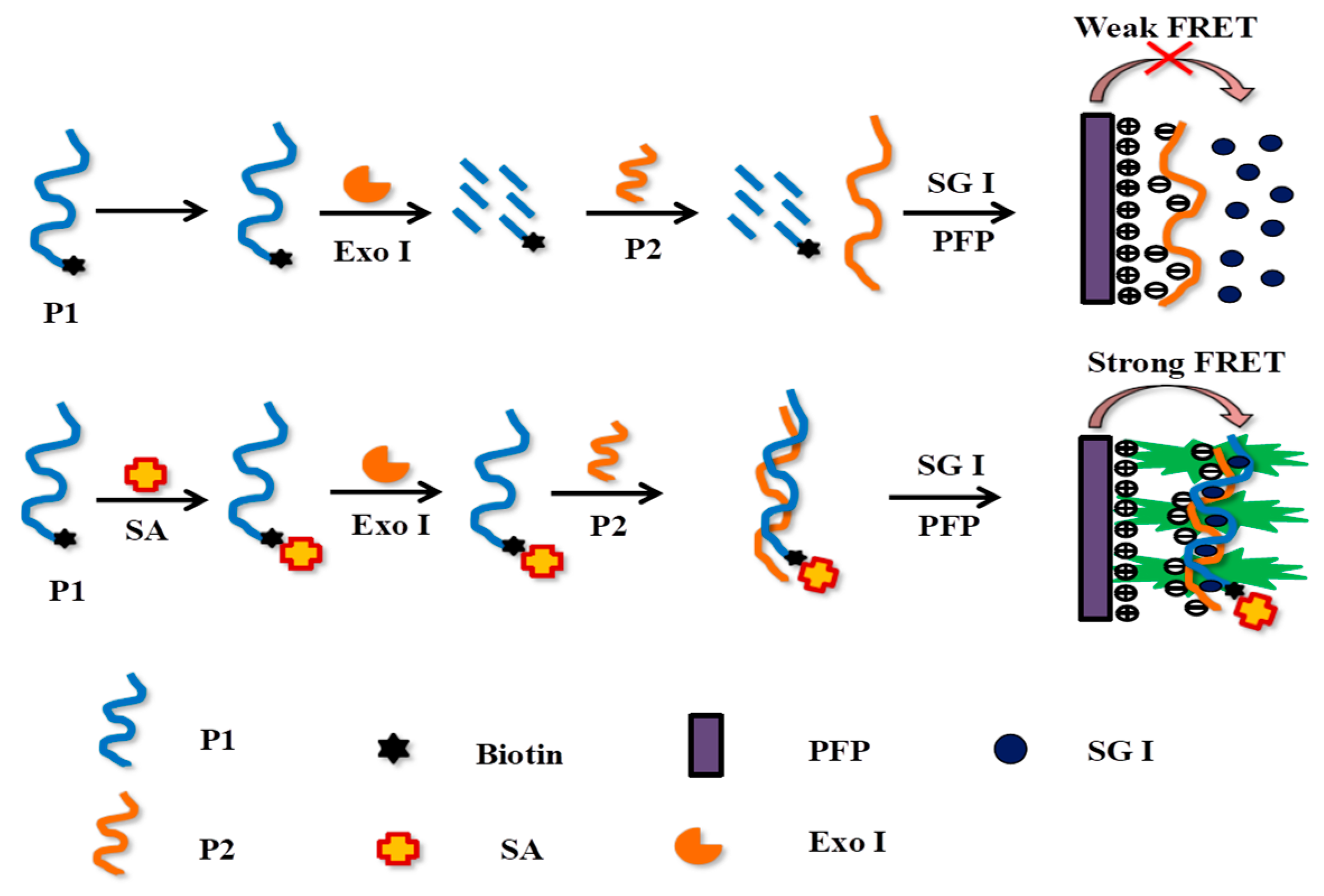
3.1. Experimental Principles
3.2. Feasibility Assessment of the SA Detection Assay
3.3. Optimization of Experimental Conditions
3.4. Quantitative Detection of SA
3.5. Selectivity of SA Assay
3.6. Application of the Proposed Assay in Biological Systems
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, C.; Xiang, X.; Liu, Y.; Zhou, G.; Ji, X.; He, Z. Dual-color determination of protein via terminal protection of small-molecule-linked DNA and the enzymolysis of exonuclease III. Biosens. Bioelectron. 2014, 58, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Deng, Z.; Ma, C.; Zhao, H.; Wu, K.; Wang, K. A sensitive fluorescence method for the detection of streptavidin based on target-induced DNA machine amplification. Anal. Methods 2018, 10, 1870–1874. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, H.; Chen, Y.; Liu, Y. A fluorescent biosensor for protein detection based on poly(thymine)-templated copper nanoparticles and terminal protection of small molecule-linked DNA. Biosens. Bioelectron. 2015, 74, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, C.; Chen, Y.; Luo, F.; Wang, Y.; Chen, X. Terminal protection of a small molecule-linked loop DNA probe for turn-on label-free fluorescence detection of proteins. Biosens. Bioelectron. 2016, 83, 97–101. [Google Scholar] [CrossRef]
- Xiang, X.; Shi, J.; Huang, F.; Zheng, M.; Deng, Q.; Xu, J. MoS2 nanosheet-based fluorescent biosensor for protein detection via terminal protection of small-molecule-linked DNA and exonuclease III-aided DNA recycling amplification. Biosens. Bioelectron. 2015, 74, 227–232. [Google Scholar] [CrossRef]
- Xu, F.; Yang, T.; Chen, Y. Quantification of microRNA by DNA-peptide probe and liquid chromatography-tandem mass spectrometry-based quasi-targeted proteomics. Anal. Chem. 2016, 88, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Focsan, M.; Campu, A.; Craciun, A.M.; Potara, M.; Leordean, C.; Maniu, D.; Astilean, S. A simple and efficient design to improve the detection of biotin-streptavidin interaction with plasmonic nanobiosensors. Biosens. Bioelectron. 2016, 86, 728–735. [Google Scholar] [CrossRef]
- Zhuang, Y.; Chiang, P.; Wang, C.; Tan, K. Environment-sensitive fluorescent turn-on probes targeting hydrophobic ligand-binding domains for selective protein detection. Angew. Chem. Int. Ed. 2013, 52, 8124–8128. [Google Scholar] [CrossRef]
- Taunton, J.; Hassig, C.A.; Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996, 272, 408–411. [Google Scholar] [CrossRef]
- Andrei, P.D.; Maxim, V.B.; Michael, U.M.; Sergey, N.K. Selection of Smart Small-Molecule Ligands: The Proof of Principle. Anal. Chem. 2009, 81, 490–494. [Google Scholar]
