A Modified 3D-QSAR Model Based on Ideal Point Method and Its Application in the Molecular Modification of Plasticizers with Flame Retardancy and Eco-Friendliness
Abstract
1. Introduction
2. Materials and Methods
2.1. Sources of Data
 contained in PAEs is 2, while the CFT values of other halogen-free functional groups are 0. M refers to the molecular weight of monomers in the polymer, and here refers to the relative molecular weight of PAEs.
contained in PAEs is 2, while the CFT values of other halogen-free functional groups are 0. M refers to the molecular weight of monomers in the polymer, and here refers to the relative molecular weight of PAEs.2.2. Multi-Effect Comprehensive Evaluation Method for PAEs’ Flammability, Biotoxicity, and Enrichment—Ideal Point Method
2.3. Construction of PAEs’ Flammability, Biotoxicity, and Enrichment Multi-Effect 3D-QSAR Model
2.4. Evaluation of Eco-Friendliness, Stability, and Insulation of PAE Derivatives
3. Results
3.1. Construction and Verification of Multi-Effect 3D-QSAR Model of PAEs’ Flammability, Biotoxicity, and Enrichment
3.2. Molecular Modification and Evaluation Based on PAEs’ 3D-QSAR Model
3.3. Evaluation of Eco-Friendliness, Stability, and Insulation of PAE Derivatives
3.4. Mechanism Analysis of PAE Derivatives’ Improved Multi-Effect and Flammability, Biotoxicity, Enrichment Effects
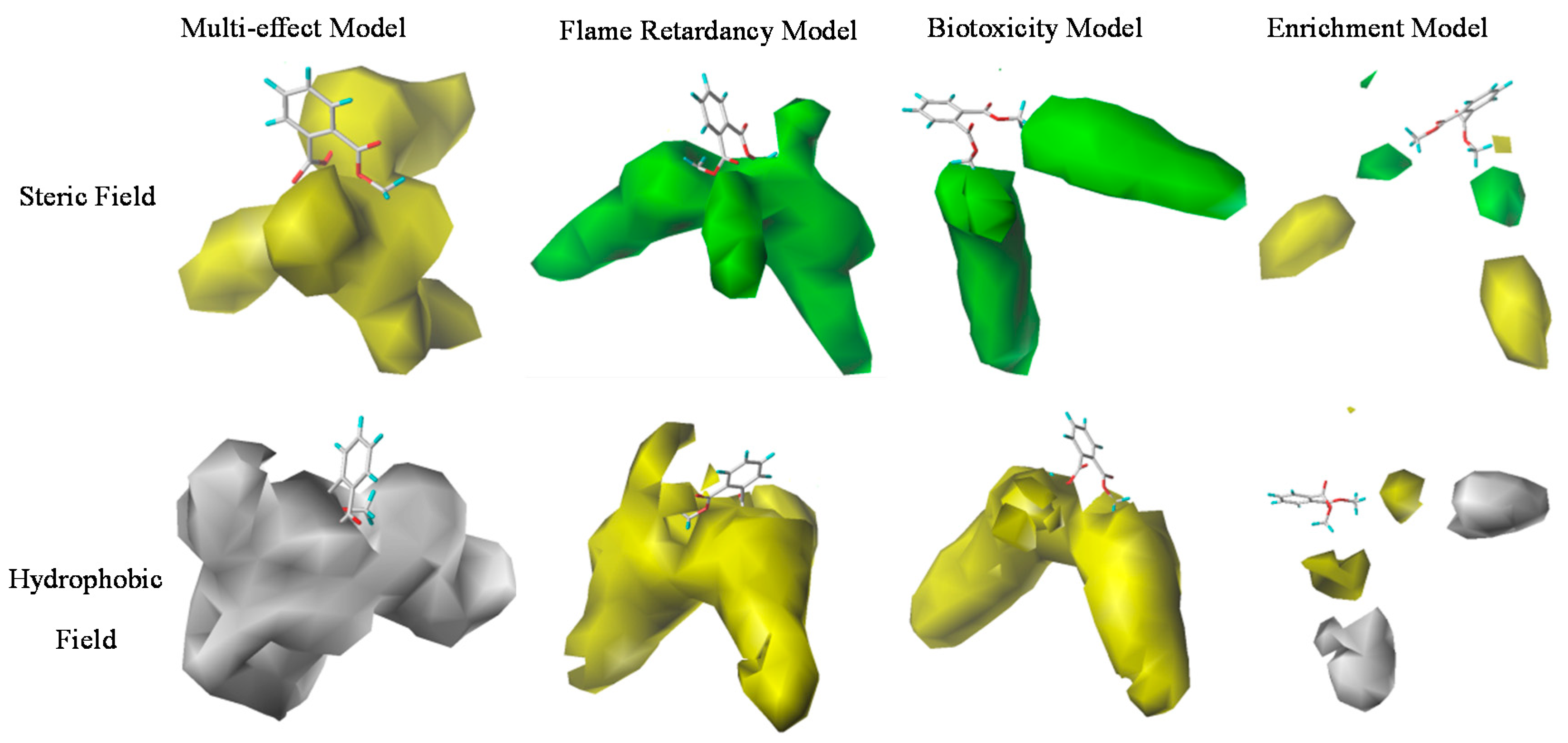
3.4.1. Mechanism Analysis of PAE Derivatives’ Improved Multiple Effects Based on the Three-Dimension Contour Maps
3.4.2. Mechanism Analysis of PAE Derivatives’ Improved Multiple Effects Based on Modified Group Properties
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shrivastava, A. Plastic Properties and Testing. In Introduction to Plastics Engineering; Elsevier BV: Amsterdam, The Netherlands, 2018; Volume 3, pp. 49–110. [Google Scholar]
- Yu, G.; Ma, C.; Li, J. Flame retardant effect of cytosine pyrophosphate and pentaerythritol on polypropylene. Compos. Part B Eng. 2020, 180, 107520. [Google Scholar] [CrossRef]
- Beaugendre, A.; Lemesle, C.; Bellayer, S.; Degoutin, S.; Duquesne, S.; Casetta, M.; Pierlot, C.; Jaime, F.; Kim, T.; Jimenez, M. Flame retardant and weathering resistant self-layering epoxy-silicone coatings for plastics. Prog. Org. Coat. 2019, 136, 105269. [Google Scholar] [CrossRef]
- Smith, R.; Georlette, P.; Finberg, I.; Reznick, G. Development of environmentally friendly multifunctional flame retardants for commodity and engineering plastics. Polym. Degrad. Stab. 1996, 54, 167–173. [Google Scholar] [CrossRef]
- Ribeiro, F.; O’Brien, J.; Galloway, T.; Thomas, K.V. Accumulation and fate of nano- and micro-plastics and associated contaminants in organisms. TrAC Trends Anal. Chem. 2019, 111, 139–147. [Google Scholar] [CrossRef]
- Xu, S.; Ma, J.; Ji, R.; Pan, K.; Miao, A.-J. Microplastics in aquatic environments: Occurrence, accumulation, and biological effects. Sci. Total. Environ. 2020, 703, 134699. [Google Scholar] [CrossRef]
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as contaminants in the marine environment: A review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef]
- Llorca, M.; Álvarez-Muñoz, D.; Ábalos, M.; Rodríguez-Mozaz, S.; Santos, L.H.; León, V.M.; Campillo, J.A.; Martínez-Gómez, C.; Abad, E.; Farré, M. Microplastics in Mediterranean coastal area: Toxicity and impact for the environment and human health. Trends Environ. Anal. Chem. 2020, 27, e00090. [Google Scholar] [CrossRef]
