Upconversion Nanoparticles Encapsulated with Molecularly Imprinted Amphiphilic Copolymer as a Fluorescent Probe for Specific Biorecognition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of LiYF4: Yb3+/Tm3+@LiYF4:Yb3+ Core/Shell UCNPs
2.3. Synthesis of Amphiphilic Random Copolymer Poly(MAA-co-OD)
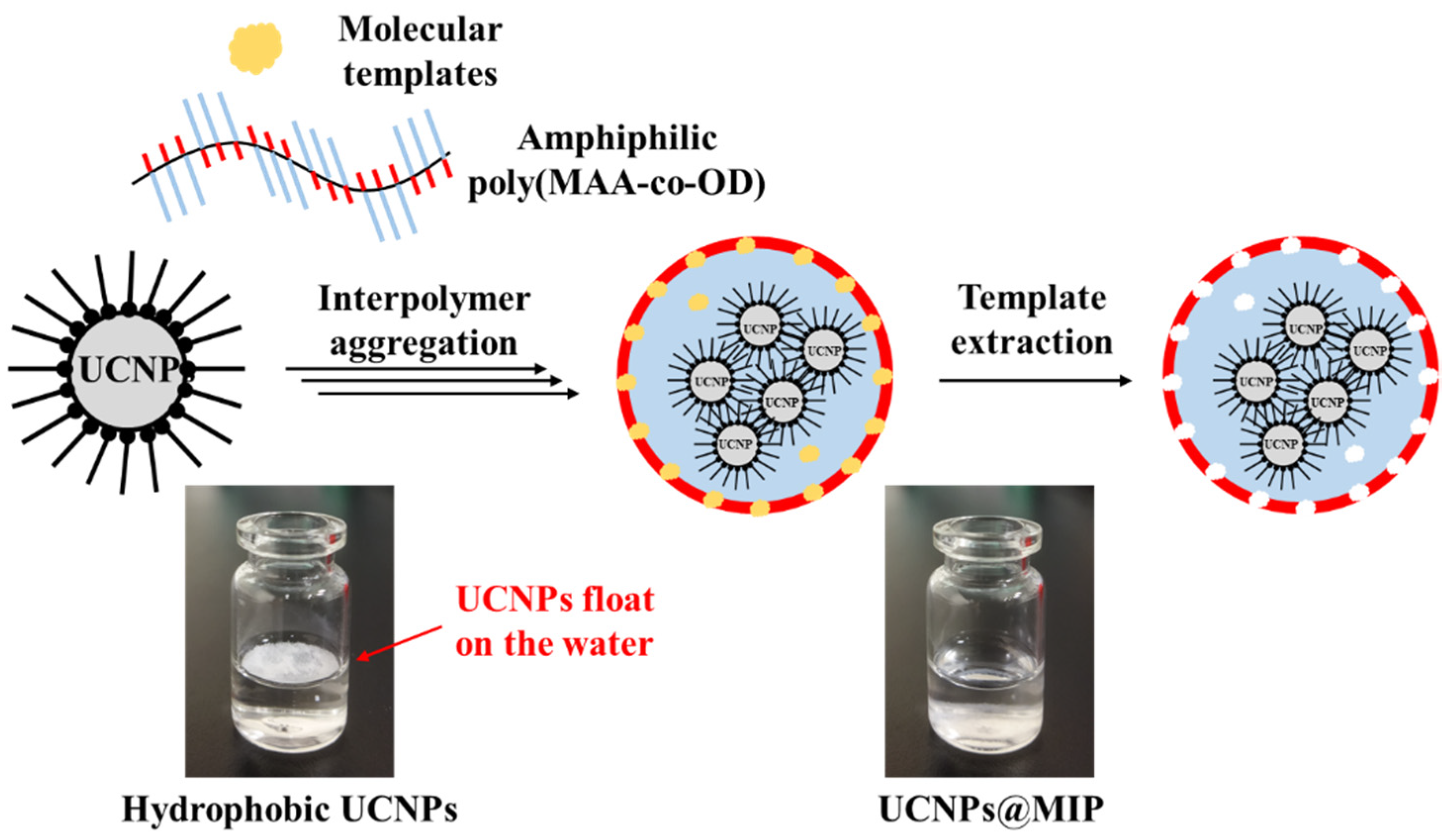
2.4. Preparation of UCNPs@MIP
2.5. Characterization of UCNPs and UCNPs@MIP
2.6. Application in Biorecognition
3. Results and Discussion
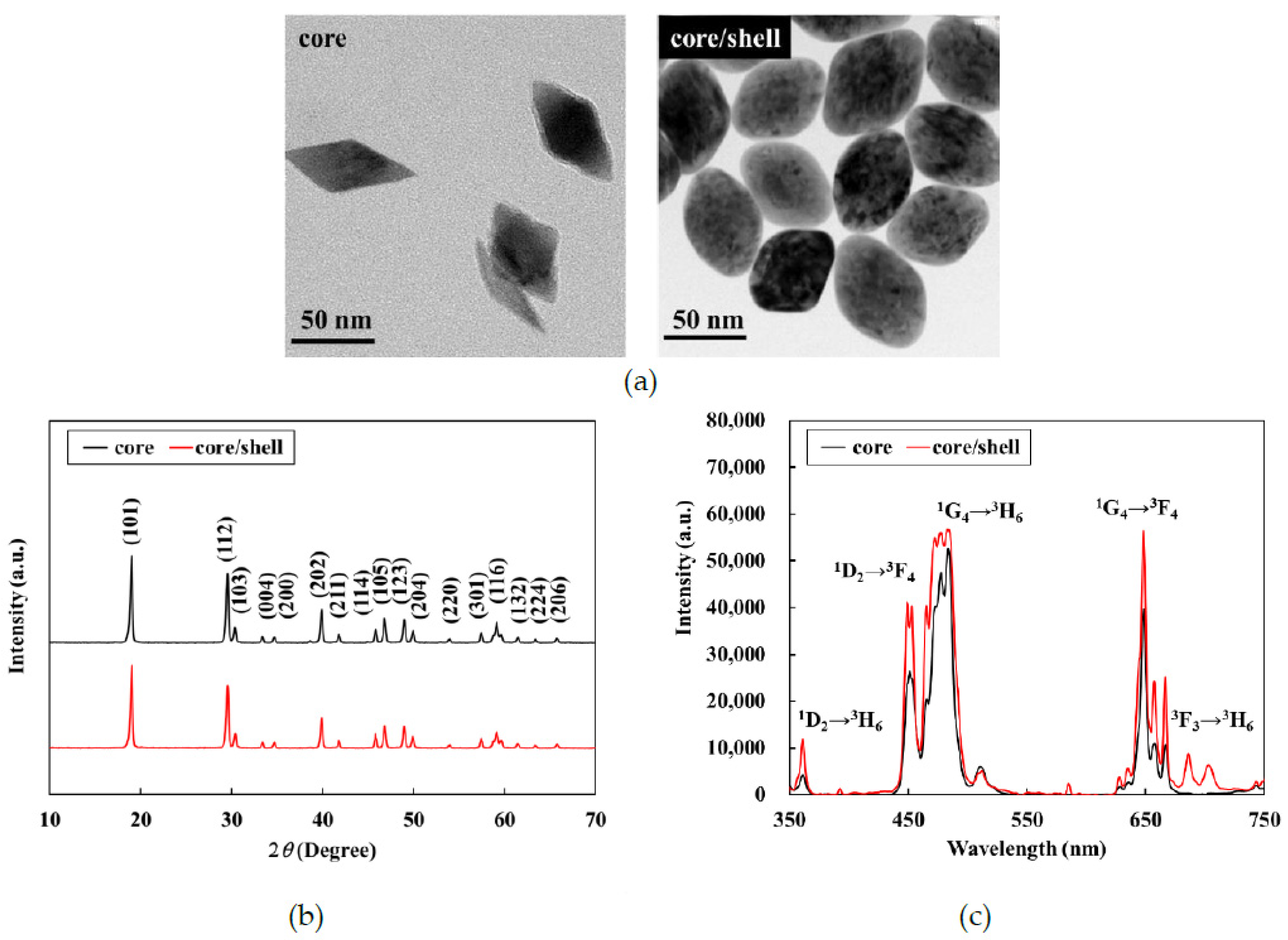
3.1. Characterization of the UCNPs
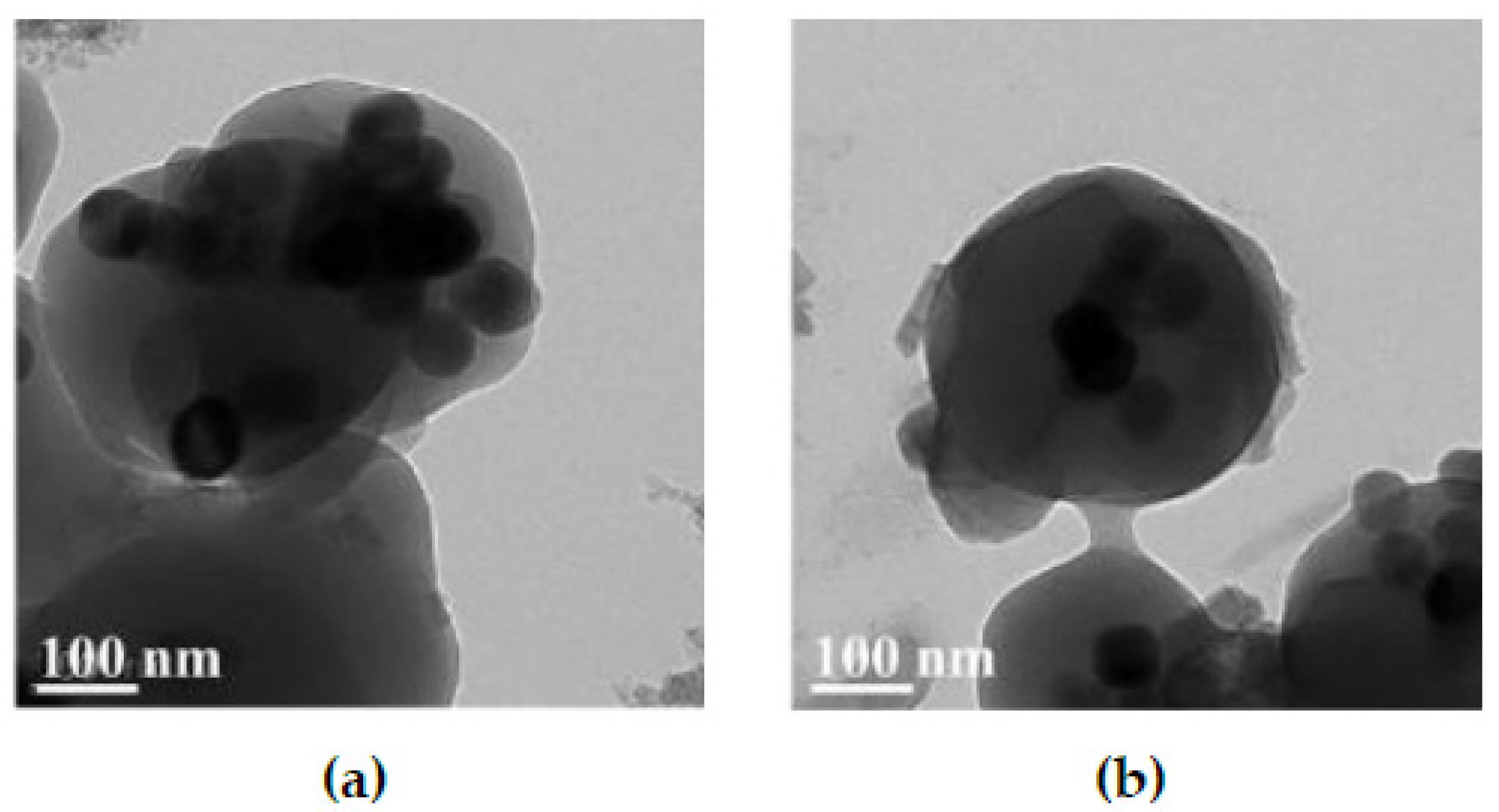
3.2. Hydrophilicity of UCNPs@MIP
3.3. Equilibrium Binding of UCNPs@MIP and UCNPs@NIP
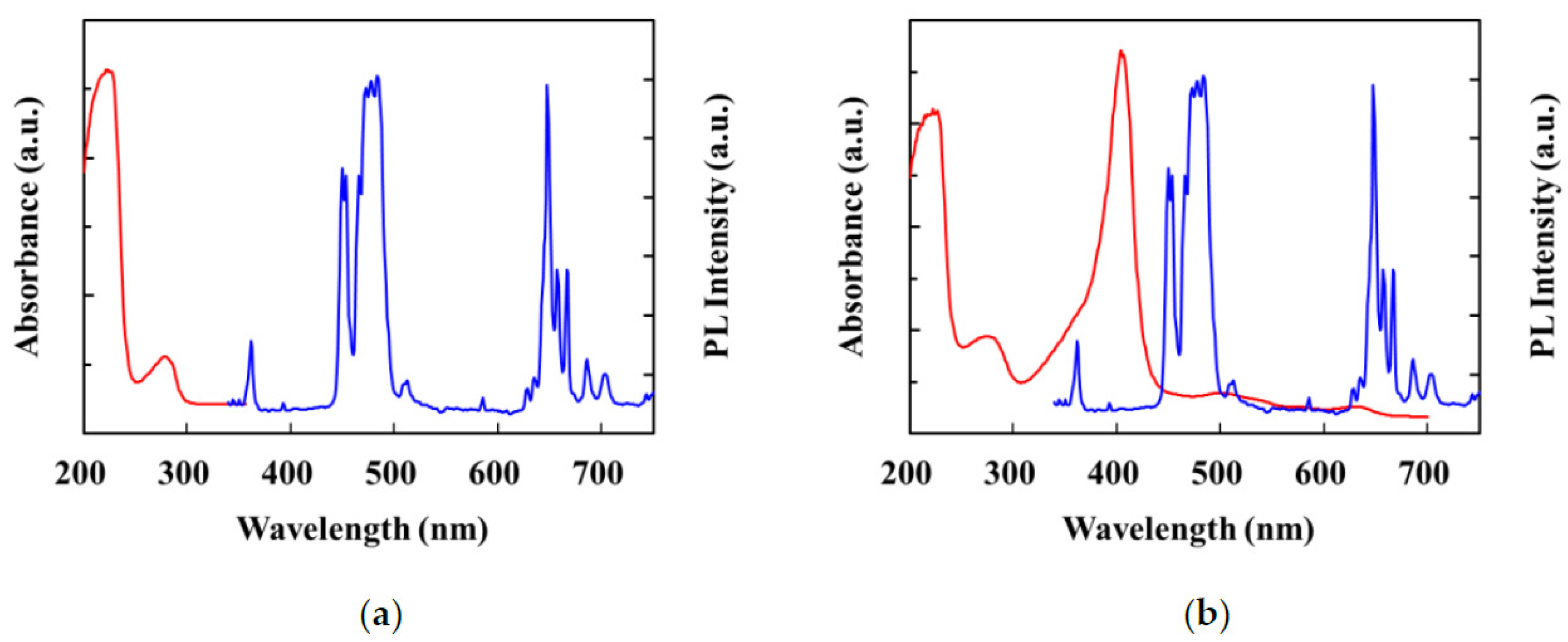
3.4. Effect of Template Molecules on Fluorescence Quenching
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zheng, K.; Loh, K.Y.; Wang, Y.; Chen, Q.; Fan, J.; Jung, T.; Nam, S.H.; Suh, Y.D.; Liu, X. Recent advances in upconversion nanocrystals: Expanding the kaleidoscopic toolbox for emerging applications. Nano Today 2019, 29, 100797. [Google Scholar] [CrossRef]
- Bünzli, J.C.G. Lanthanide photonics: Shaping the nanoworld. Trends Chem. 2019, 1, 751. [Google Scholar] [CrossRef]
- Luther, D.C.; Huang, R.; Jeon, T.; Zhang, X.; Lee, Y.W.; Nagaraj, H.; Rotello, V.M. Delivery of drugs, proteins, and nucleic acids using inorganic nanoparticles. Adv. Drug Deliv. Rev. 2020, 156, 188. [Google Scholar] [CrossRef]
