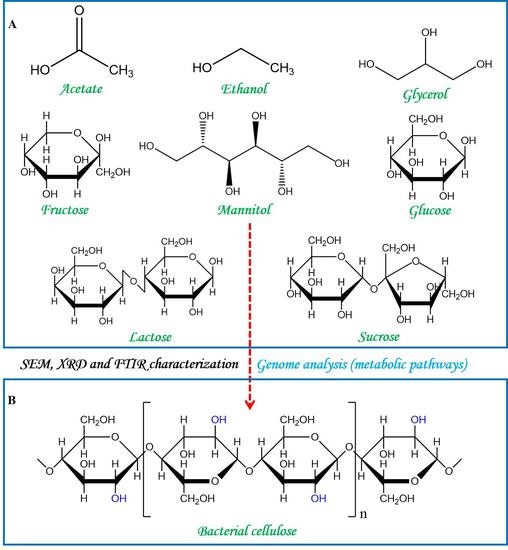
Insights into Bacterial Cellulose Biosynthesis from Different Carbon Sources and the Associated Biochemical Transformation Pathways in Komagataeibacter sp. W1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Micro-Organism, Culture Media and Cultivation
2.2. BC Purification and Yield Calculation
2.3. BC Characterization
2.4. Analysis of Biochemical Transformation Pathways Associated with Carbon Source Metabolisms
2.5. Statistical Analysis
3. Results and Discussions
3.1. BC Production from Various Carbon Sources
3.2. BC Morphology and Microfibril Analysis by SEM Observation
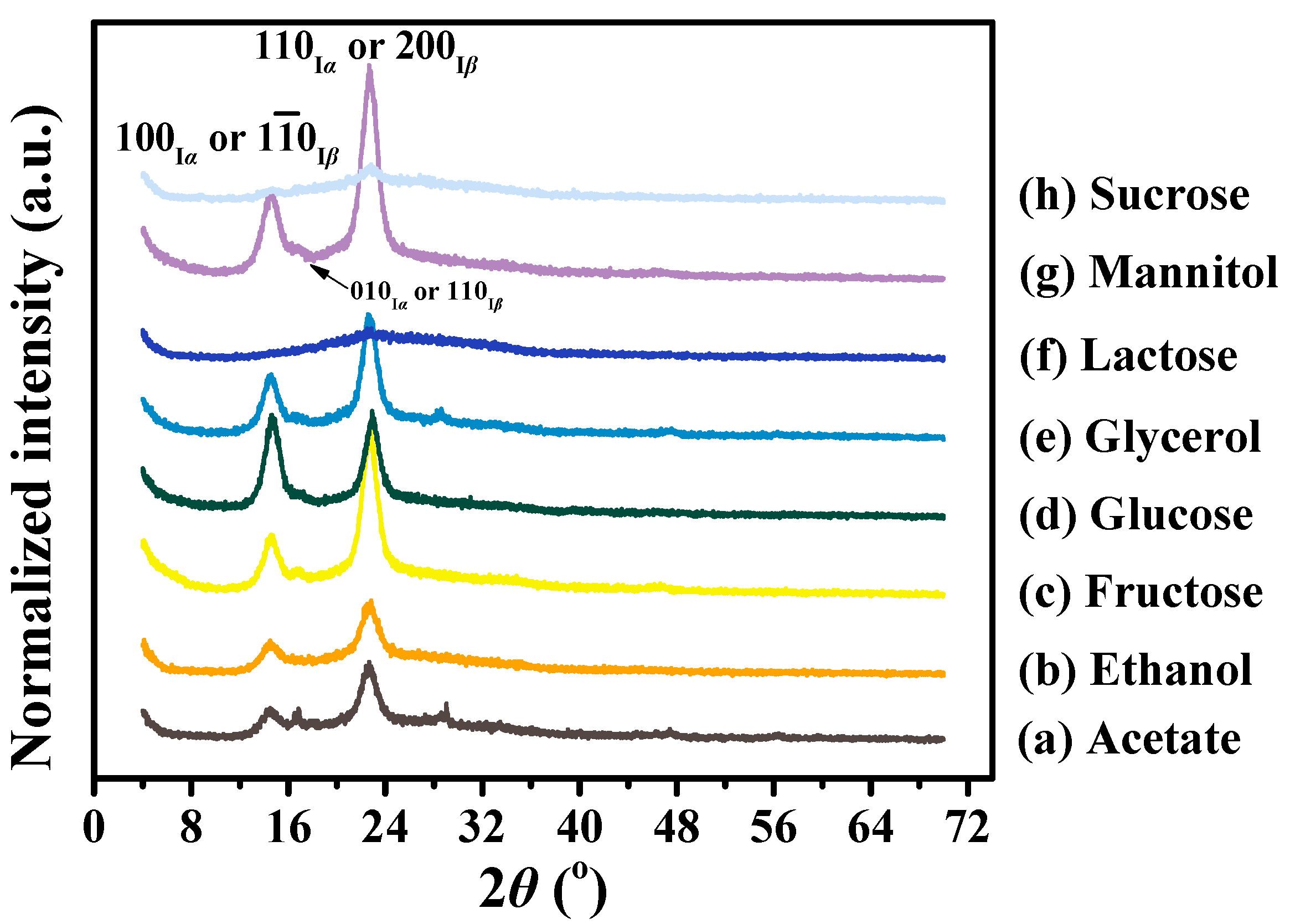
3.3. Crystalline Differences of BC Membranes Produced from Various Carbon Sources
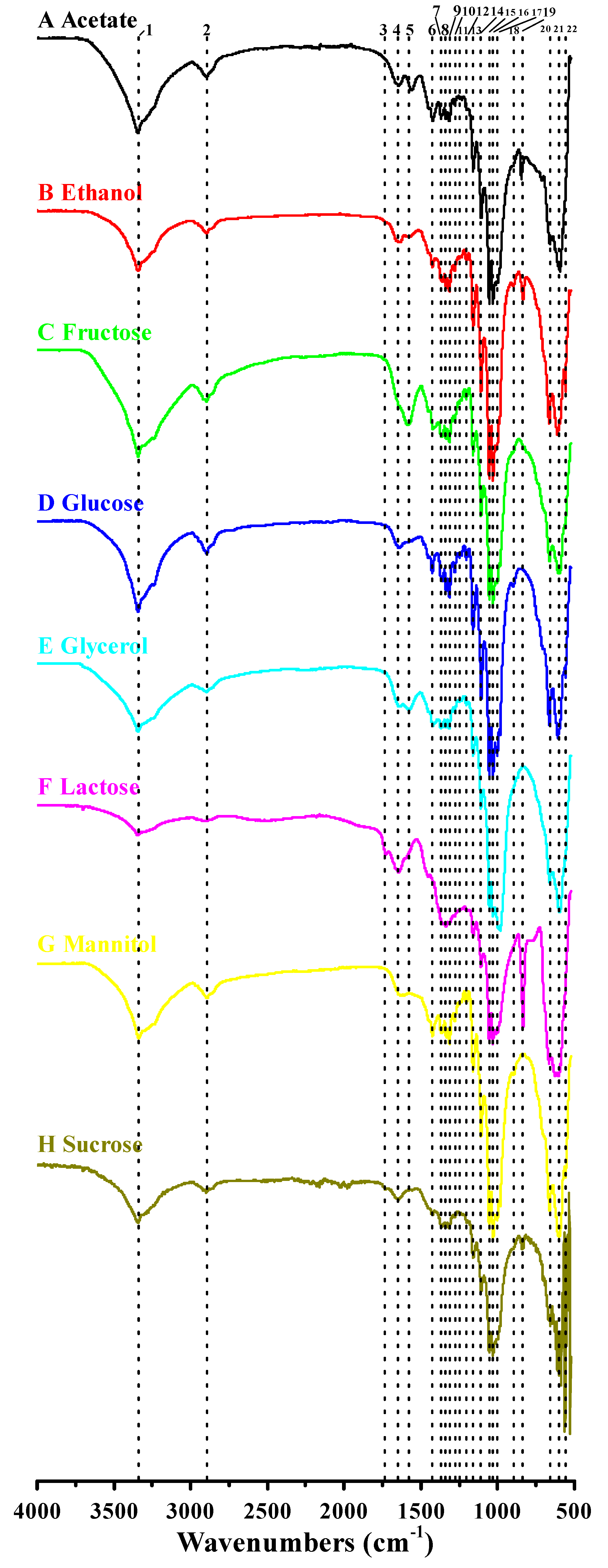
3.4. Functional Groups and Cellulose Types Characterization Based on FTIR Analysis
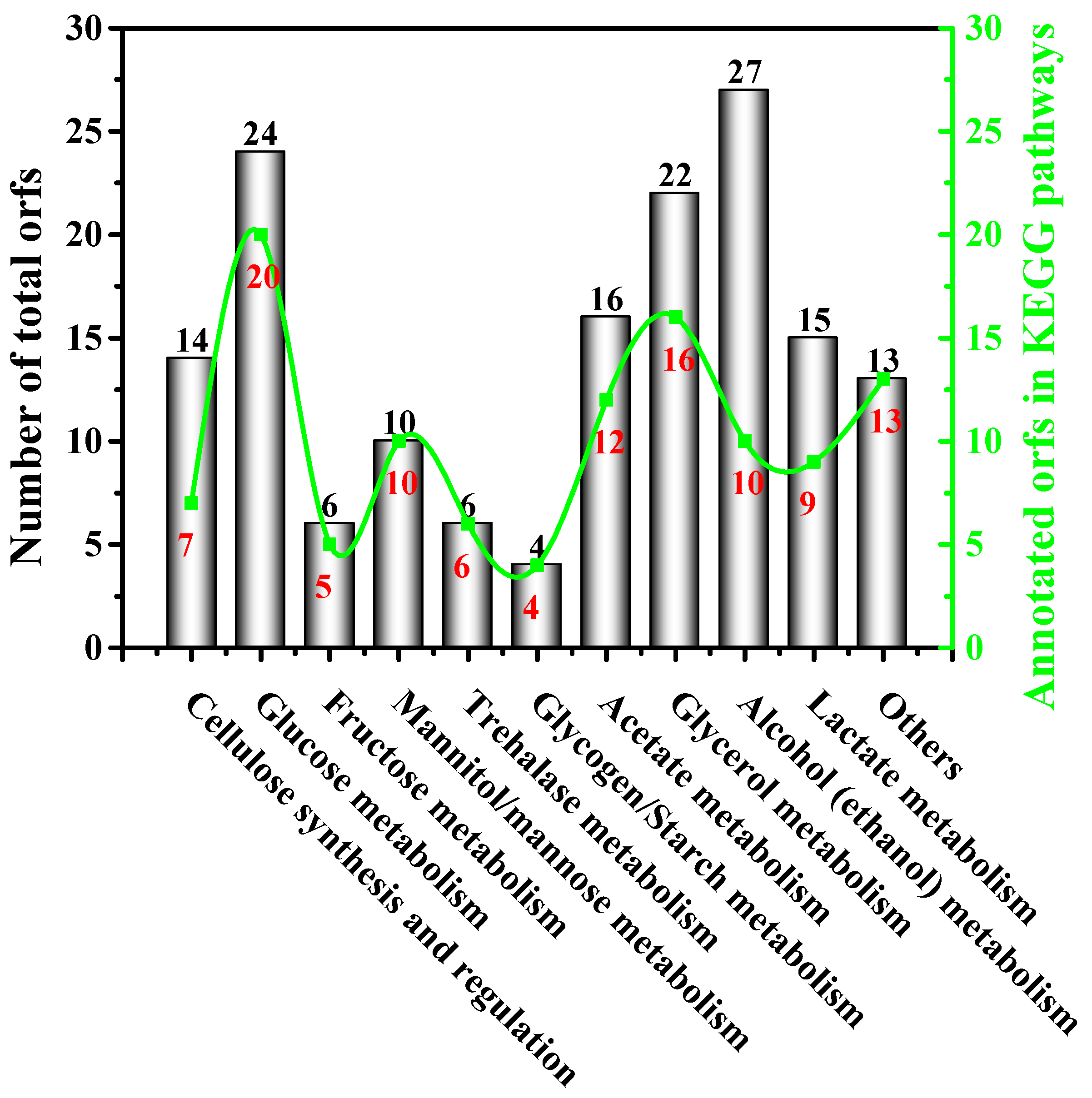
3.5. Insights into the Biochemical Pathways for Carbon Sources Utilization and BC Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Velásquez-Riaño, M.; Bojacá, V. Production of bacterial cellulose from alternative low-cost substrates. Cellulose 2017, 24, 2677–2698. [Google Scholar] [CrossRef]
- Molina-Ramírez, C.; Castro, M.; Osorio, M.; Torres-Taborda, M.; Gómez, B.; Zuluaga, R.; Gómez, C.; Gañán, P.; Rojas, O.J.; Castro, C. Effect of different carbon sources on bacterial nanocellulose production and structure using the low pH resistant strain Komagataeibacter medellinensis. Materials 2017, 10, 639. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-S.; Han, Y.-H.; Ye, Y.-X.; Shi, X.-X.; Xiang, P.; Chen, D.-L.; Li, M. Physicochemical characterization of high-quality bacterial cellulose produced by Komagataeibacter sp. strain W1 and identification of the associated genes in bacterial cellulose production. RSC Adv. 2017, 7, 45145–45155. [Google Scholar] [CrossRef]