- Amereh, F.; Babaei, M.; Eslami, A.; Fazelipour, S.; Rafiee, M. The emerging risk of exposure to nano(micro)plastics on endocrine disturbance and reproductive toxicity: From a hypothetical scenario to a global public health challenge. Environ. Pollut. 2020, 261, 114158. [Google Scholar] [CrossRef]
- Abayomi, O.A.; Range, P.; Al-Ghouti, M.A.; Obbard, J.P.; Almeer, S.H.; Ben-Hamadou, R. Microplastics in coastal environments of the Arabian Gulf. Mar. Pollut. Bull. 2017, 124, 181–188. [Google Scholar] [CrossRef]
- Wang, W.; Ge, J.; Yu, X. Bioavailability and toxicity of microplastics to fish species: A review. Ecotoxicol. Environ. Saf. 2020, 189, 109913. [Google Scholar] [CrossRef]
- Lu, L.; Wan, Z.; Luo, T.; Fu, Z.; Jin, Y. Polystyrene microplastics induce gut microbiota dysbiosis and hepatic lipid metabolism disorder in mice. Sci. Total. Environ. 2018, 631–632, 449–458. [Google Scholar] [CrossRef]
- Wu, F.; Wang, Y.; Leung, J.Y.S.; Huang, W.; Zeng, J.; Tang, Y.; Chen, J.; Shi, A.; Yu, X.; Xu, X.; et al. Accumulation of microplastics in typical commercial aquatic species: A case study at a productive aquaculture site in China. Sci. Total. Environ. 2020, 708, 135432. [Google Scholar] [CrossRef]
- Youli, Q.; Long, J.; Yu, L. Theoretical support for the enhancement of infrared spectrum signals by derivatization of phthalic acid esters using a pharmacophore model. Spectrosc. Lett. 2018, 51, 155–162. [Google Scholar] [CrossRef]
- Jia, P.; Zhang, M.; Hu, L.; Feng, G.; Bo, C.; Zhou, Y. Synthesis and Application of Environmental Castor Oil Based Polyol Ester Plasticizers for Poly(vinyl chloride). ACS Sustain. Chem. Eng. 2015, 3, 2187–2193. [Google Scholar] [CrossRef]
- Liepins, R.; Pearce, E.M. Chemistry and Toxicity of Flame Retardants for Plastics. Environ. Health Perspect. 1976, 17, 55–63. [Google Scholar] [CrossRef]
- Wang, F.; Pan, S.; Zhang, P.; Fan, H.; Chen, Y.; Yan, J. Synthesis and Application of Phosphorus-containing Flame Retardant Plasticizer for Polyvinyl Chloride. Fibers Polym. 2018, 19, 1057–1063. [Google Scholar] [CrossRef]
- Kumar, A.; T’Ien, J.S. A computational study of low oxygen flammability limit for thick solid slabs. Combust. Flame 2006, 146, 366–378. [Google Scholar] [CrossRef]
- Fromme, H.; Küchler, T.; Otto, T.; Pilz, K.; Müller, J.; Wenzel, A. Occurrence of phthalates and bisphenol A and F in the environment. Water Res. 2002, 36, 1429–1438. [Google Scholar] [CrossRef]
- Jobling, S.; Reynolds, T.; White, R.; Parker, M.G.; Sumpter, J.P. A variety of environmentally persistent chemicals, including some phthalate plasticizers, are weakly estrogenic. Environ. Health Perspect. 1995, 103, 582–587. [Google Scholar] [CrossRef]
- Poopal, R.-K.; Zhang, J.; Zhao, R.; Ramesh, M.; Ren, Z. Biochemical and behavior effects induced by diheptyl phthalate (DHpP) and Diisodecyl phthalate (DIDP) exposed to zebrafish. Chemosphere 2020, 252, 126498. [Google Scholar] [CrossRef]
- Marx, J.L. Phthalic Acid Esters: Biological Impact Uncertain. Science 1972, 178, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.; Cho, S.; Lee, H.; Deb, G.; Lee, Y.; Kwon, T.; Kong, I. Effect of cytoplasmic lipid content on in vitro developmental efficiency of bovine IVP embryos. Theriogenology 2009, 72, 584–589. [Google Scholar] [CrossRef]
