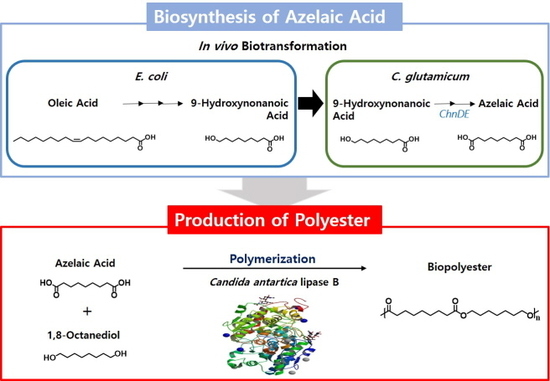
Multi-Step Enzymatic Synthesis of 1,9-Nonanedioic Acid from a Renewable Fatty Acid and Its Application for the Enzymatic Production of Biopolyesters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
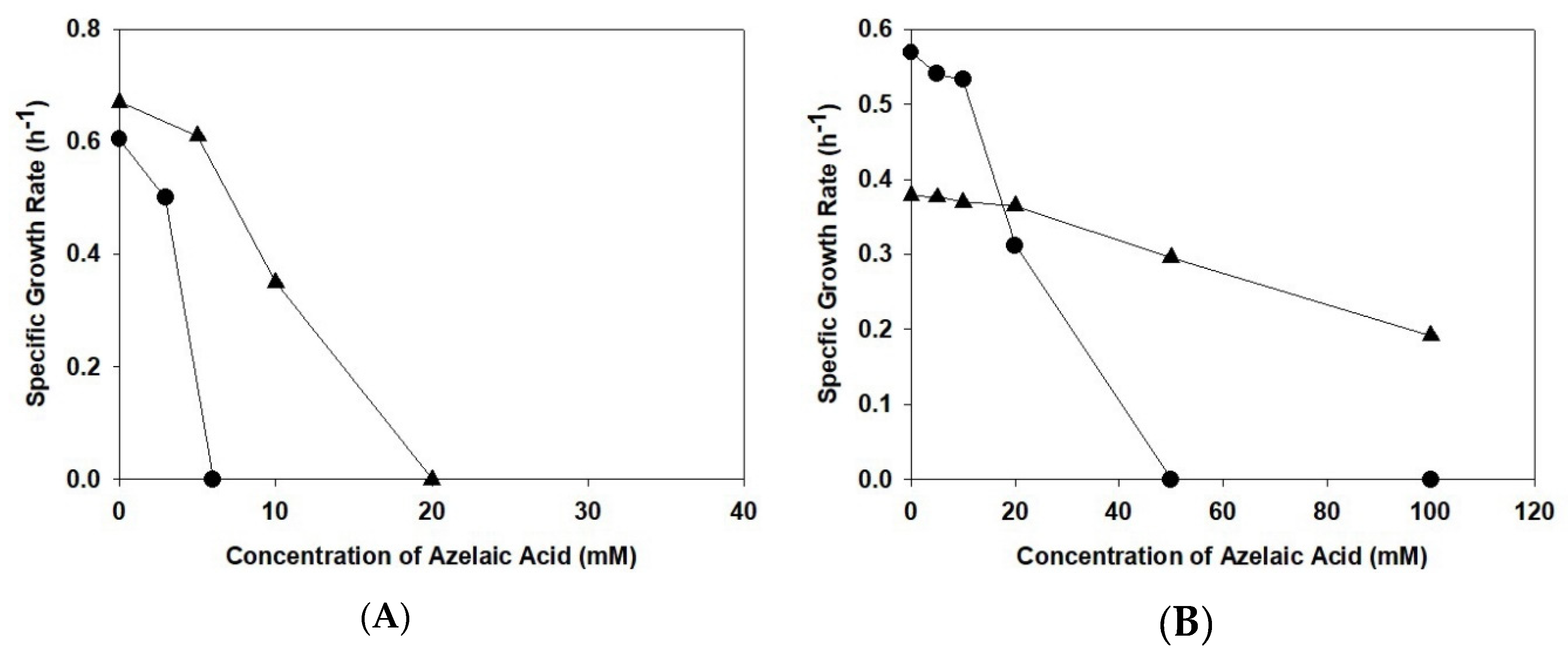
2.2. Azelaic Acid Tolerance Assay
2.3. Chemicals and Reagents
2.4. Plasmid Construction
2.5. Whole-Cell Biotransformation
2.6. Polymerization of Azelaic Acid for the Synthesis of Bio-Based Polyesters
2.7. GC/MS Analysis
2.8. Gel Permeation Chromatography Analysis
2.9. Characterization of Polyesters Using Fourier-Transform Infrared Spectroscopy and 1H-NMR Analysis
3. Results and Discussion
3.1. Construction of the Recombinant C. Glutamicum-Based Biocatalyst
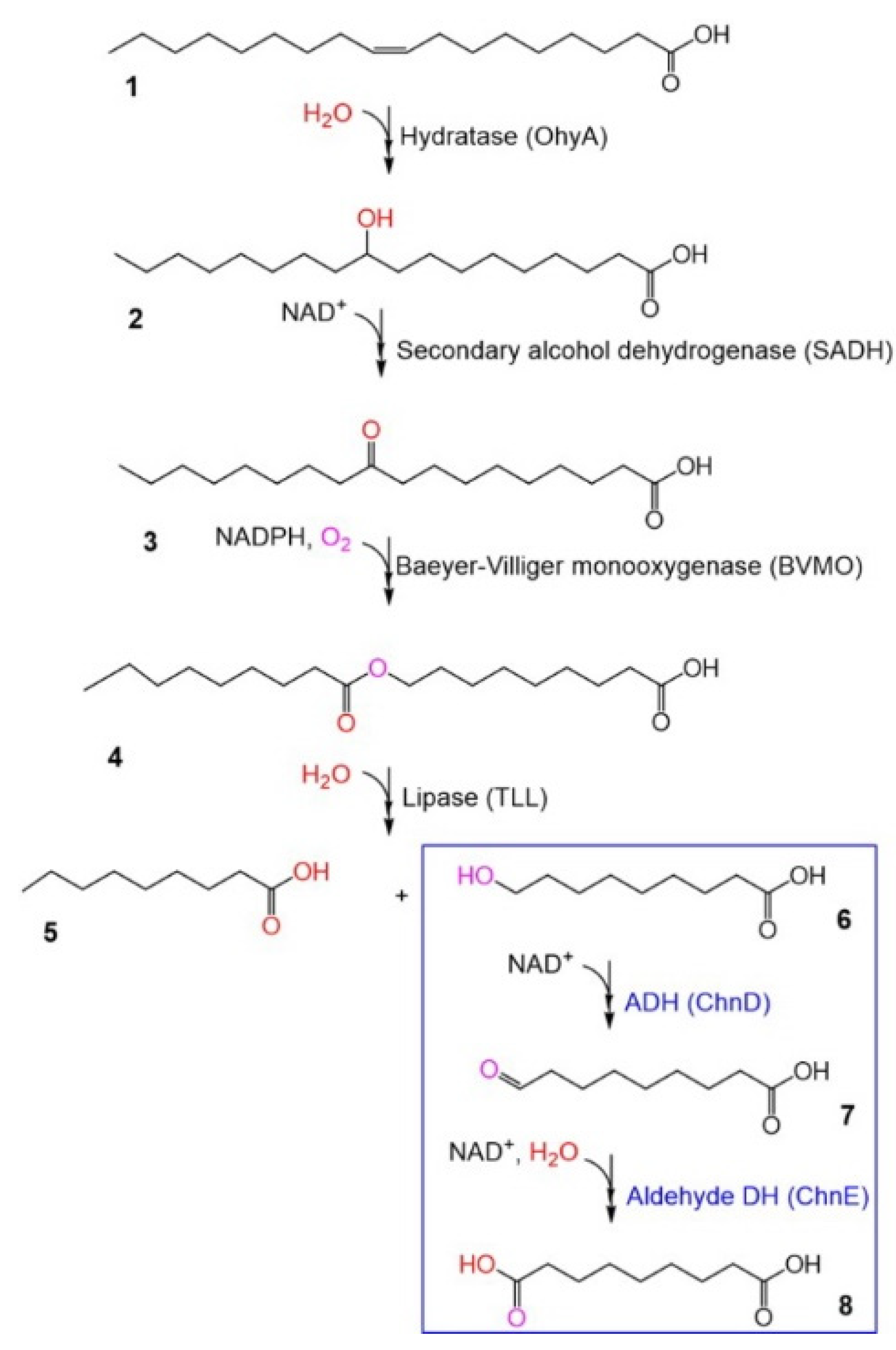
3.2. Biosynthesis of 1,9-Nonanedioic Acid from 9-Hydroxynonanoic Acid
3.3. Production of Polyesters from α,ω-Diacids and α,ω-Diols
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Meier, M.A.R.; Metzger, J.O.; Schubert, U.S. Plant Oil Renewable Resources as Green Alternatives in Polymer Science. Chem. Soc. Rev. 2007, 36, 1788–1802. [Google Scholar] [CrossRef] [PubMed]
- Biermann, U.; Bornscheuer, U.; Meier, M.A.; Metzger, J.O.; Schafer, H.J. Oils and Fats as Renewable Raw Materials in Chemistry. Angew. Chem. Int. Ed. Engl. 2011, 50, 3854–3871. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture. World Agricultural Supply and Demand Estimates; United States Department of Agriculture: Washington, DC, USA, 2015.
- Enamala, M.K.; Enamala, S.; Chavali, M.; Donepudi, J.; Yadavalli, R.; Kolapalli, B.; Aradhyula, T.V.; Velpuri, J.; Kuppam, C. Production of Biofuels from Microalgae—A Review on Cultivation, Harvesting, Lipid Extraction, and Numerous Applications of Microalgae. Renew. Sustain. Energy Rev. 2018, 94, 49–68. [Google Scholar] [CrossRef]
- De Bhowmick, G.; Koduru, L.; Sen, R. Metabolic Pathway Engineering Towards Enhancing Microalgal Lipid Biosynthesis for Biofuel Application—A Review. Renew. Sustain. Energy Rev. 2015, 50, 1239–1253. [Google Scholar] [CrossRef]
