Effects of Different Denaturants on Properties and Performance of Soy Protein-Based Adhesive
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Cross-Linker Triglycidylamine (CA)
2.3. Preparation of Adhesive
2.4. Surface Hydrophobicity Measurement
2.5. Residual Rate Test
2.6. Three-Ply Plywood Preparation and Evaluation
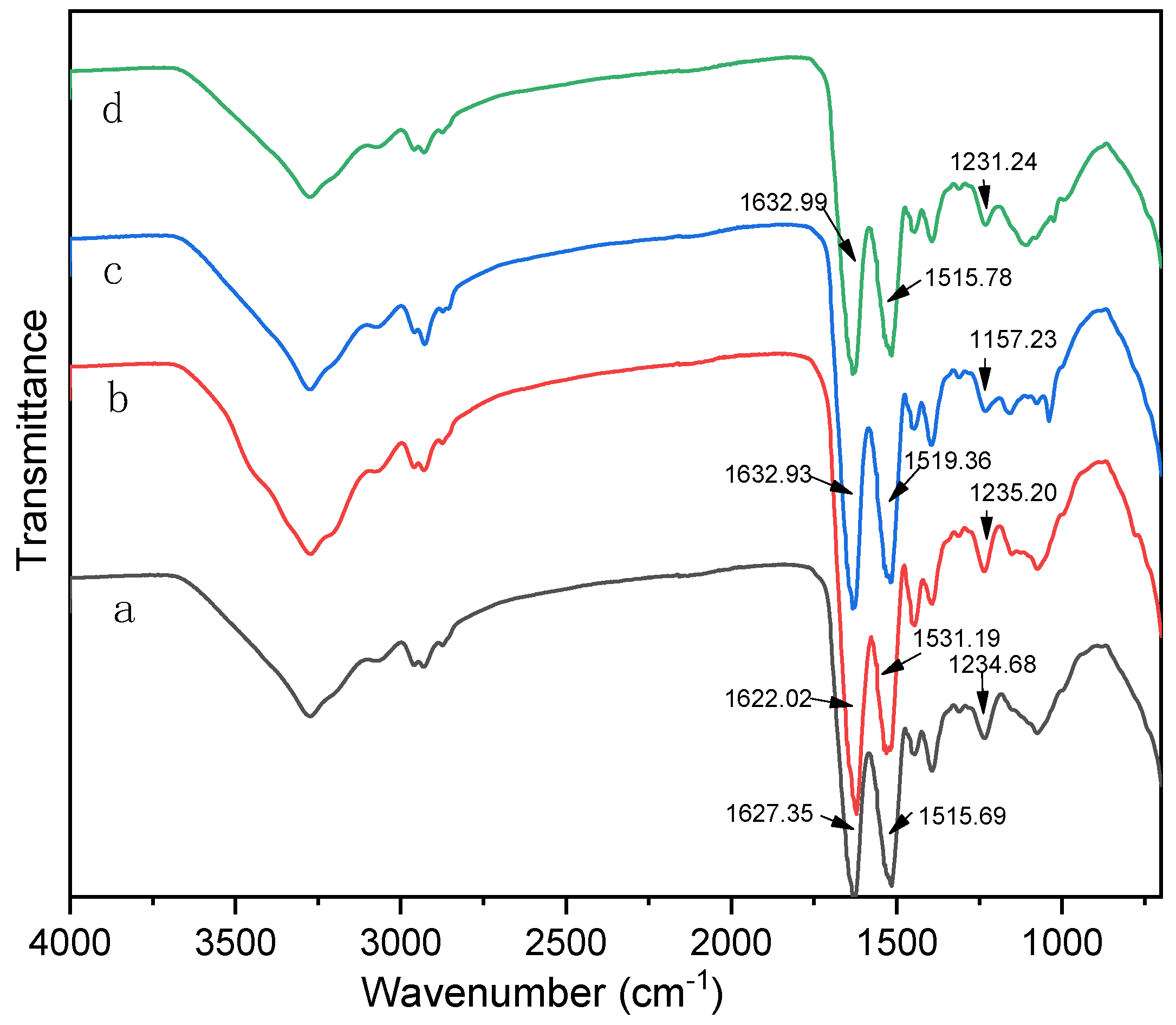
2.7. Fourier Transform Infrared (FTIR) Spectroscopy
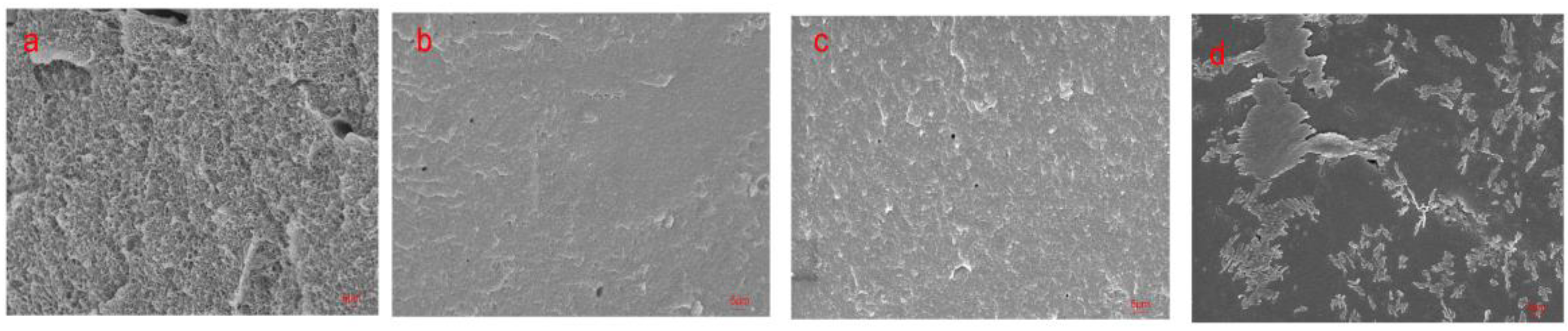
2.8. Scanning Electron Microscopy (SEM)
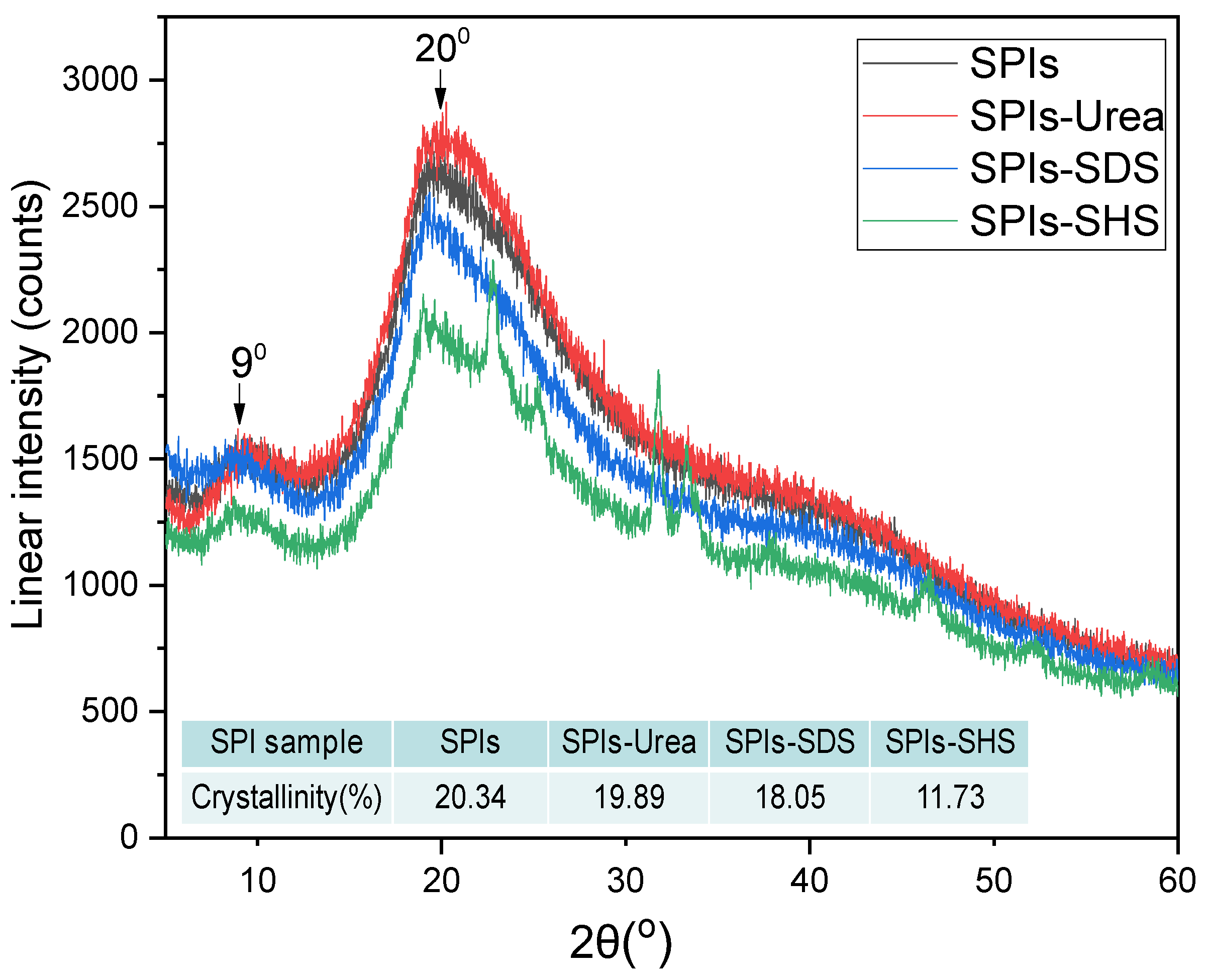
2.9. X-ray Diffraction (XRD)
2.10. Apparent Viscosity
2.11. Statistical Analysis
3. Results and Discussion
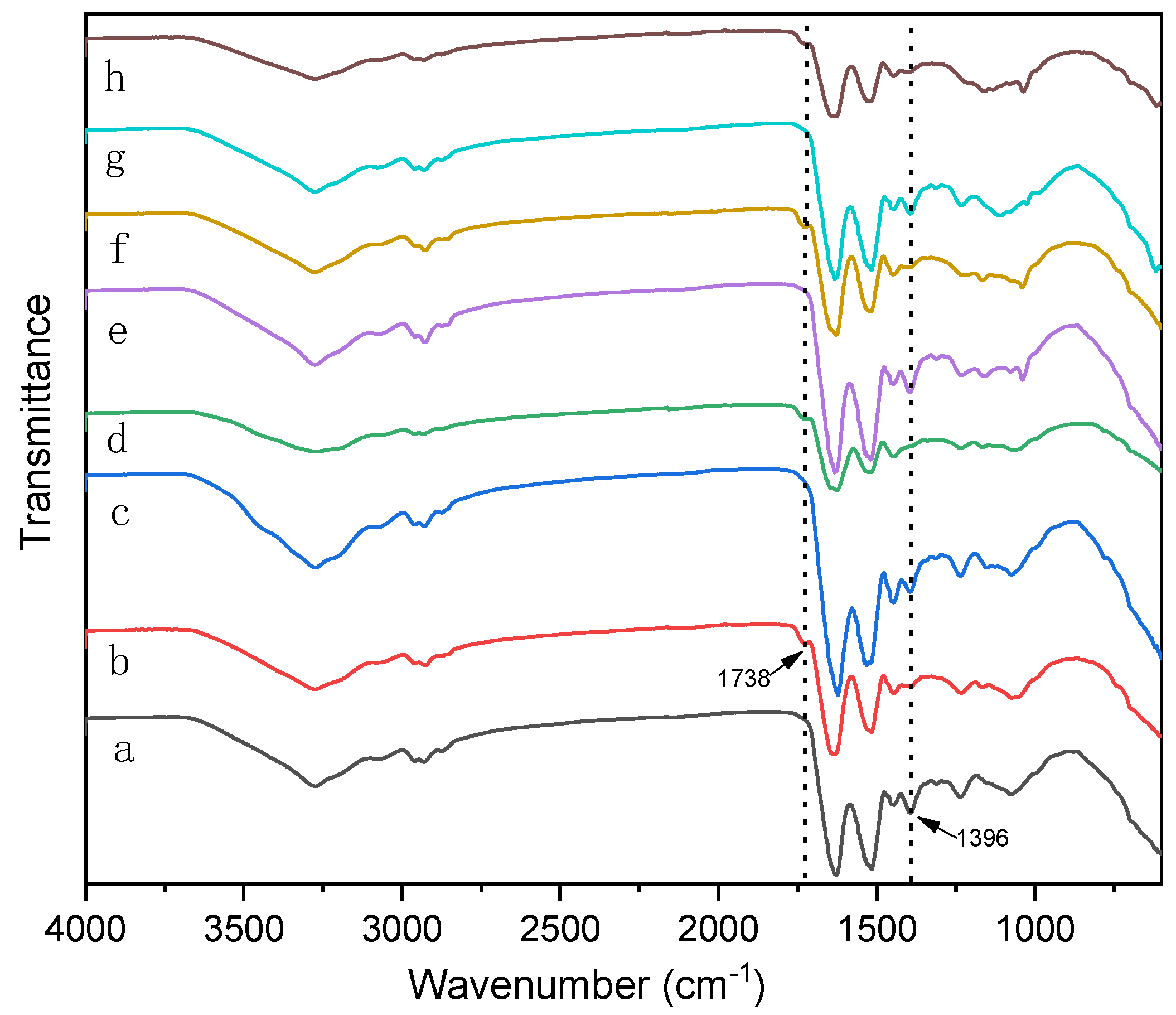
3.1. The effects of Denaturing Agent on SPI Adhesive
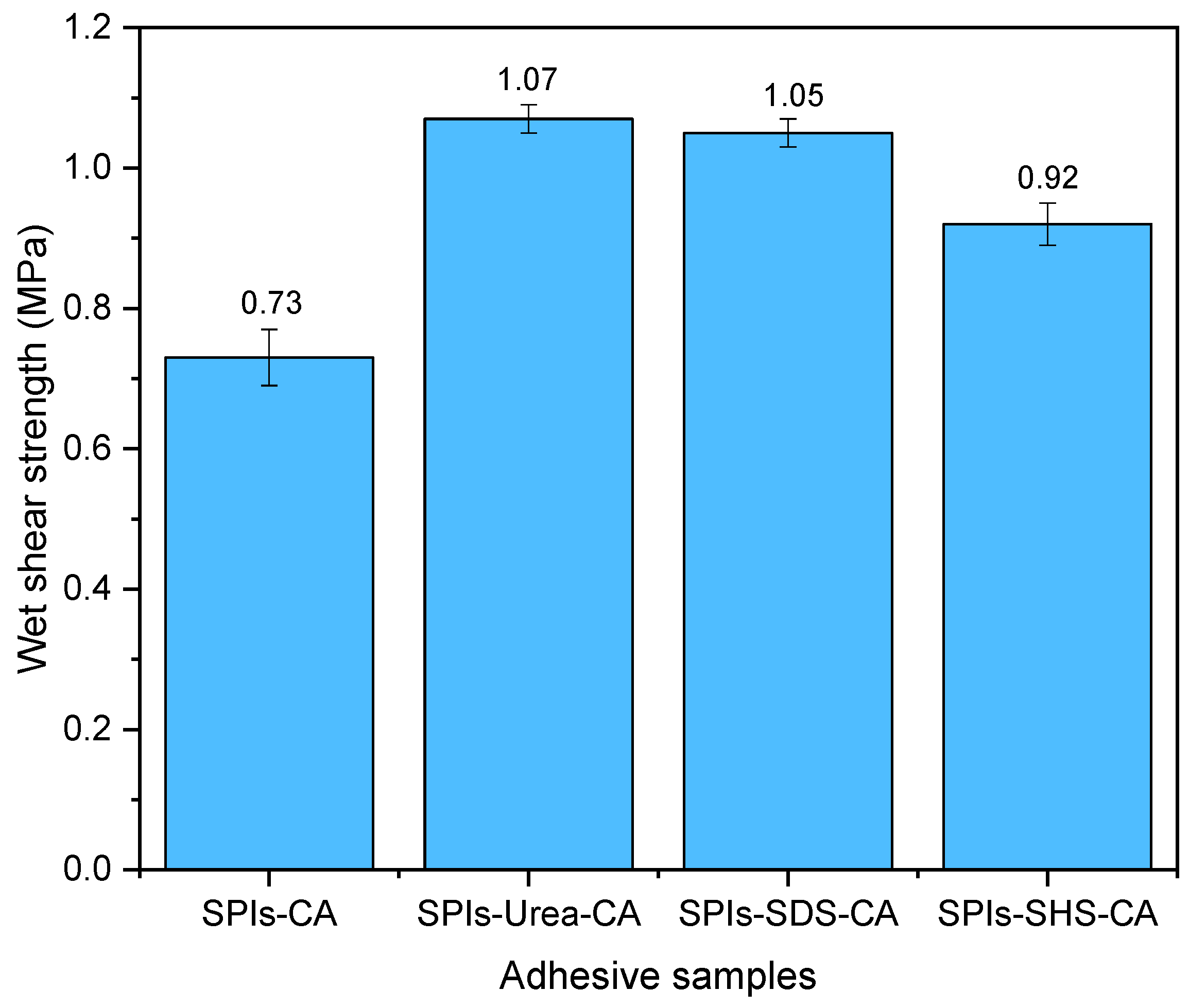
3.2. The effects of Denaturing Agent on SPI/CA Adhesive