- Wu, N.; Niu, F.; Lang, W.; Yu, J.; Fu, G. Synthesis of reactive phenylphosphoryl glycol ether oligomer and improved flame retardancy and mechanical property of modified rigid polyurethane foams. Mater. Des. 2019, 181, 107929. [Google Scholar] [CrossRef]
- Vahabi, H.; Kandola, B.K.; Saeb, M. Flame Retardancy Index for Thermoplastic Composites. Polymers 2019, 11, 407. [Google Scholar] [CrossRef] [PubMed]
- Vahabi, H.; Laoutid, F.; Movahedifar, E.; Khalili, R.; Rahmati, N.; Vagner, C.; Cochez, M.; Brison, L.; Ducos, F.; Ganjali, M.R.; et al. Description of complementary actions of mineral and organic additives in thermoplastic polymer composites by Flame Retardancy Index. Polym. Adv. Technol. 2019, 30, 2056–2066. [Google Scholar] [CrossRef]
- Li, Q.; Qiu, Y.; Li, Y. Molecular Design of Environment-Friendly PAE Derivatives Based on 3D-QSAR Assisted with a Comprehensive Evaluation Method Combining Toxicity and Estrogen Activities. Water Air Soil Pollut. 2020, 231, 1–13. [Google Scholar] [CrossRef]
- Xu, Y.L.; Wang, Y.H.; Xia, G.L. Practical Flame Retardant Technology for High Polymer Materials, 1st ed.; Chemical Industry Press: Beijing, China, 1987; pp. 78–103. [Google Scholar]
- Donckels, B.M.; De Pauw, D.J.; Vanrolleghem, P.; De Baets, B. An ideal point method for the design of compromise experiments to simultaneously estimate the parameters of rival mathematical models. Chem. Eng. Sci. 2010, 65, 1705–1719. [Google Scholar] [CrossRef]
- Nilsson, L.M.; Carter, R.E.; Sterner, O.; Liljefors, T. Structure-Activity Relationships for Unsaturated Dialdehydes 2. A PLS Correlation of Theoretical Descriptors for Six Compounds with Mutagenic Activity in the Ames Salmonella Assay. Quant. Struct. Relationships 1988, 7, 84–91. [Google Scholar] [CrossRef]
- Chang, M.-W.; Lin, C.-J. Leave-One-Out Bounds for Support Vector Regression Model Selection. Neural Comput. 2005, 17, 1188–1222. [Google Scholar] [CrossRef]
- Gu, W.; Chen, Y.; Zhang, L.; Li, Y. Prediction of octanol-water partition coefficient for polychlorinated naphthalenes through three-dimensional QSAR models. Hum. Ecol. Risk Assess. Int. J. 2016, 23, 1–16. [Google Scholar] [CrossRef]
- Mahapatra, M.K.; Bera, K.; Singh, D.V.; Kumar, R.; Kumar, M. In silico modelling and molecular dynamics simulation studies of thiazolidine based PTP1B inhibitors. J. Biomol. Struct. Dyn. 2017, 36, 1195–1211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mao, J.; Li, W.; Koike, K.; Wang, J. Improved 3D-QSAR prediction by multiple-conformational alignment: A case study on PTP1B inhibitors. Comput. Biol. Chem. 2019, 83, 107134. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhao, Y.; Li, Q.; Li, Y. Highly biodegradable fluoroquinolone derivatives designed using the 3D-QSAR model and biodegradation pathways analysis. Ecotoxicol. Environ. Saf. 2020, 191, 110186. [Google Scholar] [CrossRef]
- Gissi, A.; Lombardo, A.; Roncaglioni, A.; Gadaleta, D.; Mangiatordi, G.F.; Nicolotti, O.; Benfenati, E. Evaluation and comparison of benchmark QSAR models to predict a relevant REACH endpoint: The bioconcentration factor (BCF). Environ. Res. 2015, 137, 398–409. [Google Scholar] [CrossRef]
- Gu, W.; Zhao, Y.; Li, Q.; Li, Y. Environmentally friendly polychlorinated naphthalenes (PCNs) derivatives designed using 3D-QSAR and screened using molecular docking, density functional theory and health-based risk assessment. J. Hazard. Mater. 2019, 363, 316–327. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Babu, S.; Sohn, H.; Madhavan, T. Computational Analysis of CRTh2 receptor antagonist: A Ligand-based CoMFA and CoMSIA approach. Comput. Biol. Chem. 2015, 56, 109–121. [Google Scholar] [CrossRef]