- Yi, Z.; Luo, Z.; Qin, X.; Chen, Q.; Liu, X. Lanthanide-activated nanoparticles: A toolbox for bioimaging, therapeutics, and neuromodulation. Acc. Chem. Res. 2020, 53, 2692. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Kankala, R.K.; Liu, C.G.; Wang, S.B.; Chen, A.Z.; Zhang, Y. Lanthanides-doped near-infrared active upconversion nanocrystals: Upconversion mechanisms and synthesis. Coord. Chem. Rev. 2021, 438, 213870. [Google Scholar] [CrossRef]
- Suter II, J.D.; Pekas, N.J.; Berry, M.T.; May, P.S. Real-time-monitoring of the synthesis of β-NaYF4:17% Yb,3% Er nanocrystals using NIR-to-visible upconversion luminescence. J. Phys. Chem. C 2014, 118, 13238. [Google Scholar] [CrossRef]
- Grzyb, T.; Kamiński, P.; Przybylska, D.; Tymiński, A.; Sanz-Rodríguezb, F.; Gonzalez, P.H. Manipulation of up-conversion emission in NaYF4 core@shell nanoparticles doped by Er3+, Tm3+, or Yb3+ ions by excitation wavelength—Three ions—Plenty of possibilities. Nanoscale 2021, 13, 7322. [Google Scholar] [CrossRef]
- Muhr, V.; Wilhelm, S.; Hirsch, T.; Wolfbeis, O.S. Upconversion nanoparticles: From hydrophobic to hydrophilic surfaces. Acc. Chem. Res. 2014, 47, 3481. [Google Scholar] [CrossRef]
- Chen, Y.; D’Amario, C.; Gee, A.; Duong, H.T.T.; Shimoni, O.; Valenzuela, S.M. Dispersion stability and biocompatibility of four ligand-exchanged NaYF4: Yb, Er upconversion nanoparticles. Acta Biomater. 2020, 102, 384. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.L.; Sharma, P.K.; Singh, A.; Kumar, A.; Shankar, K.R.; Singh, Y.; Garg, N.; Krishnan, V. Amine-functionalized, porous silica-coated NaYF4:Yb/Er upconversion nanophosphors for efficient delivery of doxorubicin and curcumin. Mater. Sci. Eng. C 2019, 96, 86. [Google Scholar] [CrossRef]