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, M.; Luo, J.; Shi, R.; Zhang, J.; Gao, Q.; Li, J. Improved Adhesion Performance of Soy Protein-Based Adhesives with a Larch Tannin-Based Resin. Polymers 2017, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liu, C.; Luo, J.; Li, X.; Chen, L.; Wang, W.; Li, J. Effects of resin open time and melamine addition on cold pre-pressing performance of a urea–formaldehyde resin. Eur. J. Wood Wood Prod. 2018, 76, 1253–1261. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.; Chen, M.; Gao, Q.; Li, J. A High-Performance and Low-Cost Soy Flour Adhesive with a Hydroxymethyl Melamine Prepolymer. Polymers 2018, 10, 909. [Google Scholar] [CrossRef] [PubMed]
- Kaith, B.S.; Jindal, R.; Bhatia, J.K. Morphological and thermal evaluation of soy protein concentrate on graft copolymerization with ethylmethacrylate. J. Appl. Polym. Sci. 2011, 120, 2183–2190. [Google Scholar] [CrossRef]
- Pradyawong, S.; Qi, G.; Sun, X.S.; Wang, D. Laccase/TEMPO-modified lignin improved soy-protein-based adhesives: Adhesion performance and properties. Int. J. Adhes. Adhes. 2019, 91, 116–122. [Google Scholar] [CrossRef]
- Wang, F.; Wang, J.; Chu, F.; Wang, C.; Jin, C.; Wang, S.; Pang, J. Combinations of soy protein and polyacrylate emulsions as wood adhesives. Int. J. Adhes. Adhes. 2018, 82, 160–165. [Google Scholar] [CrossRef]
- Roman, J.K.; Wilker, J.J. Cooking Chemistry Transforms Proteins into High-Strength Adhesives. J. Am. Chem. Soc. 2019, 141, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Luo, J.; Li, X.; Yi, Z.; Gao, Q.; Li, J. Soybean meal-based wood adhesive enhanced by ethylene glycol diglycidyl ether and diethylenetriamine. Ind. Crop. Prod. 2015, 74, 613–618. [Google Scholar] [CrossRef]
- Luo, J.; Luo, J.; Yuan, C.; Zhang, W.; Li, J.; Gao, Q.; Chen, H. An eco-friendly wood adhesive from soy protein and lignin: performance properties. RSC Adv. 2015, 5, 100849–100855. [Google Scholar] [CrossRef]