- Hirokazu, S.; Chie, Y.; Seiji, K.; Takashi, K.; Makoto, H.; Mitsuhiro, M.; Yuichi, O.; Yu, H.; Yoshiko, M. Biotinylation of silicon and nickel surfaces and detection of streptavidin as biosensor. Langmuir 2013, 29, 9457–9463. [Google Scholar]
- Ban, F.; Shi, H.; Feng, C.; Mao, X.; Yin, Y.; Zhu, X. A one-pot strategy for the detection of proteins based on sterically and allosterically tunable hybridization chain reaction. Biosens. Bioelectron. 2016, 86, 219–224. [Google Scholar] [CrossRef]
- Zhou, R.; Xu, C.; Dong, J.; Wang, G. Labeling-free fluorescent detection of DNA hybridization through FRET from pyrene excimer to DNA intercalator SYBR green I. Biosens. Bioelectron. 2015, 65, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, C.; Li, Z.; Wu, K. An exonuclease-assisted fluorescence sensor for assaying alkaline phosphatase based on SYBR Green I. Mol. Cell. Probes 2019, 45, 26–30. [Google Scholar] [CrossRef]
- Meng, F.; Xu, H.; Yao, X.; Qin, X.; Jiang, T.; Gao, S.; Zhang, Y.; Yang, D.; Liu, X. Mercury detection based on label-free and isothermal enzyme-free amplified fluorescence platform. Talanta 2017, 162, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, Y.; Liu, Y.; Guo, Y. Water-Soluble Conjugated Polymer as a Platform for Adenosine Deaminase Sensing Based on Fluorescence Resonance Energy Transfer Technique. Anal. Chem. 2014, 86, 6433–6438. [Google Scholar] [CrossRef]
- Lian, S.; Liu, C.; Zhang, X.; Wang, H.; Li, Z. Detection of T4 polynucleotide kinase activity based on cationic con-jugated polymer-mediated fluorescence resonance energy transfer. Biosens. Bioelectron. 2015, 66, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Kim, H.Y.; Kim, S.; Park, K.S.; Park, H.G. A simple and sensitive detection of small molecule-protein interactions based on terminal protection-mediated exponential strand displacement amplification. Analyst 2018, 143, 2023–2028. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, G.; Mallik, A.K.; Qiu, H. A WS2 nanosheet-based nanosensor for the ultrasensitive detection of small molecule–protein interaction via terminal protection of small molecule-linked DNA and Nt.BstNBI-assisted recycling amplification. J. Mater. Chem. B 2016, 4, 5161–5166. [Google Scholar] [CrossRef]
- Wang, R.; Xu, X.; Li, P.; Wang, Y.; Jiang, W. Terminal protection-mediated autocatalytic cascade amplification coupled with graphene oxide fluorescence switch for sensitive and rapid detection of folate receptor. Talanta 2017, 174, 684–688. [Google Scholar] [CrossRef]
- Huang, D.; Wu, Z.; Yu, R.; Jiang, J. Small molecule-linked programmable DNA for washing-free imaging of cell surface biomarkers. Talanta 2018, 190, 429–435. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jiao, B. Detection of biotin-streptavidin interactions via terminal protection of small molecule linked DNA and the formation of fluorescent DNA-templated silver nanoclusters. Microchim. Acta 2016, 183, 3183–3189. [Google Scholar] [CrossRef]