- Wu, L.; Yu, J.; Chen, L.; Yang, D.; Zhang, S.; Han, L.; Ban, M.; He, L.; Xu, Y.; Zhang, Q. A general and facile approach to disperse hydrophobic nanocrystals in water with enhanced long-term stability. J. Mater. Chem. C 2017, 5, 3065. [Google Scholar] [CrossRef]
- Su, Q.; Zhou, M.T.; Zhou, M.Z.; Sun, Q.; Ai, T.; Su, Y. Microscale self-assembly of upconversion nanoparticles driven by block copolymer. Front. Chem. 2020, 8, 836. [Google Scholar] [CrossRef]
- Han, G.M.; Li, H.; Huang, X.X.; Kong, D.M. Simple synthesis of carboxyl-functionalized upconversion nanoparticles for biosensing and bioimaging applications. Talanta 2016, 147, 207. [Google Scholar] [CrossRef]
- Vijayan, A.N.; Liu, Z.; Zhao, H.; Zhang, P. Nicking enzyme-assisted signal-amplifiable Hg2+ detection using upconversion nanoparticles. Anal. Chim. Acta 2019, 1072, 75. [Google Scholar] [CrossRef] [PubMed]
- Fernando, P.U.A.I.; Glasscott, M.W.; Pokrzywinski, K.; Fernando, B.M.; Kosgei, G.K.; Moores, L.C. Analytical methods incorporating molecularly imprinted polymers (MIPs) for the quantification of microcystins: A mini-review. Crit. Rev. Anal. Chem. 2021, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.D.; Silva, W.R.; Mizaikoff, B. Molecularly imprinted materials for biomedical sensing. Med. Devices Sens. 2021, 4, e10166. [Google Scholar] [CrossRef]
- Gao, M.; Gao, Y.; Chen, G.; Huang, X.; Xu, X.; Lv, J.; Wang, J.; Xu, D.; Liu, G. Recent advances and future trends in the detection of contaminants by molecularly imprinted polymers in food samples. Front. Chem. 2020, 8, 616326. [Google Scholar] [CrossRef]