- Liu, S.; Sun, S. Combined QSAR/QSPR, Molecular Docking, and Molecular Dynamics Study of Environmentally Friendly PBDEs with Improved Insulating Properties. Chem. Res. Chin. Univ. 2019, 35, 478–484. [Google Scholar] [CrossRef]
- Hennig, C.; Cooper, D.M. Brief communication: The relation between standard error of the estimate and sample size of histomorphometric aging methods. Am. J. Phys. Anthropol. 2011, 145, 658–664. [Google Scholar] [CrossRef]
- Halder, A.; Saha, A.; Jha, T. Exploring QSAR and pharmacophore mapping of structurally diverse selective matrix metalloproteinase-2 inhibitors. J. Pharm. Pharmacol. 2013, 65, 1541–1554. [Google Scholar] [CrossRef]
- Khan, N.; Halim, S.A.; Khan, W.; Zafar, S.K.; Ul-Haq, Z. In-silico designing and characterization of binding modes of two novel inhibitors for CB1 receptor against obesity by classical 3D-QSAR approach. J. Mol. Graph. Model. 2019, 89, 199–214. [Google Scholar] [CrossRef]
- Nazari, M.; Tabatabai, S.A.; Rezaee, E. 2D & 3D-QSAR Study on Novel Piperidine and Piperazine Derivatives as Acetylcholinesterase Enzyme Inhibitors. Curr. Comput. Drug Des. 2018, 14, 391–397. [Google Scholar] [CrossRef]
- Khabeev, N.S.; Shagapov, V.; Yumagulova, Y.; Bailey, S. Evolution of vapor pressure at contact with liquid. Acta Astronaut. 2015, 114, 147–151. [Google Scholar] [CrossRef]
- Wania, F. Assessing the Potential of Persistent Organic Chemicals for Long-Range Transport and Accumulation in Polar Regions. Environ. Sci. Technol. 2003, 37, 1344–1351. [Google Scholar] [CrossRef]

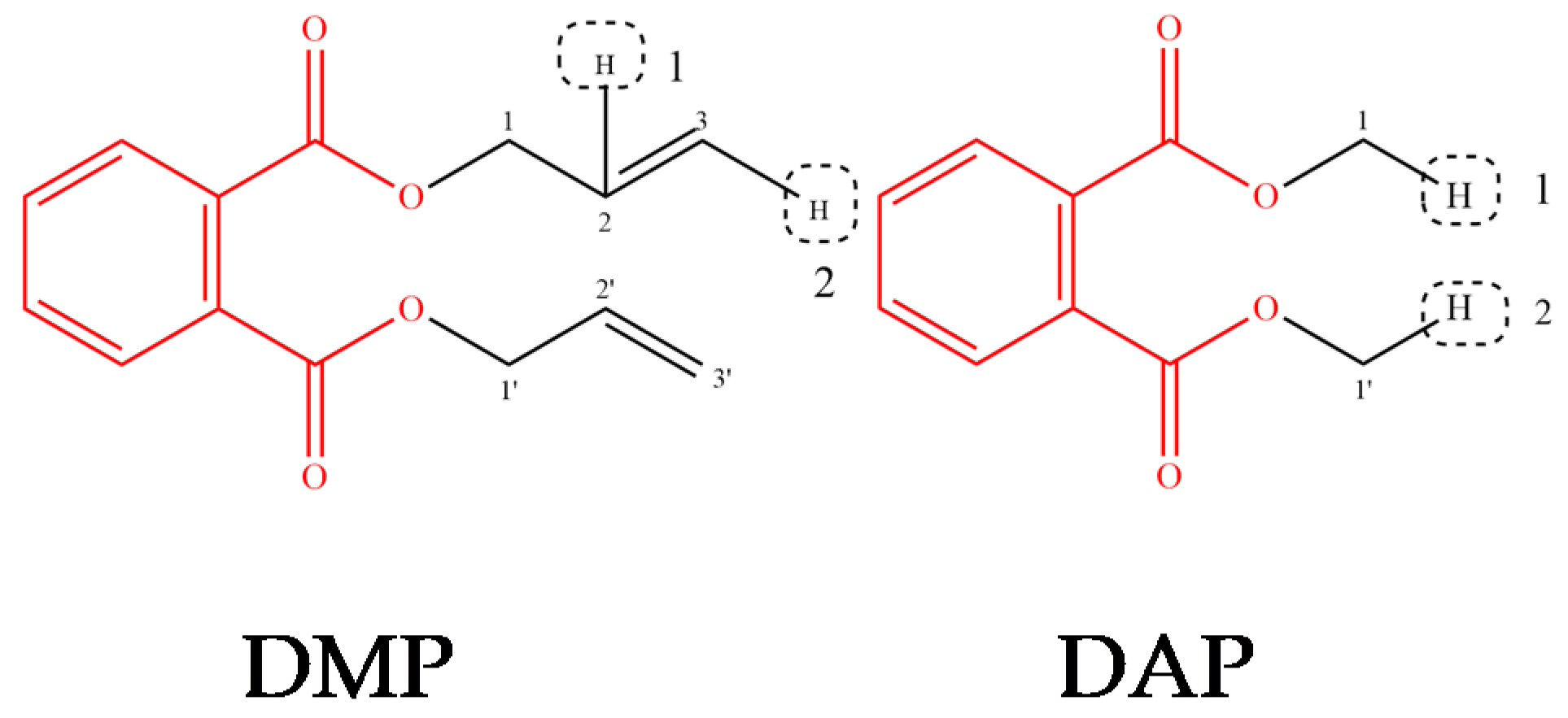
| Molecule | LOI (%) | Fish LC50 (mg/L) | logBCF | logHL | logKOA | Z |
|---|---|---|---|---|---|---|
| dimethyl phthalate (DMP) | 22.44 | 40.822 | 0.723 | 3.617 | 6.694 | 0.371 |
| diisopropyl phthalate (DIPrP) | 21.34 | 4.568 | 1.534 | 3.179 | 7.376 | 0.599 |
| diethyl phthalate (DEP) | 21.82 | 12.471 | 1.264 | 3.156 | 7.505 | 0.526 |
| di-n-butyl phthalate (DBP) | 20.95 | 1.113 | 2.636 | 2.734 | 8.631 | 0.712 |
| di-isopentyl phthalate (DIPP) | 20.63 | 0.398 | 3.260 | 2.904 | 9.504 | 0.794 |
| benzyl butyl phthalate (BBP) | 20.57 | 0.911 | 2.788 | 2.915 | 9.018 | 0.760 |
| diisobutyl phthalate (DIBP) | 20.95 | 1.356 | 2.379 | 3.182 | 8.412 | 0.692 |
| di-n-octyl phthalate (DNOP) | 19.96 | 0.008 | 2.988 | 2.655 | 12.079 | 0.845 |
| diundecyl phthalate (DUP) | 19.52 | 0.000183 | 1.330 | 1.398 | 14.068 | 0.818 |
| di-isoheptyl phthalate (DIHP) | 20.15 | 0.028 | 3.255 | 2.501 | 11.122 | 0.844 |
| ditridecyl phthalate (DTDP) | 19.31 | 0.0000146 | 0.964 | 0.924 | 15.535 | 0.841 |
| di-n-hexyl phthalate (DHP) | 20.37 | 0.095 | 2.793 | 1.639 | 9.799 | 0.784 |
| di-n-pentyl phthalate (DPP) | 20.63 | 0.327 | 2.001 | 3.063 | 9.674 | 0.703 |
| diallyl phthalate (DAP) | 21.40 | 5.323 | 1.798 | 3.895 | 8.032 | 0.605 |
| di-iso-hexyl-phthalates (DIHxP) | 20.37 | 0.116 | 3.908 | 2.623 | 10.239 | 0.880 |
| di-n-propyl phthalate (DPrP) | 21.34 | 3.749 | 1.825 | 3.362 | 8.053 | 0.617 |
| dimethoxyethyl phthalate (DMEP) | 20.90 | 124.130 | 0.402 | 9.544 | 9.766 | 0.311 |
| Molecule | Z | Pred. | Relative Error (%) |
|---|---|---|---|
| DMP a | 0.371 | 0.369 | −0.54 |