- Gallegos, A.M.A.; Carrera, S.H.; Parra, R.; Keshavarz, T.; Iqbal, H.M.N. Bacterial cellulose: A sustainable source to develop value-added products—A review. BioResources 2016, 11, 5641–5655. [Google Scholar] [CrossRef]
- Klemm, D.; Heublein, B.; Fink, H.-P.; Bohn, A. Cellulose: Fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. 2005, 44, 3358–3393. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Chen, W.; Fan, Y.; Zheng, K.; Jin, K.; Yu, H.; Buehler, M.J.; Kaplan, D.L. Biopolymer nanofibrils: Structure, modeling, preparation, and applications. Prog. Polym. Sci. 2018, 85, 1–56. [Google Scholar] [CrossRef]
- Khosravi-Darani, K.; Koller, M.; Akramzadeh, N.; Mortazavian, A.M. Bacterial nanocellulose: Biosynthesis and medical application. Biointerface Res. Appl. Chem. 2016, 6, 1511–1516. [Google Scholar]
- Huang, X.; Zhan, X.; Wen, C.; Xu, F.; Luo, L. Amino-functionalized magnetic bacterial cellulose/activated carbon composite for Pb2+ and methyl orange sorption from aqueous solution. J. Mater. Sci. Technol. 2018, 34, 855–863. [Google Scholar] [CrossRef]
- Jin, X.; Xiang, Z.; Liu, Q.; Chen, Y.; Lu, F. Polyethyleneimine-bacterial cellulose bioadsorbent for effective removal of copper and lead ions from aqueous solution. Bioresour. Technol. 2017, 244, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Reiniati, I.; Hrymak, A.N.; Margaritis, A. Recent developments in the production and applications of bacterial cellulose fibers and nanocrystals. Crit. Rev. Biotechnol. 2017, 37, 510–524. [Google Scholar] [CrossRef] [PubMed]
- Krasteva, P.V.; Bernal-Bayard, J.; Travier, L.; Martin, F.A.; Kaminski, P.-A.; Karimova, G.; Fronzes, R.; Ghigo, J.-M. Insights into the structure and assembly of a bacterial cellulose secretion system. Nat. Commun. 2018, 8, 2065. [Google Scholar] [CrossRef] [PubMed]
- Singhsa, P.; Narain, R.; Manuspiya, H. Physical structure variations of bacterial cellulose produced by different Komagataeibacter xylinus strains and carbon sources in static and agitated conditions. Cellulose 2018, 25, 1571–1581. [Google Scholar] [CrossRef]