- Hu, X.; Cao, Y.; Tian, Y.; Qi, Y.; Fang, G.; Wang, S. A molecularly imprinted fluorescence nanosensor based on upconversion metal-organic frameworks for alpha-cypermethrin specific recognition. Microchim. Acta 2020, 187, 632. [Google Scholar] [CrossRef]
- Elmasry, M.R.; Tawfik, S.M.; Kattaev, N.; Lee, Y.I. Ultrasensitive detection and removal of carbamazepine in wastewater using UCNPs functionalized with thin-shell MIPs. Microchem. J. 2021, 170, 106674. [Google Scholar] [CrossRef]
- Yu, Q.; He, C.; Li, Q.; Zhou, Y.; Duan, N.; Wu, S. Fluorometric determination of acetamiprid using molecularly imprinted upconversion nanoparticles. Mikrochim. Acta 2020, 187, 222. [Google Scholar] [CrossRef]
- Nagao, C.; Sawamoto, M.; Terashima, T. Molecular imprinting on amphiphilic folded polymers for selective molecular recognition in water. J. Polym. Sci. 2020, 58, 215. [Google Scholar] [CrossRef] [Green Version]
- Liras, M.; González-Béjar, M.; Peinado, E.; Francés-Soriano, L.; Pérez-Prieto, J.; Quijada-Garrido, I.; García, O. Thin amphiphilic polymer-capped upconversion nanoparticles: Enhanced emission and thermoresponsive properties. Chem. Mater. 2014, 26, 4014. [Google Scholar] [CrossRef]
- Chien, H.W.; Yang, C.H.; Tsai, M.T.; Wang, T.L. Photoswitchable spiropyran-capped hybrid nanoparticles based on UV-emissive and dual-emissive upconverting nanocrystals for bioimaging. J. Photochem. Photobiol. A 2020, 392, 112303. [Google Scholar] [CrossRef]
- Chien, H.W.; Wu, C.H.; Yang, C.H.; Wang, T.L. Multiple doping effect of LiYF4:Yb3+/Er3+/Ho3+/Tm3+@LiYF4:Yb3+ core/shell nanoparticles and its application in Hg2+ sensing detection. J. Alloys Compd. 2019, 806, 272–282. [Google Scholar] [CrossRef]