- Maltsev, Y.I.; Konovalenko, T.V.; Barantsova, I.A.; Maltseva, K.I.; Maltseva, K.I. Prospects of Using Algae in Biofuel Production. Regul. Mech. Biosyst. 2017, 8, 455–460. [Google Scholar] [CrossRef]
- Diaz, A.; Katsarava, R.; Puiggali, J. Synthesis, Properties and Applications of Biodegradable Polymers Derived from Diols and Dicarboxylic Acids: From Polyesters to Poly (ester amide) s. Int. J. Mol. Sci. 2014, 15, 7064–7123. [Google Scholar] [CrossRef]
- Jiang, Y.; Loos, K. Enzymatic Synthesis of Biobased Polyesters and Polyamides. Polymers 2016, 8, 243. [Google Scholar] [CrossRef]
- Seo, E.J.; Kang, C.W.; Woo, J.M.; Jang, S.; Yeon, Y.J.; Jung, G.Y.; Park, J.B. Multi-level Engineering of Baeyer-Villiger Monooxygenase-Based Escherichia coli Biocatalysts for the Production of C9 Chemicals from Oleic Acid. Metab. Eng. 2019, 54, 137–144. [Google Scholar] [CrossRef]
- Seo, E.J.; Yeon, Y.J.; Seo, J.H.; Lee, J.H.; Bongol, J.P.; Oh, Y.; Park, J.M.; Lim, S.M.; Lee, C.G.; Park, J.B. Enzyme/Whole-Cell Biotransformation of Plant Oils, Yeast Derived Oils, and Microalgae Fatty Acid Methyl Esters into n-Nonanoic Acid, 9-Hydroxynonanoic Acid, and 1,9-Nonanedioic Acid. Bioresour. Technol. 2018, 251, 288–294. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Lipid Modification by Enzymes and Engineered Microbes; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Yeon, Y.J.; Park, J.-B. Regiospecific Conversion of Lipids and Fatty Acids Through Enzymatic Cascade Reactions. In Lipid Modification by Enzymes and Engineered Microbes; Elsevier: Amsterdam, The Netherlands, 2018; pp. 139–155. [Google Scholar]
- Song, J.W.; Jeon, E.Y.; Song, D.H.; Jang, H.Y.; Bornscheuer, U.T.; Oh, D.K.; Park, J.B. Multistep Enzymatic Synthesis of Long-chain α,ω-Dicarboxylic and ω-Hydroxycarboxylic Acids from Renewable Fatty Acids and Plant Oils. Angew. Chem. Int. Ed. Engl. 2013, 52, 2534–2537. [Google Scholar] [CrossRef]
- Song, J.-W.; Lee, J.-H.; Bornscheuer, U.T.; Park, J.-B. Microbial Synthesis of Medium-Chain α,ω-Dicarboxylic Acids and ω-Aminocarboxylic Acids from Renewable Long-Chain Fatty Acids. Adv. Synth. Catal. 2014, 356, 1782–1788. [Google Scholar] [CrossRef]