- Mikkelsen, D.; Flanagan, B.M.; Dykes, G.A.; Gidley, M.J. Influence of different carbon sources on bacterial cellulose production by Gluconacetobacter xylinus strain ATCC 53524. J. Appl. Microbiol. 2009, 107, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Keshk, S.M.A.S.; Sameshima, K. Evaluation of different carbon sources for bacterial cellulose production. Afr. J. Biotechnol. 2005, 4, 478–482. [Google Scholar]
- Kose, R.; Sunagawa, N.; Yoshida, M.; Tajima, K. One-step production of nanofibrillated bacterial cellulose (NFBC) from waste glycerol using Gluconacetobacter intermedius NEDO-01. Cellulose 2013, 20, 2971–2979. [Google Scholar] [CrossRef]
- Çoban, E.; Biyik, H. Effect of various carbon and nitrogen sources on cellulose synthesis by Acetobacter lovaniensis HBB5. Afr. J. Biotechnol. 2011, 10, 5346–5354. [Google Scholar]
- Gatenholm, P.; Höglund, K.; Johannesson, S.; Puchades, M.; Brackmann, C.; Enejder, A.; Olsson, L. Effect of Cultivation Conditions on the Structure and Morphological Properties of BNC Biomaterials with a Focus on Vascular Grafts. In Bacterial NanoCellulose-A Sophisticated Multifunctional Material, 1st ed.; Gama, M., Gatenholm, P., Klemm, D., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 20–42. [Google Scholar]
- Yunoki, S.; Osada, Y.; Kono, H.; Takai, M. Role of ethanol in improvement of bacterial cellulose production: Analysis using 13C-labeled carbon sources. Food Sci. Technol. Res. 2004, 10, 307–313. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Huang, C.; Guo, H.-J.; Xiong, L.; Luo, J.; Wang, B.; Chen, X.-F.; Lin, X.-Q.; Chen, X.-D. Beneficial effect of acetic acid on the xylose utilization nd bacterial cellulose production by Gluconacetobacter xylinus. Indian J. Microbiol. 2014, 54, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Jung, J.Y.; Park, Y.H. Cellulose production by Gluconacetobacter hansenii in a medium containing ethanol. Biotechnol. Lett. 2003, 25, 2055–2059. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, Y.; Chi, Y.; Xu, N.; Yao, W.; Sun, B. Effects of alcohols on bacterial cellulose production by Acetobacter xylinum 186. World J. Microb. Biot. 2011, 27, 2281–2285. [Google Scholar] [CrossRef]