- Mo, X.; Sun, X.; Wang, D. Thermal properties and adhesion strength of modified soybean storage proteins. J. Am. Oil Chem. Soc. 2004, 81, 395–400. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, B.; Fan, B.; Gao, Z.; Shi, J. Liquefaction of soybean protein and its effects on the properties of soybean protein adhesive. Pigment Resin Technol. 2017, 46, 399–407. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, Y.; Zhu, W.; Zhang, W.; Gao, Q.; Li, J. Improve the Performance of Soy Protein-Based Adhesives by a Polyurethane Elastomer. Polymers 2018, 10, 1016. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Li, Y.; Li, F.; Ou, Y.; Lin, Q.; Chen, N. Development of Defatted Soy Flour-Based Adhesives by Acid Hydrolysis of Carbohydrates. Polymers 2017, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, C.; Sun, X.S. Improved water resistance in undecylenic acid (UA)-modified soy protein isolate (SPI)-based adhesives. Ind. Crop. Prod. 2015, 74, 577–584. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Chen, M.; Luo, J.; Li, X.; Gao, Q.; Li, J. Preparation and characterization of a soy protein-based high-performance adhesive with a hyperbranched cross-linked structure. Chem. Eng. J. 2018, 354, 11. [Google Scholar] [CrossRef]
- Eslah, F.; Jonoobi, M.; Faezipour, M.; Ashori, A. Chemical modification of soybean flour-based adhesives using acetylated cellulose nanocrystals. Polym. Compos. 2018, 39, 3618–3625. [Google Scholar] [CrossRef]
- Schmid, M.; Prinz, T.K.; Stabler, A.; Sangerlaub, S. Effect of Sodium Sulfite, Sodium Dodecyl Sulfate, and Urea on the Molecular Interactions and Properties of Whey Protein Isolate-Based Films. Front. Chem. 2016, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhao, M. Physical and chemical modification of SPI as a potential means to enhance small peptide contents and antioxidant activity found in hydrolysates. Innov. Food Sci. Emerg. Technol. 2010, 11, 677–683. [Google Scholar] [CrossRef]
- Connolly, J.M.; Alferiev, I.; Eidelman, N.; Sacks, M.; Palmatory, E.; Kronsteiner, A.; DeFelice, S.; Xu, J.; Ohri, R.; Narula, N. Triglycidylamine Crosslinking of Porcine Aortic Valve Cusps or Bovine Pericardium Results in Improved Biocompatibility, Biomechanics, and Calcification Resistance. Am. J. Pathol. 2005, 166, 1–13. [Google Scholar] [CrossRef]
- Mozafarpour, R.; Koocheki, A.; Milani, E.; Varidi, M. Extruded soy protein as a novel emulsifier: Structure, interfacial activity and emulsifying property. Food Hydrocoll. 2019, 93, 361–373. [Google Scholar] [CrossRef]
- Test Methods of Evaluating the Poperties of Wood-Based Panels and Surface Decorated Wood-Based Panels; GB/T 17657; Standardization Administration of The People’s Republic of China: Beijing, China, 2013.