- Seo, J.H.; Lee, S.M.; Lee, J.; Park, J.B. Adding Value to Plant Oils and Fatty Acids: Biological Transformation of Fatty Acids into ω-Hydroxycarboxylic, α,ω-Dicarboxylic, and ω-Aminocarboxylic acids. J. Biotechnol. 2015, 216, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Van Nuland, Y.M.; Eggink, G.; Weusthuis, R.A. Application of AlkBGT and AlkL from Pseudomonas putida GPo1 for Selective Alkyl Ester ω-Oxyfunctionalization in Escherichia coli. Appl. Environ. Microbiol. 2016, 82, 3801–3807. [Google Scholar] [CrossRef] [PubMed]
- Schrewe, M.; Julsing, M.K.; Lange, K.; Czarnotta, E.; Schmid, A.; Buhler, B. Reaction and Catalyst Engineering to Exploit Kinetically Controlled Whole-Cell Multistep Biocatalysis for Terminal FAME Oxyfunctionalization. Biotechnol. Bioeng. 2014, 111, 1820–1830. [Google Scholar] [CrossRef]
- Koppireddi, S.; Seo, J.-H.; Jeon, E.-Y.; Chowdhury, P.S.; Jang, H.-Y.; Park, J.-B.; Kwon, Y.-U. Combined Biocatalytic and Chemical Transformations of Oleic Acid to ω-Hydroxynonanoic Acid and α,ω-Nonanedioic Acid. Adv. Synth. Catal. 2016, 358, 3084–3092. [Google Scholar] [CrossRef]
- Jeon, E.-Y.; Seo, J.-H.; Kang, W.-R.; Kim, M.-J.; Lee, J.-H.; Oh, D.-K.; Park, J.-B. Simultaneous Enzyme/Whole-Cell Biotransformation of Plant Oils into C9 Carboxylic Acids. ACS Catal. 2016, 6, 7547–7553. [Google Scholar] [CrossRef]
- Cha, H.-J.; Seo, E.-J.; Song, J.-W.; Jo, H.-J.; Kumar, A.R.; Park, J.-B. Simultaneous Enzyme/Whole-Cell Biotransformation of C18 Ricinoleic Acid into (R)-3-Hydroxynonanoic Acid, 9-Hydroxynonanoic Acid, and 1,9-Nonanedioic Acid. Adv. Synth. Catal. 2018, 360, 696–703. [Google Scholar] [CrossRef]
- Jang, H.-Y.; Singha, K.; Kim, H.-H.; Kwon, Y.-U.; Park, J.-B. Chemo-Enzymatic Synthesis of 11-Hydroxyundecanoic Acid and 1, 11-Undecanedioic Acid from Ricinoleic Acid. J. Green Chem. 2016, 18, 1089–1095. [Google Scholar] [CrossRef]
- Sudheer, P.D.V.N.; Yun, J.; Chauhan, S.; Kang, T.J.; Choi, K.-Y. Screening, Expression, and Characterization of Baeyer-Villiger Monooxygenases for the Production of 9-(Nonanoyloxy)nonanoic Acid from Oleic Acid. Biotechnol. Bioprocess Eng. 2018, 22, 717–724. [Google Scholar] [CrossRef]