| DIPrP a | 0.599 | 0.599 | 0.00 |
| DEP b | 0.526 | 0.563 | 7.03 |
| DBP a | 0.712 | 0.703 | −1.26 |
| DIPP b | 0.794 | 0.583 | −26.57 |
| BBP a | 0.760 | 0.760 | 0.00 |
| DIBP a | 0.692 | 0.692 | 0.00 |
| DNOP b | 0.845 | 0.752 | −11.01 |
| DUP a | 0.818 | 0.818 | 0.00 |
| DIHP a | 0.844 | 0.844 | 0.00 |
| DTDP b | 0.841 | 0.814 | −3.21 |
| DHP a | 0.784 | 0.784 | 0.00 |
| DPP a | 0.703 | 0.707 | 0.57 |
| DAP a | 0.605 | 0.605 | 0.00 |
| DIHxP a | 0.880 | 0.880 | 0.00 |
| DPrP a | 0.617 | 0.624 | 1.13 |
| DMEP a | 0.311 | 0.311 | 0.00 |
| Model | n | q2 | r2 | SEE | F | r2pred | SEP | S | E | H | D | A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CoMSIA | 10 | 0.616 | 1 | 0.008 | 507.314 | 0.66 | 0.16 | 22.90% | 14.00% | 44.90% | 0.00% | 11.20% |
| 3D-QSAR Model | Template Molecule | q2 | n | r2 | F | SEE | r2pred | SEP |
|---|---|---|---|---|---|---|---|---|
| Flammability | DMP | 0.580 | 10 | 1.000 | 9275.112 | 0.010 | 0.901 | 0.346 |
| Biotoxicity | DTDP | 0.832 | 6 | 0.999 | 1375.319 | 0.077 | 0.997 | 0.169 |
| Concentration | DIHxP | 0.526 | 6 | 0.999 | 673.700 | 0.054 | 0.868 | 1.096 |
| Degradability | DAP | 0.747 | 4 | 0.958 | 39.680 | 0.222 | 0.724 | 0.494 |
| Mobility | DTDP | 0.756 | 4 | 0.998 | 162.553 | 0.342 | 0.993 | 0.311 |
| 3D-QSAR Model | S (%) | E (%) | H (%) | D (%) | A (%) |
|---|---|---|---|---|---|
| Flammability | 30.7 | 15.9 | 39.6 | 0 | 13.7 |
| Biotoxicity | 31.0 | 13.2 | 44.4 | 0.0 | 11.3 |
| Concentration | 32.4 | 15.3 | 41.9 | 0.0 | 10.3 |
| Degradability | 35.7 | 12.9 | 47.5 | 0.0 | 3.9 |
| Mobility | 32.5 | 13.7 | 42.1 | 0.0 | 11.6 |
| Flammability Model | Biotoxicity Model | Concentration Model | Degradability Model | Mobility Model | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecule | LOI (%) | Pred. | Relative Error (%) | Molecule | logLC50 | Pred. | Relative Error (%) | Molecule | logBCF | Pred. | Relative Error (%) | Molecule | logHL | Pred. | Relative Error (%) | Molecule | logKOA | Pred. | Relative Error (%) |
| DMP a | 22.44 | 22.440 | 0.00 | DMP a | 1.61 | 1.601 | −0.56 | DMP a | 0.723 | 0.742 | 2.63 | DMP a | 3.617 | 3.622 | 0.14 | DMP a | 6.694 | 6.737 | 0.64 |
| DIPrP a | 21.34 | 21.341 | 0.00 | DIPrP a | 0.66 | 0.679 | 2.88 | DIPrP a | 1.534 | 1.573 | 2.54 | DIPrP a | 3.179 | 3.381 | 6.35 | DIPrP a | 7.376 | 7.66 | 3.85 |
| DEP a | 21.82 | 21.820 | 0.00 | DEP a | 1.10 | 1.075 | −2.27 | DEP a | 1.264 | 1.248 | −1.27 | DEP b | 3.156 | 3.363 | 6.56 | DEP b | 7.505 | 7.72 | 2.86 |
| DBP a | 20.95 | 20.943 | −0.03 | DBP a | 0.05 | 0.105 | 110.00 | DBP b | 2.636 | 2.333 | −11.49 | DBP a | 2.734 | 3.073 | 12.40 | DBP b | 8.631 | 8.548 | −0.96 |
| DIPP b | 20.63 | 20.778 | 0.72 | DIPP a | −0.40 | −0.428 | 7.00 | DIPP a | 3.26 | 3.259 | −0.03 | DIPP b | 2.904 | 3.251 | 11.95 | DIPP a | 9.504 | 9.597 | 0.98 |
| BBP a | 20.57 | 20.570 | 0.00 | BBP a | −0.04 | −0.053 | 32.50 | BBP a | 2.788 | 2.798 | 0.36 | BBP b | 2.915 | 3.266 | 12.04 | BBP b | 9.018 | 9.074 | 0.62 |
| DIBP a | 20.95 | 20.950 | 0.00 | DIBP a | 0.13 | 0.130 | 0.00 | DIBP a | 2.379 | 2.403 | 1.01 | DIBP a | 3.182 | 3.137 | −1.41 | DIBP a | 8.412 | 8.607 | 2.32 |
| DNOP b | 19.96 | 20.124 | 0.82 | DNOP a | −2.10 | −2.102 | 0.10 | DNOP b | 2.988 | 2.532 | −15.26 | DNOP a | 2.655 | 2.787 | 4.97 | DNOP a | 12.079 | 11.845 | −1.94 |