- Huang, C.; Yang, X.-Y.; Xiong, L.; Guo, H.-J.; Luo, J.; Wang, B.; Zhang, H.-R.; Lin, X.-Q.; Chen, X.-D. Evaluating the possibility of using acetone-butanol-ethanol (ABE) fermentation wastewater for bacterial cellulose production by Gluconacetobacter xylinus. Lett. Appl. Microbiol. 2015, 60, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Kiziltas, E.E.; Kiziltas, A.; Gardner, D.J. Synthesis of bacterial cellulose using hot water extracted wood sugars. Carbohydr. Polym. 2015, 124, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-I.; Jeong, J.-H.; Lee, O.-M.; Park, G.-T.; Kim, K.-K.; Park, H.-C.; Lee, S.-M.; Kim, Y.-G.; Son, H.-J. Influence of glycerol on production and structural–physical properties of cellulose from Acetobacter sp. V6 cultured in shake flasks. Bioresour. Technol. 2010, 101, 3602–3608. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-Q.; Mikkelsen, D.; Lopez-Sanchez, P.; Wang, D.; Martinez-Sanz, M.; Gilbert, E.P.; Flanagan, B.M.; Gidley, M.J. Characterisation of bacterial cellulose from diverse Komagataeibacter strains and their application to construct plant cell wall analogues. Cellulose 2017, 24, 1211–1226. [Google Scholar] [CrossRef]
- Chen, S.-Q.; Lopez-Sanchez, P.; Wang, D.; Mikkelsen, D.; Gidley, M.J. Mechanical properties of bacterial cellulose synthesised by diverse strains of the genus Komagataeibacter. Food Hydrocolloids 2018, 81, 87–95. [Google Scholar] [CrossRef]
- Hestrin, S.; Schramm, M. Synthesis of cellulose by Acetobacter xylinum. 2. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem. J. 1954, 58, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Mohammadkazemi, F.; Azin, M.; Ashori, A. Production of bacterial cellulose using different carbon sources and culture media. Carbohydr. Polym. 2015, 117, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Segal, L.; Creely, J.J.; Martin, A.E., Jr.; Conrad, C.M. An empirical method for estimating the degree of crystallinity of native cellulose using the X-ray diffractometer. Text. Res. J. 1959, 29, 786–794. [Google Scholar] [CrossRef]