- Steffen-Munsberg, F.; Vickers, C.; Thontowi, A.; Schätzle, S.; Meinhardt, T.; Svedendahl Humble, M.; Land, H.; Berglund, P.; Bornscheuer, U.T.; Höhne, M. Revealing the Structural Basis of Promiscuous Amine Transaminase Activity. J. ChemCatChem 2013, 5, 154–157. [Google Scholar] [CrossRef]
- Jiang, Y.; Woortman, A.J.J.; Alberda van Ekenstein, G.O.R.; Loos, K. A Biocatalytic Approach Towards Sustainable Furanic–Aliphatic Polyesters. Polym. Chem. 2015, 6, 5198–5211. [Google Scholar] [CrossRef]
- Pellis, A.; Comerford, J.W.; Weinberger, S.; Guebitz, G.M.; Clark, J.H.; Farmer, T.J. Enzymatic Synthesis of Lignin Derivable Pyridine Based Polyesters for the Substitution of Petroleum Derived Plastics. Nat. Commun. 2019, 10, 1762. [Google Scholar] [CrossRef] [PubMed]
- Maniar, D.; Hohmann, K.F.; Jiang, Y.; Woortman, A.J.J.; van Dijken, J.; Loos, K. Enzymatic Polymerization of Dimethyl 2,5-Furandicarboxylate and Heteroatom Diamines. ACS Omega 2018, 3, 7077–7085. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Maniar, D.; Woortman, A.J.; Alberda van Ekenstein, G.O.; Loos, K. Enzymatic Polymerization of Furan-2,5-Dicarboxylic Acid-Based Furanic-Aliphatic Polyamides as Sustainable Alternatives to Polyphthalamides. Biomacromolecules 2015, 16, 3674–3685. [Google Scholar] [CrossRef] [PubMed]
- Zhenova, A.; Pellis, A.; Milescu, R.A.; McElroy, C.R.; White, R.J.; Clark, J.H. Solvent Applications of Short-Chain Oxymethylene Dimethyl Ether Oligomers. ACS Sustain. Chem. Eng. 2019, 7, 14834–14840. [Google Scholar] [CrossRef]
- Cha, H.-J.; Park, J.-B.; Park, S. Esterification of Secondary Alcohols and Multi-hydroxyl Compounds by Candida antarctica Lipase B and Subtilisin. Biotechnol. Bioprocess Eng. 2019, 24, 41–47. [Google Scholar] [CrossRef]
- Moreno, A.; Lligadas, G.; Ronda, J.C.; Galià, M.; Cádiz, V. Linear and Branched Acetal Polymers from Castor Oil via Acetal Metathesis Polymerization. Eur. Polym. J. 2018, 108, 348–356. [Google Scholar] [CrossRef]
- Turunc, O.; Meier, M.A. Fatty Acid Derived Monomers and Related Polymers via Thiol-ene (Click) Additions. Macromol. Rapid Commun. 2010, 31, 1822–1826. [Google Scholar] [CrossRef]