- EL-Sharif, H.F.; Yapati, H.; Kalluru, S.; Reddy, S.M. Highly selective BSA imprinted polyacrylamide hydrogels facilitated by a metal-coding MIP approach. Acta Biomater. 2015, 28, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, H.W.; Tsai, M.T.; Yang, C.H.; Lee, R.H.; Wang, T.L. Interaction of LiYF4:Yb3+/Er3+/Ho3+/Tm3+@LiYF4:Yb3+ upconversion nanoparticles, molecularly imprinted polymers, and templates. RSC Adv. 2020, 10, 35600. [Google Scholar] [CrossRef]
- Wu, N.; Wei, Y.; Pan, L.; Yang, X.; Qi, H.; Gao, Q.; Zhang, C. Lateral flow immunostrips for the sensitive and rapid determination of 8-hydroxy-2′-deoxyguanosine using upconversion nanoparticles. Microchim. Acta 2020, 187, 377. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, M.; Gao, Z.; Liu, X.; Gao, X.; Ma, T.; Lu, X.; Li, J. Upconversion nanoparticles capped with molecularly imprinted polymer as fluorescence probe for the determination of ractopamine in water and pork. Food Anal. Methods 2017, 10, 2964. [Google Scholar] [CrossRef]
- Zhang, Q.; Yan, B. Hydrothermal synthesis and characterization of LiREF4 (RE = Y, Tb−Lu) nanocrystals and their core−shell nanostructures. Inorg. Chem. 2010, 49, 6834. [Google Scholar] [CrossRef]
- Wen, S.; Zhou, J.; Schuck, P.J.; Suh, Y.D.; Schmidt, T.W.; Jin, D. Future and challenges for hybrid upconversion nanosystems. Nat. Photonics 2019, 13, 828. [Google Scholar] [CrossRef]
- Tessitore, G.; Mandl, G.A.; Brik, M.G.; Park, W.; Capobianco, J.A. Recent insights into upconverting nanoparticles: Spectroscopy, modeling, and routes to improved luminescence. Nanoscale 2019, 11, 12015. [Google Scholar] [CrossRef]