- Yamamoto, H.; Horii, F.; Hirai, A. In situ crystallization of bacterial cellulose II. Influences of different polymeric additives on the formation of celluloses Iα and Iβ at the early stage of incubation. Cellulose 1996, 3, 229–242. [Google Scholar] [CrossRef]
- Saxena, I.M.; Malcolm Brown, R.J. Biosynthesis of Bacterial Cellulose. In Bacterial NanoCellulose-A Sophisticated Multifunctional Material, 1st ed.; Gama, M., Gatenholm, P., Klemm, D., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 1–18. [Google Scholar]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a role for bacterial cellulose in environmental interactions: Lessons learned from diverse biofilm-producing Proteobacteria. Front. Microbiol. 2015, 6, 1282. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.; Zuluaga, R.; Putaux, J.-L.; Caro, G.; Mondragon, I.; Gañán, P. Structural characterization of bacterial cellulose produced by Gluconacetobacter swingsii sp. from Colombian agroindustrial wastes. Carbohydr. Polym. 2011, 84, 96–102. [Google Scholar] [CrossRef]
- Gullo, M.; China, S.L.; Falcone, P.M.; Giudici, P. Biotechnological production of cellulose by acetic acid bacteria: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 6885–6898. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.; Mayer, R.; Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol. Mol. Biol. Rev. 1991, 55, 35–58. [Google Scholar]
- Ramana, K.V.; Tomar, A.; Singh, L. Effect of various carbon and nitrogen sources on cellulose synthesis by Acetobacter xylinum. World J. Microb. Biotechnol. 2000, 16, 245–248. [Google Scholar] [CrossRef]