- Valverde, C.; Lligadas, G.; Ronda, J.C.; Galià, M.; Cádiz, V. PEG-Modified Poly(10,11-dihydroxyundecanoic acid) Amphiphilic Copolymers. Grafting versus Macromonomer Copolymerization Approaches Using CALB. Eur. Polym. J. 2018, 109, 179–190. [Google Scholar] [CrossRef]
- Liu, G.; Kong, X.; Wan, H.; Narine, S. Production of 9-Hydroxynonanoic Acid from Methyl Oleate and Conversion into Lactone Monomers for the Synthesis of Biodegradable Polylactones. Biomacromolecules 2008, 9, 949–953. [Google Scholar] [CrossRef]
- Iwaki, H.; Hasegawa, Y.; Teraoka, M.; Tokuyama, T.; Bergeron, H.; Lau, P.C. Identification of a Transcriptional Activator (ChnR) and a 6-Oxohexanoate Dehydrogenase (ChnE) in the Cyclohexanol Catabolic Pathway in Acinetobacter sp. Strain NCIMB 9871 and Localization of the Genes that Encode Them. J. Appl. Environ. Microbiol. 1999, 65, 5158–5162. [Google Scholar]
- Riesenberg, D. High-Cell-Density Cultivation of Escherichia coli. Curr. Opin. Biotechnol. 1991, 2, 380–384. [Google Scholar] [CrossRef]
- Lee, B.; Lee, S.; Kim, H.; Jeong, K.; Park, J.; Lee, E.; Lee, J. Biotransformation of Oleic Acid into 10-Ketostearic Acid by Recombinant Corynebacterium glutamicum-Based Biocatalyst. Biotechnol. Lett. 2015, 37, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, S.; Cho, S.; Yang, J.; Jeong, K.; Park, J.; Lee, J. Bioprocess Engineering to Produce 9-(Nonanoyloxy) nonanoic Acid by a Recombinant Corynebacterium glutamicum-Based Biocatalyst. J. Ind. Microbiol. Biotechnol. 2017, 44, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yang, J.; Cho, S.; Jeong, K.; Park, J.; Lee, J. Microbial Synthesis of Undec-9-enoic acid, Heptyl Ester from Renewable Fatty Acids using Recombinant Corynebacterium glutamicum-based Whole-Cell Biocatalyst. J. Process Biochem. 2018, 66, 61–69. [Google Scholar] [CrossRef]
- Park, J.-U.; Jo, J.-H.; Kim, Y.-J.; Chung, S.-S.; Lee, J.-H.; Lee, H.-H. Construction of Heat-Inducible Expression Vector of Corynebacterium glutamicum and C. ammoniagenes: Fusion of λ Operator with Promoters Isolated from C. ammoniagenes. J. Microbiol. Biotechnol. 2008, 18, 639–647. [Google Scholar]