| DUP a | 19.52 | 19.512 | −0.04 | DUP a | −3.74 | −3.850 | 2.94 | DUP a | 1.33 | 1.363 | 2.48 | DUP a | 1.398 | 1.309 | −6.37 | DUP b | 14.068 | 14.352 | 2.02 |
| DIHP a | 20.15 | 20.150 | 0.00 | DIHP a | −1.55 | −1.556 | 0.39 | DIHP a | 3.255 | 3.226 | −0.89 | DIHP a | 2.501 | 2.408 | −3.72 | DIHP a | 11.122 | 10.857 | −2.38 |
| DTDP a | 19.31 | 19.316 | 0.03 | DTDP a | −2.10 | −2.102 | 0.10 | DTDP a | 0.964 | 0.932 | −3.32 | DTDP a | 0.924 | 0.976 | 5.63 | DTDP a | 15.535 | 15.427 | −0.70 |
| DHP a | 20.37 | 20.376 | 0.03 | DHP a | −1.02 | −1.066 | 4.51 | DHP a | 2.793 | 2.84 | 1.68 | DHP b | 1.639 | 2.543 | 55.16 | DHP a | 9.799 | 10.219 | 4.29 |
| DPP b | 20.63 | 20.622 | −0.04 | DPP b | −0.49 | −0.469 | −4.29 | DAP b | 1.798 | 1.835 | 2.06 | DPP a | 3.063 | 2.82 | −7.93 | DPP a | 9.674 | 9.377 | −3.07 |
| DAP a | 21.40 | 21.399 | 0.00 | DAP b | 0.73 | 0.504 | −30.96 | DIHxP a | 3.908 | 3.894 | −0.36 | DAP a | 3.895 | 3.608 | −7.37 | DAP b | 8.032 | 8.079 | 0.59 |
| DIHxP b | 20.37 | 20.784 | 2.03 | DPrP b | 0.57 | 0.671 | 17.72 | DPrP a | 1.825 | 1.745 | −4.38 | DIHxP a | 2.623 | 2.7 | 2.94 | DIHxP a | 10.239 | 9.886 | −3.45 |
| DPrP a | 21.34 | 21.344 | 0.02 | DMEP a | 2.09 | 2.134 | 2.11 | DPrP a | 3.362 | 3.31 | −1.55 | DPrP a | 8.053 | 7.707 | −4.30 | ||||
| DMEP a | 20.90 | 20.900 | 0.00 | DMEP a | 9.766 | 10.049 | 2.90 | ||||||||||||
| Molecule | Z | Decreasing (%) | LOI | Increasing (%) | logLC50 | Increasing (%) | logBCF | Decreasing (%) |
|---|---|---|---|---|---|---|---|---|
| DMP | 0.371 | - | 22.44 | - | 1.61 | - | 0.723 | - |
| DMP-1-CH2NO2 | 0.085 | 77.09 | 23.39 | 4.23 | 1.76 | 9.50 | 0.161 | 77.73 |
| DMP-1-NO2 | 0.234 | 36.93 | 22.92 | 2.13 | 1.98 | 22.67 | 0.372 | 48.55 |
| DMP-1-CH3-2-CH2NO2 | 0.145 | 60.92 | 23.12 | 3.03 | 2.29 | 42.48 | 0.353 | 51.18 |
| DMP-1-CH3-2-NO2 | 0.292 | 21.29 | 22.66 | 0.98 | 2.17 | 34.97 | 0.556 | 23.10 |
| DAP | 0.605 | - | 21.40 | - | 0.73 | - | 1.798 | - |
| DAP-1-CH2NO2 | 0.325 | 46.28 | 22.23 | 3.86 | 2.28 | 212.47 | 1.007 | 43.99 |
| DAP-1-SH | 0.519 | 14.21 | 21.82 | 1.94 | 1.13 | 54.52 | 1.360 | 24.36 |
| DAP-2-CH2NO2 | 0.279 | 53.88 | 22.52 | 5.24 | 2.27 | 210.41 | 0.162 | 90.99 |
| DAP-2-NO2 | 0.293 | 51.57 | 22.36 | 4.50 | 1.94 | 165.89 | 0.183 | 89.82 |
| DAP-2-SH | 0.557 | 7.93 | 21.64 | 1.13 | 0.94 | 28.63 | 1.604 | 10.79 |
| DAP-1-NO2-2-CH2C6H5 | 0.301 | 50.25 | 22.65 | 5.83 | 1.66 | 127.81 | 1.182 | 34.26 |
| DAP-1-NO2-2-CH2CH3 | 0.365 | 39.67 | 22.57 | 5.49 | 1.88 | 157.95 | 1.482 | 17.58 |
| DAP-1-NO2-2-CH2NO2 | 0.164 | 72.89 | 23.18 | 8.30 | 2.55 | 249.45 | 1.324 | 26.36 |
| DAP-1-NO2-2-CH3 | 0.471 | 22.15 | 21.65 | 1.15 | 0.92 | 26.30 | 0.830 | 53.84 |
| DAP-1-NO2-2-CH=CH2 | 0.447 | 26.12 | 22.18 | 3.63 | 2.03 | 177.81 | 0.696 | 61.29 |
| DAP-1-NO2-2-NO2 | −0.172 | 128.43 | 23.35 | 9.12 | 3.20 | 338.22 | 0.761 | 57.68 |
| DAP-1-NO2-2-OCH3 | 0.340 | 43.80 | 22.54 | 5.32 | 1.83 | 150.96 | 1.401 | 22.08 |
| DAP-1-NO2-2-SH | 0.497 | 17.85 | 21.72 | 1.47 | 0.96 | 30.96 | 1.557 | 13.40 |
| DAP-2-CH=CH2-1-CH2NO2 | 0.353 | 41.65 | 21.63 | 1.09 | 0.95 | 30.41 | 0.898 | 50.06 |
| DAP-2-CH=CH2-1-CH3 | 0.529 | 12.56 | 21.56 | 0.74 | 0.90 | 23.70 | 1.366 | 24.03 |
| DAP-2-CH=CH2-1-NO2 | 0.399 | 34.05 | 22.53 | 5.27 | 1.67 | 128.49 | 1.166 | 35.15 |
| DAP-2-CH=CH2-1-OCH3 | 0.602 | 0.50 | 21.79 | 1.80 | 0.94 | 28.08 | 1.393 | 22.53 |
| DAP-2-CH=CH2-1-SH | 0.543 | 10.25 | 21.59 | 0.89 | 0.84 | 14.38 | 1.459 | 18.85 |