- Ago, M.; Yamane, C.; Hattori, M.; Ono, H.; Okajima, K. Characterization of morphology and physical strength for bacterial cellulose produced by an Enterobacter sp. Sen-i Gakkaishi 2006, 62, 42–46. [Google Scholar] [CrossRef]
- Tanskul, S.; Amornthatre, K.; Jaturonlak, N. A new cellulose-producing bacterium, Rhodococcus sp. MI 2: Screening and optimization of culture conditions. Carbohydr. Polym. 2013, 92, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Hassan, E.A.; Abdelhady, H.M.; El-Salam, S.S.A.; Abdullah, S.M. The characterization of bacterial cellulose produced by Acetobacter xylinum and Komgataeibacter saccharovorans under optimized fermentation conditions. Brit. Microbiol. Res. J. 2015, 9, 1–13. [Google Scholar] [CrossRef]
- Kojima, Y.; Seto, A.; Tonouchi, N.; Tsuchida, T.; Yoshinaga, F. High rate production in static culture of bacterial cellulose from sucrose by a newly isolated Acetobacter strain. Biosci. Biotech. Biochem. 1997, 61, 1585–1586. [Google Scholar] [CrossRef]
- Tabaii, M.J.; Emtiazi, G. Comparison of bacterial cellulose production among different strains and fermented media. Appl. Food Biotechnol. 2016, 3, 35–41. [Google Scholar]
- Ishihara, M.; Matsunaga, M.; Hayashi, N.; Tišler, V. Utilization of d-xylose as carbon source for production of bacterial cellulose. Enzyme Microb. Technol. 2002, 31, 986–991. [Google Scholar] [CrossRef]
- Recouvreux, D.O.S.; Carminatti, C.A.; Pitlovanciv, A.K.; Rambo, C.R.; Porto, L.M.; Antônio, R.V. Cellulose biosynthesis by the beta-proteobacterium, Chromobacterium violaceum. Curr. Microbiol. 2008, 57, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Hungund, B.S.; Gupta, S.G. Production of bacterial cellulose from Enterobacter amnigenus GH-1 isolated from rotten apple. World J. Microb. Biotechnol. 2010, 26, 1823–1828. [Google Scholar] [CrossRef]
- Trovatti, E.; Serafim, L.S.; Freire, C.S.R.; Silvestre, A.J.D.; Neto, C.P. Gluconacetobacter sacchari: An efficient bacterial cellulose cell-factory. Carbohydr. Polym. 2011, 86, 1417–1420. [Google Scholar] [CrossRef]