- Fang, Y.; Liu, L.; Zhang, F. Exploiting lanthanide-doped upconversion nanoparticles with core/shell structures. Nano Today 2019, 25, 68. [Google Scholar]
- Shi, L.; Hu, J.; Wu, X.; Zhan, S.; Hu, S.; Tang, Z.; Chen, M.; Liu, Y. Upconversion core/shell nanoparticles with lowered surface quenching for fluorescence detection of Hg2+ ions. Dalton Trans. 2018, 47, 16445. [Google Scholar] [CrossRef] [PubMed]
- Ohshio, M.; Ishihara, K.; Yusa, S. Self-association behavior of cell membrane-inspired amphiphilic random copolymers in water. Polymers 2019, 11, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.W.; Jeans, C.W.; Brain, K.R.; Allender, C.J.; Hlady, V.; Britt, D.W. From 3D to 2D: A review of the molecular imprinting of proteins. Biotechnol. Prog. 2006, 22, 1474. [Google Scholar] [CrossRef]
- Baler, K.; Martin, O.A.; Carignano, M.A.; Ameer, A.G.; Vila, J.A.; Szleifer, I. Electrostatic unfolding and interactions of albumin driven by pH Changes: A molecular dynamics study. J. Phys. Chem. B 2014, 118, 921. [Google Scholar] [CrossRef]
- Erickson, H.P. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 2009, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- van Oss, C.J. Macroscopic and microscopic aspects of repulsion versus attraction in adsorption and adhesion in water. Interface Sci. Technol. 2008, 16, 167. [Google Scholar]
- Sapsford, K.E.; Berti, L.; Medintz, I.L. Materials for fluorescence resonance energy transfer analysis: Beyond traditional donor–acceptor combinations. Angew. Chem. Int. Ed. Engl. 2006, 45, 4562–4589. [Google Scholar] [CrossRef]