- Zhao, J.; Hu, S.; Chen, X.; Zhang, B.; Wang, K.; Liu, X. A colorimetric method for the detection of folate receptor based on terminal protection-assisted cascade signal amplification. Sens. Actuat. B-Chem. 2014, 202, 1243–1247. [Google Scholar] [CrossRef]
- Bai, D.; Ji, D.; Shang, J.; Hu, Y.; Gao, J.; Lin, Z.; Ge, J.; Li, Z. A rapid biosensor for highly sensitive protein detection based on G-quadruplex-Thioflavin T complex and terminal protection of small molecule-linked DNA. Sensor Actuat. B-Chem. 2017, 252, 1146–1152. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, J.; Ma, G.; Jia, H. A Cationic Conjugated Polymer-Based Fluorescence Resonance Energy Transfer Strategy for Telomerase Activity Assay Combining with Signal Amplification of Hybridization Chain Reaction. Anal. Chem. 2019, 47, 1006–1013. [Google Scholar]
- Huang, Y.Z.; Zhang, L.N.; Zhang, S.B.; Zhao, P.N.; Li, L.; Ge, S.G.; Yu, J.H. Paper-based electrochemiluminescence determination of streptavidin using reticular DNA-functionalized PtCu nanoframes and analyte-triggered DNA walker. Mikrochim. Acta 2020, 187, 530. [Google Scholar] [CrossRef] [PubMed]





| Material | LOD (nM) | Dynamic Range (nM) | Reference |
|---|---|---|---|
| CuNP | 0.1 | 0.5–1000 | 3 |
| FAM | 0.08 | 0.15–12 | 1 |
| Copper nanocluster | 0.47 | 1–200 | 2 |
| Gold nanoparticle | 5 | 5–50 | 7 |
| SYBR Green I | 0.016 | 0.01–0.1 | 18 |
| SYBR Green I | 0.4 | 0.4–200 | 4 |
| SYBR Green I | 0.068 | 0.1–20 | This work |
| Sample | Added (nM) | Found (nM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 5 | 4.87 ± 0.51 | 97.48 | 9.9 |
| 2 | 8 | 8.31 ± 0.63 | 103.85 | 6.88 |
| 3 | 15 | 13.57 ± 0.50 | 90.49 | 3.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, T.; Yan, Y.; Tang, Z.; Liu, X.; Ma, C. Detection of Streptavidin Based on Terminal Protection and Cationic Conjugated Polymer-Mediated Fluorescence Resonance Energy Transfer. Polymers 2021, 13, 725. https://doi.org/10.3390/polym13050725
Hu T, Yan Y, Tang Z, Liu X, Ma C. Detection of Streptavidin Based on Terminal Protection and Cationic Conjugated Polymer-Mediated Fluorescence Resonance Energy Transfer. Polymers. 2021; 13(5):725. https://doi.org/10.3390/polym13050725
Chicago/Turabian StyleHu, Tingting, Ying Yan, Zhenwei Tang, Xinfa Liu, and Changbei Ma. 2021. "Detection of Streptavidin Based on Terminal Protection and Cationic Conjugated Polymer-Mediated Fluorescence Resonance Energy Transfer" Polymers 13, no. 5: 725. https://doi.org/10.3390/polym13050725
APA StyleHu, T., Yan, Y., Tang, Z., Liu, X., & Ma, C. (2021). Detection of Streptavidin Based on Terminal Protection and Cationic Conjugated Polymer-Mediated Fluorescence Resonance Energy Transfer. Polymers, 13(5), 725. https://doi.org/10.3390/polym13050725