- Luo, J.; Li, L.; Luo, J.; Li, X.; Li, K.; Gao, Q. A High Solid Content Bioadhesive Derived from Soybean Meal and Egg White: Preparation and Properties. J. Polym. Environ. 2016, 25, 948–959. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.; Ren, J.; Zhao, M. Effects of ultrasound pretreatment on the enzymatic hydrolysis of soy protein isolates and on the emulsifying properties of hydrolysates. J. Agric. Food Chem. 2011, 59, 2600–2609. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, X.; Liu, J.; Cui, Q.; Wang, X.; Chen, S.; Jiang, L. Relationship between Molecular Flexibility and Emulsifying Properties of Soy Protein Isolate-Glucose Conjugates. J. Agric. Food Chem. 2019, 67, 4089–4097. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Y.; Li, X.; Luo, J.; Gao, Q.; Li, J. “Green” bio-thermoset resins derived from soy protein isolate and condensed tannins. Ind. Crop. Prod. 2017, 108, 363–370. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, Y.; Han, Y.; Chen, M.; Zhang, W.; Gao, Q.; Li, J. The Effect of Enzymolysis on Performance of Soy Protein-Based Adhesive. Molecules 2018, 23, 2752. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Li, X.; Luo, J.; Gao, Q.; Li, J. A high-performance bio-adhesive derived from soy protein isolate and condensed tannins. RSC Adv. 2017, 7, 21226–21233. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Luo, J.; Li, K.; Gao, Q.; Li, J. A Novel Eco-friendly Blood Meal-based Bio-adhesive: Preparation and Performance. J. Polym. Environ. 2017, 26, 607–615. [Google Scholar] [CrossRef]
- Xu, L.; Liu, Y.; Yang, M.; Cao, W.; Zhang, H.; Xia, N.; Li, T.; Zhao, X. Properties of soy protein isolate/nano-silica bilayer films during storage. J. Food Process Eng. 2018, 42, e12984. [Google Scholar] [CrossRef]
- Luo, J.; Luo, J.; Li, X.; Li, K.; Gao, Q.; Li, J. Toughening improvement to a soybean meal-based bioadhesive using an interpenetrating acrylic emulsion network. J. Mater. Sci. 2016, 51, 9330–9341. [Google Scholar] [CrossRef]


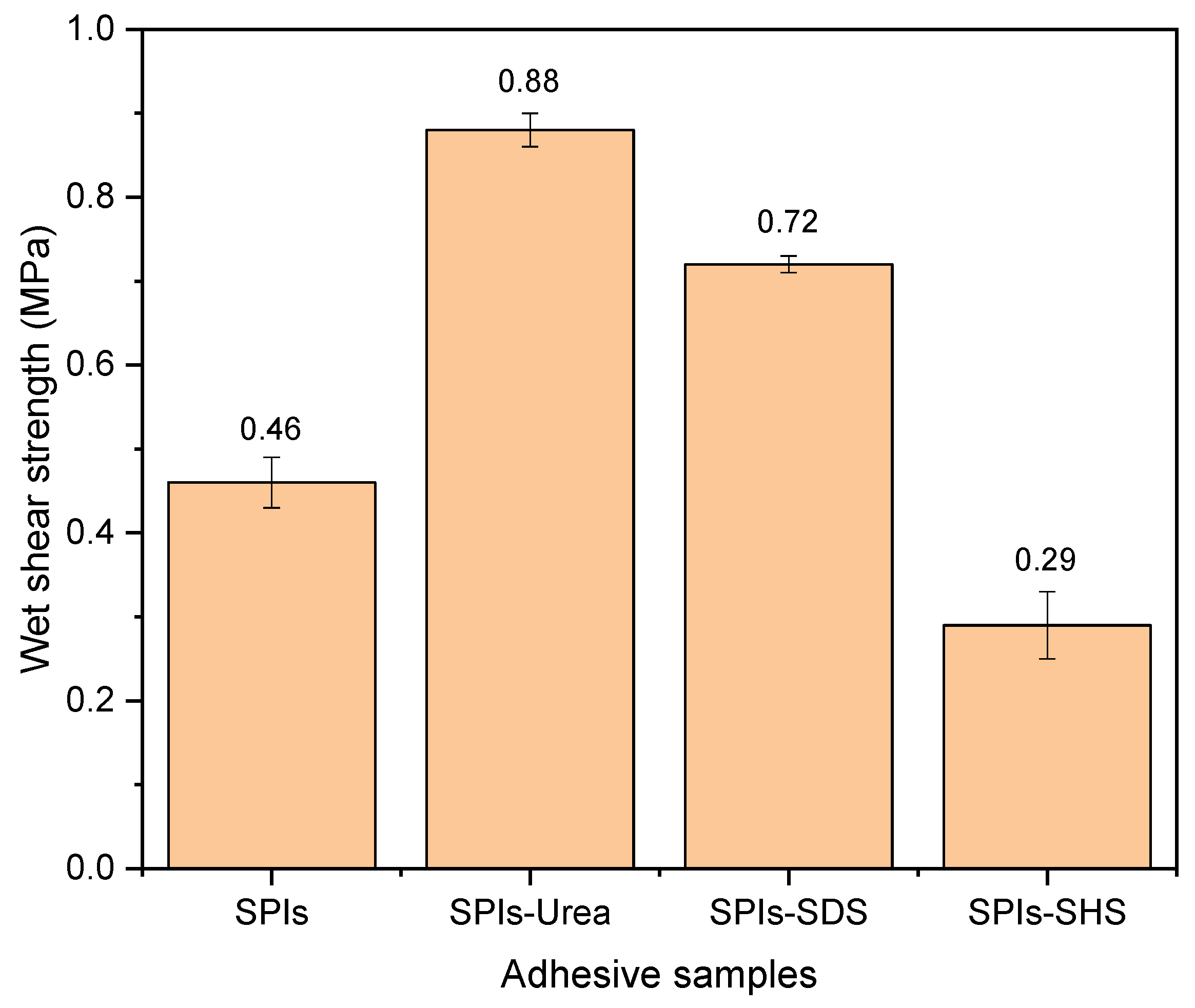


| Amino Acid Name | SPI |
|---|---|
| Tyrosine | 3.25% |