- Bagheri, A.; Arandiyan, H.; Boyer, C.; Lim, M. Lanthanide-doped upconversion nanoparticles: Emerging intelligent light-activated drug delivery systems. Adv. Sci. 2016, 3, 1500437. [Google Scholar] [CrossRef] [Green Version]
- Suryawanshi, V.D.; Walekar, L.S.; Gore, A.H.; Anbhule, P.V.; Kolekar, G.B. Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin. J. Pharm. Anal. 2016, 6, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmoud, S.S.; Amal, E.I. Analysis of retinal b-wave by fourier transformation due to ammonia exposure and the role of blood erythrocytes. Rom. J. Biophys. 2010, 20, 269. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chien, H.-W.; Yang, C.-H.; Shih, Y.-T.; Wang, T.-L. Upconversion Nanoparticles Encapsulated with Molecularly Imprinted Amphiphilic Copolymer as a Fluorescent Probe for Specific Biorecognition. Polymers 2021, 13, 3522. https://doi.org/10.3390/polym13203522
Chien H-W, Yang C-H, Shih Y-T, Wang T-L. Upconversion Nanoparticles Encapsulated with Molecularly Imprinted Amphiphilic Copolymer as a Fluorescent Probe for Specific Biorecognition. Polymers. 2021; 13(20):3522. https://doi.org/10.3390/polym13203522
Chicago/Turabian StyleChien, Hsiu-Wen, Chien-Hsin Yang, Yan-Tai Shih, and Tzong-Liu Wang. 2021. "Upconversion Nanoparticles Encapsulated with Molecularly Imprinted Amphiphilic Copolymer as a Fluorescent Probe for Specific Biorecognition" Polymers 13, no. 20: 3522. https://doi.org/10.3390/polym13203522
APA StyleChien, H.-W., Yang, C.-H., Shih, Y.-T., & Wang, T.-L. (2021). Upconversion Nanoparticles Encapsulated with Molecularly Imprinted Amphiphilic Copolymer as a Fluorescent Probe for Specific Biorecognition. Polymers, 13(20), 3522. https://doi.org/10.3390/polym13203522