| Molecule | logHL | Decreasing (%) | logKOA | Increasing (%) | Freq. | Energy Gap (a.u.) | Increasing (%) |
|---|---|---|---|---|---|---|---|
| DAP | 3.895 | 0.00 | 8.032 | 0.00 | 17.30 | 0.203 | 0.00 |
| DAP-2-CH2NO2 | 3.875 | 0.51 | 6.282 | −21.79 | 9.38 | 0.195 | −3.89 |
| DAP-1-NO2-2-CH2C6H5 | 3.643 | 6.47 | 6.902 | −14.07 | 12.33 | 0.154 | −23.84 |
| DAP-1-NO2-2-CH2CH3 | 3.678 | 5.57 | 7.121 | −11.34 | 12.94 | 0.156 | −22.97 |
| DAP-1-NO2-2-CH2NO2 | 3.624 | 6.96 | 6.936 | −13.65 | 7.75 | 0.154 | −23.91 |
| DAP-1-NO2-2-NO2 | 3.620 | 7.06 | 7.087 | −11.77 | 21.69 | 0.127 | −37.42 |
| DAP-1-NO2-2-OCH3 | 3.527 | 9.45 | 7.523 | −6.34 | 11.70 | 0.175 | −13.40 |
| DAP-2-CH=CH2-1-NO2 | 3.434 | 11.84 | 7.748 | −3.54 | 20.13 | 0.162 | −20.26 |
| Site 1 | Site 2 | Site 1 and 2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | Molecule | Volume | Increasing (%) | logP | Increasing (%) | Coupling Value (%) | Volume | Increasing (%) | logP | Increasing (%) | Coupling Value (%) | Volume | Increasing (%) | logP | Increasing (%) | Coupling Value (%) |
| DMP | 1.0 | 0.00 | 1.54 | 0 | 0.00 | 1.0 | 0.00 | 1.54 | 0.00 | 0.00 | 2.0 | 0.00 | 1.54 | 0.00 | 0.00 | |
| DAP | 1.0 | 0.00 | 3.06 | 0 | 0.00 | 1.0 | 0.00 | 3.06 | 0.00 | 0.00 | 2.0 | 0.00 | 3.06 | 0.00 | 0.00 | |
| –CH3 | DMP-1-CH3-2-CH2NO2 | 15.0 | 1400.00 | 60.0 | 5900.00 | 75.0 | 3650.00 | 1.30 | −15.58 | 828.85 | ||||||
| DMP-1-CH3-2-NO2 | 15.0 | 1400.00 | 46.0 | 4500.00 | 61.0 | 2950.00 | 0.84 | −45.45 | 655.14 | |||||||
| DAP-1-NO2-2-CH3 | 46.0 | 4500.00 | 15.0 | 1400.00 | 61.0 | 2950.00 | 2.77 | −9.48 | 671.29 | |||||||
| DAP-2-CH=CH2-1-CH3 | 27.0 | 2600.00 | 15.0 | 1400.00 | 42.0 | 2000.00 | 3.75 | 22.55 | 468.12 | |||||||
| –CH2CH3 | DAP-1-NO2-2-CH2CH3 | 46.0 | 4500.00 | 29.1 | 2810.00 | 75.1 | 3655.00 | 3.18 | 3.92 | 838.76 | ||||||
| –CH2C6H5 | DAP-1-NO2-2-CH2C6H5 | 46.0 | 4500.00 | 91.1 | 9010.00 | 137.1 | 6755.00 | 4.37 | 42.81 | 1566.12 | ||||||
| –NO2 | DMP-1-NO2 | 46.0 | 4500.00 | 0.56 | −63.64 | 1001.93 | ||||||||||
| DMP-1-CH3-2-NO2 | 15.0 | 1400.00 | 46.0 | 4500.00 | 61.0 | 2950.00 | 0.84 | −45.45 | 655.14 | |||||||
| DAP-2-NO2 | 46.0 | 4500.00 | 2.93 | −4.25 | 1028.59 | |||||||||||
| DAP-1-NO2-2-CH2C6H5 | 46.0 | 4500.00 | 91.1 | 9010.00 | 137.1 | 6755.00 | 4.37 | 42.81 | 1566.12 | |||||||
| DAP-1-NO2-2-CH2CH2CH3 | 46.0 | 4500.00 | 43.1 | 4210.00 | 89.1 | 4355.00 | 3.60 | 17.65 | 1005.22 | |||||||
| DAP-1-NO2-2-CH2CH3 | 46.0 | 4500.00 | 29.1 | 2810.00 | 75.1 | 3655.00 | 3.18 | 3.92 | 838.76 | |||||||
| DAP-1-NO2-2-CH2NO2 | 46.0 | 4500.00 | 60.0 | 5900.00 | 106.0 | 5200.00 | 2.38 | −22.22 | 1180.82 | |||||||
| DAP-1-NO2-2-CH3 | 46.0 | 4500.00 | 15.0 | 1400.00 | 61.0 | 2950.00 | 2.77 | −9.48 | 671.29 | |||||||
| DAP-1-NO2-2-CH=CH2 | 46.0 | 4500.00 | 27.0 | 2600.00 | 73.0 | 3550.00 | 2.91 | −4.90 | 810.75 | |||||||
| DAP-1-NO2-2-NO2 | 46.0 | 4500.00 | 46.0 | 4500.00 | 92.0 | 4500.00 | 2.28 | −25.49 | 1019.05 | |||||||
| DAP-1-NO2-2-OCH3 | 46.0 | 4500.00 | 31.0 | 3000.00 | 77.0 | 3750.00 | 1.96 | −35.95 | 842.61 | |||||||
| DAP-1-NO2-2-SH | 46.0 | 4500.00 | 33.1 | 3210.00 | 79.1 | 3855.00 | 2.43 | −20.59 | 873.55 | |||||||
| DAP-2-CH=CH2-1-NO2 | 27.0 | 2600.00 | 46.0 | 4500.00 | 73.0 | 3550.00 | 2.91 | −4.90 | 810.75 | |||||||
| –CH2NO2 | DMP-1-CH2NO2 | 60.0 | 5900.00 | 1.01 | −34.42 | 1335.65 | ||||||||||
| DMP-1-CH3-2-CH2NO2 | 15.0 | 1400.00 | 60.0 | 5900.00 | 75.0 | 3650.00 | 1.30 | −15.58 | 828.85 | |||||||