- Yim, S.S.; An, S.J.; Kang, M.; Lee, J.; Jeong, K.J. Isolation of Fully Synthetic Promoters for High-Level Gene Expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 2013, 110, 2959–2969. [Google Scholar] [CrossRef]
- Pellis, A.; Comerford, J.W.; Maneffa, A.J.; Sipponen, M.H.; Clark, J.H.; Farmer, T.J. Elucidating Enzymatic Polymerisations: Chain-Length Selectivity of Candida antarctica Lipase B Towards Various Aliphatic Diols and Dicarboxylic Acid Diesters. Eur. Polym. J. 2018, 106, 79–84. [Google Scholar] [CrossRef]
- Mahapatro, A.; Kalra, B.; Kumar, A.; Gross, R.A. Lipase-Catalyzed Polycondensations: Effect of Substrates and Solvent on Chain Formation, Dispersity, and End-Group Structure. Biomacromolecules 2003, 4, 544–551. [Google Scholar] [CrossRef]
- Pyo, S.-H.; Nuszkiewicz, K.; Persson, P.; Lundmark, S.; Hatti-Kaul, R. Lipase-Mediated Synthesis of Six-Membered Cyclic Carbonates from Trimethylolpropane and Dialkyl Carbonates: Influence of Medium Engineering on Reaction Selectivity. J. Mol. Catal. 2011, 73, 67–73. [Google Scholar] [CrossRef]
- Pyo, S.-H.; Persson, P.; Lundmark, S.; Hatti-Kaul, R. Solvent-Free Lipase-Mediated Synthesis of Six-Membered Cyclic Carbonates from Trimethylolpropane and Dialkyl Carbonates. Green Chem. 2011, 13, 976–982. [Google Scholar] [CrossRef]
- Hosseini, S.M.; Kim, S.M.; Sayed, M.; Younesi, H.; Bahramifar, N.; Park, J.H.; Pyo, S.-H. Lipase-Immobilized Chitosan-Crosslinked Magnetic Nanoparticle as a Biocatalyst for Ring Opening Esterification of Itaconic Anhydride. Biochem. Eng. J. 2019, 143, 141–150. [Google Scholar] [CrossRef]
- Sheldon, R.A. Fundamentals of Green Chemistry: Efficiency in Reaction Design. Chem. Soc. Rev. 2012, 41, 1437–1451. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Meng, X.; Zhou, H.; Liu, Y.; Secundo, F.; Liu, Y. Enzyme Stability and Activity in Non-Aqueous Reaction Systems: A Mini Review. Catalysts 2016, 6, 32. [Google Scholar] [CrossRef]
- Azim, H.; Dekhterman, A.; Jiang, Z.; Gross, R.A. Candida antarctica Lipase B-Catalyzed Synthesis of Poly (butylene succinate): Shorter Chain Building Blocks Also Work. Biomacromolecules 2006, 7, 3093–3097. [Google Scholar] [CrossRef] [PubMed]