| Glycine | 3.54% |
| Leucine | 6.81% |
| Proline | 4.54% |
| Alanine | 3.73% |
| Phenylalanine | 4.43% |
| Valine | 5.37% |
| Isoleucine | 3.97% |
| Serine | 4.53% |
| Threonine | 3.33% |
| Aspartic acid | 10% |
| Glutamic acid | 16.65% |
| Histidine | 2.22% |
| Arginine | 6.74% |
| Lysine | 5.37% |
| Methionine | 1.03% |
| Sample | Formulation | |||
|---|---|---|---|---|
| SPI | Deionized Water | Denaturing Agents | CA | |
| 1 SPI | 15 g | 85 g | ||
| 2 SPI-Urea | 15 g | 85 g | 1 g Urea | |
| 3 SPI-SDS | 15 g | 85 g | 1 g SDS | |
| 4 SPI-SHS | 15 g | 85 g | 1 g SHS | |
| 5 SPI-CA | 15 g | 85 g | 5 g CA | |
| 6 SPI-Urea-CA | 15 g | 85 g | 1 g Urea | 5 g CA |
| 7 SPI-SDS-CA | 15 g | 85 g | 1 g SDS | 5 g CA |
| 8 SPI-SHS-CA | 15 g | 85 g | 1 g SHS | 5 g CA |
| Samples | SPI | SPI-Urea | SPI-SDS | SPI-SHS |
|---|---|---|---|---|
| Initial viscosity (mPa·s) | 53672 | 48720 | 61463 | 6381 |
| Residual rate (%) | 84.8 ± 0.05 | 83.7 ± 0.02 | 82.5 ± 0.04 | 79.4 ± 0.07 |
| Adhesive | SPI-CA | SPI-Urea-CA | SPI-SDS-CA | SPI-SHS-CA |
|---|---|---|---|---|
| Residual rate (%) | 89.3 ± 0.08 | 91.9 ± 0.05 | 91.5 ± 0.07 | 90.2 ± 0.03 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, L.; Meng, Z.; Yi, Z.; Gao, Q.; Mao, A.; Li, J. Effects of Different Denaturants on Properties and Performance of Soy Protein-Based Adhesive. Polymers 2019, 11, 1262. https://doi.org/10.3390/polym11081262
Yue L, Meng Z, Yi Z, Gao Q, Mao A, Li J. Effects of Different Denaturants on Properties and Performance of Soy Protein-Based Adhesive. Polymers. 2019; 11(8):1262. https://doi.org/10.3390/polym11081262
Chicago/Turabian StyleYue, Li, Zhang Meng, Zhang Yi, Qiang Gao, An Mao, and Jianzhang Li. 2019. "Effects of Different Denaturants on Properties and Performance of Soy Protein-Based Adhesive" Polymers 11, no. 8: 1262. https://doi.org/10.3390/polym11081262
APA StyleYue, L., Meng, Z., Yi, Z., Gao, Q., Mao, A., & Li, J. (2019). Effects of Different Denaturants on Properties and Performance of Soy Protein-Based Adhesive. Polymers, 11(8), 1262. https://doi.org/10.3390/polym11081262