| DAP-1-CH2NO2 | 60.0 | 5900.00 | 2.85 | −6.86 | 1348.02 | |||||||||||
| DAP-2-CH2NO2 | 60.0 | 5900.00 | 3.04 | −0.65 | 1350.81 | |||||||||||
| DAP-1-NO2-2-CH2NO2 | 46.0 | 4500.00 | 60.0 | 5900.00 | 106.0 | 5200.00 | 2.38 | −22.22 | 1180.82 | |||||||
| DAP-2-CH=CH2-1-CH2NO2 | 27.0 | 2600.00 | 60.0 | 5900.00 | 87.0 | 4250.00 | 3.37 | 10.13 | 977.80 | |||||||
| –SH | DAP-1-SH | 33.1 | 3210.00 | 2.55 | −16.67 | 727.61 | ||||||||||
| DAP-2-SH | 33.1 | 3210.00 | 3.09 | 0.98 | 735.53 | |||||||||||
| DAP-1-NO2-2-SH | 46.0 | 4500.00 | 33.1 | 3210.00 | 79.1 | 3855.00 | 2.43 | −20.59 | 873.55 | |||||||
| DAP-2-CH=CH2-1-SH | 27.0 | 2600.00 | 33.1 | 3210.00 | 60.1 | 2905.00 | 3.07 | 0.33 | 665.39 | |||||||
| –OCH3 | DAP-1-NO2-2-OCH3 | 46.0 | 4500.00 | 31.0 | 3000.00 | 77.0 | 3750.00 | 1.96 | −35.95 | 842.61 | ||||||
| DAP-2-CH=CH2-1-OCH3 | 27.0 | 2600.00 | 31.0 | 3000.00 | 58.0 | 2800.00 | 2.60 | −15.03 | 634.45 | |||||||
| –CH=CH2 | DAP-1-NO2-2-CH=CH2 | 46.0 | 4500.00 | 27.0 | 2600.00 | 73.0 | 3550.00 | 2.91 | −4.90 | 810.75 | ||||||
| DAP-2-CH=CH2-1-CH2NO2 | 27.0 | 2600.00 | 60.0 | 5900.00 | 87.0 | 4250.00 | 3.37 | 10.13 | 977.80 | |||||||
| DAP-2-CH=CH2-1-CH3 | 27.0 | 2600.00 | 15.0 | 1400.00 | 42.0 | 2000.00 | 3.75 | 22.55 | 468.12 | |||||||
| DAP-2-CH=CH2-1-NO2 | 27.0 | 2600.00 | 46.0 | 4500.00 | 73.0 | 3550.00 | 2.91 | −4.90 | 810.75 | |||||||
| DAP-2-CH=CH2-1-OCH3 | 27.0 | 2600.00 | 31.0 | 3000.00 | 58.0 | 2800.00 | 2.60 | −15.03 | 634.45 | |||||||
| DAP-2-CH=CH2-1-SH | 27.0 | 2600.00 | 33.1 | 3210.00 | 60.1 | 2905.00 | 3.07 | 0.33 | 665.39 | |||||||
| Group | Comprehensive Effect (%) | Group Properties (%) | Flame Retardancy (%) | Biotoxicity (%) | Concentration (%) | Weighted Comprehensive Effect (%) |
|---|---|---|---|---|---|---|
| –CH3 | 29.23 | 655.85 | 1.48 | 31.86 | 38.04 | 21.56 |
| –CH2CH3 | 39.67 | 838.76 | 5.49 | 157.95 | 17.58 | 54.86 |
| –CH2C6H5 | 50.25 | 1566.12 | 5.83 | 127.81 | 34.26 | 50.95 |
| –NO2 | 45.42 | 941.61 | 4.43 | 134.29 | 40.26 | 54.14 |
| –CH2NO2 | 58.79 | 1170.32 | 4.29 | 125.79 | 56.72 | 56.47 |
| –SH | 12.56 | 750.52 | 1.36 | 32.12 | 16.85 | 15.23 |
| –OCH3 | 22.15 | 738.53 | 3.56 | 89.52 | 22.31 | 34.97 |
| –CH=CH2 | 20.86 | 727.88 | 2.24 | 67.15 | 35.32 | 31.63 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhao, C.; Na, H. A Modified 3D-QSAR Model Based on Ideal Point Method and Its Application in the Molecular Modification of Plasticizers with Flame Retardancy and Eco-Friendliness. Polymers 2020, 12, 1942. https://doi.org/10.3390/polym12091942
Zhang H, Zhao C, Na H. A Modified 3D-QSAR Model Based on Ideal Point Method and Its Application in the Molecular Modification of Plasticizers with Flame Retardancy and Eco-Friendliness. Polymers. 2020; 12(9):1942. https://doi.org/10.3390/polym12091942
Chicago/Turabian StyleZhang, Haigang, Chengji Zhao, and Hui Na. 2020. "A Modified 3D-QSAR Model Based on Ideal Point Method and Its Application in the Molecular Modification of Plasticizers with Flame Retardancy and Eco-Friendliness" Polymers 12, no. 9: 1942. https://doi.org/10.3390/polym12091942
APA StyleZhang, H., Zhao, C., & Na, H. (2020). A Modified 3D-QSAR Model Based on Ideal Point Method and Its Application in the Molecular Modification of Plasticizers with Flame Retardancy and Eco-Friendliness. Polymers, 12(9), 1942. https://doi.org/10.3390/polym12091942