- Byrne, F.; Forier, B.; Bossaert, G.; Hoebers, C.; Farmer, T.J.; Clark, J.H.; Hunt, A.J. 2,2,5,5-Tetramethyltetrahydrofuran (TMTHF): A Non-Polar, Non-Peroxide Forming Ether Replacement for Hazardous Hydrocarbon Solvents. Green Chem. 2017, 19, 3671–3678. [Google Scholar] [CrossRef]
- Pellis, A.; Byrne, F.P.; Sherwood, J.; Vastano, M.; Comerford, J.W.; Farmer, T.J. Safer Bio-based Solvents to Replace Toluene and Tetrahydrofuran for the Biocatalyzed Synthesis of Polyesters. Green Chem. 2019, 21, 1686–1694. [Google Scholar] [CrossRef]




| Solvent | Diacid | Diol | Mn (Da) | Mw (Da) | Polydispersity | |
|---|---|---|---|---|---|---|
| 1 | Toluene | Azelaic acid | 1,6-Hexanediol | 20,000 | 25,000 | 1.26 |
| 2 | 1,8-Octanediol | 20,000 | 25,000 | 1.27 | ||
| 3 | Diphenyl ether | Azelaic acid | 1,8-Octanediol | 21,000 | 27,000 | 1.26 |
| 4 | Sebacic acid | 1,8-Octanediol | 19,000 | 24,000 | 1.25 | |
| 5 | 1,11-Undecanedioic acid | 1,8-Octanediol | 17,000 | 21,000 | 1.21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-J.; Kang, Y.-S.; Kim, C.-Y.; Seo, E.-J.; Pyo, S.-H.; Park, J.-B. Multi-Step Enzymatic Synthesis of 1,9-Nonanedioic Acid from a Renewable Fatty Acid and Its Application for the Enzymatic Production of Biopolyesters. Polymers 2019, 11, 1690. https://doi.org/10.3390/polym11101690
Lee H-J, Kang Y-S, Kim C-Y, Seo E-J, Pyo S-H, Park J-B. Multi-Step Enzymatic Synthesis of 1,9-Nonanedioic Acid from a Renewable Fatty Acid and Its Application for the Enzymatic Production of Biopolyesters. Polymers. 2019; 11(10):1690. https://doi.org/10.3390/polym11101690
Chicago/Turabian StyleLee, Hyun-Ju, Young-Seo Kang, Chae-Yun Kim, Eun-Ji Seo, Sang-Hyun Pyo, and Jin-Byung Park. 2019. "Multi-Step Enzymatic Synthesis of 1,9-Nonanedioic Acid from a Renewable Fatty Acid and Its Application for the Enzymatic Production of Biopolyesters" Polymers 11, no. 10: 1690. https://doi.org/10.3390/polym11101690
APA StyleLee, H.-J., Kang, Y.-S., Kim, C.-Y., Seo, E.-J., Pyo, S.-H., & Park, J.-B. (2019). Multi-Step Enzymatic Synthesis of 1,9-Nonanedioic Acid from a Renewable Fatty Acid and Its Application for the Enzymatic Production of Biopolyesters. Polymers, 11(10), 1690. https://doi.org/10.3390/polym11101690