- Thorat, M.; Dastager, S. High yield production of cellulose by a Komagataeibacter rhaeticus PG2 strain isolated from pomegranate as a new host. RSC Adv. 2018, 8, 29797–29805. [Google Scholar] [CrossRef]
- Volova, T.G.; Prudnikova, S.V.; Sukovatyi, A.G.; Shishatskaya, E.I. Production and properties of bacterial cellulose by the strain Komagataeibacter xylinus B-12068. Appl. Microbiol. Biotechnol. 2018, 102, 7417–7428. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Ren, L.; Cao, Y.; Chen, X.; Tang, X. Bacterial cellulose synthesis in Kombucha by Gluconacetobacter sp and Saccharomyces sp. Adv. Mater. Res. 2012, 554–556, 1000–1003. [Google Scholar] [CrossRef]
- Amano, Y.; Ito, F.; Kanda, T. Novel cellulose producing system by microorganisms such as Acetobacter sp. J. Biol. Macromol. 2005, 5, 3–10. [Google Scholar]
- Yamanaka, S.; Watanabe, K.; Kitamura, N.; Iguchi, M.; Mitsuhashi, S.; Nishi, Y.; Uryu, M. The structure and mechanical properties of sheets prepared from bacterial cellulose. J. Mater. Sci. 1989, 24, 3141–3145. [Google Scholar] [CrossRef]
- Bi, J.C.; Liu, S.X.; Li, C.F.; Li, J.; Liu, L.X.; Deng, J.; Yang, Y.C. Morphology and structure characterization of bacterial celluloses produced by different strains in agitated culture. J. Appl. Microbiol. 2014, 117, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- French, A.D. Idealized powder diffraction patterns for cellulose polymorphs. Cellulose 2014, 21, 885–896. [Google Scholar] [CrossRef]
- Tokoh, C.; Takabe, K.; Fujita, M.; Saiki, H. Cellulose synthesized by Acetobacter xylinum in the presence of acetyl glucomannan. Cellulose 1998, 5, 249–261. [Google Scholar] [CrossRef]
- Keshk, S.; Sameshima, K. The utilization of sugar cane molasses with/without the presence of lignosulfonate for the production of bacterial cellulose. Appl. Microbiol. Biotechnol. 2006, 72, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Meza-Contreras, J.C.; Manriquez-Gonzalez, R.; Gutiérrez-Ortega, J.A.; Gonzalez-Garcia, Y. XRD and solid state 13C-NMR evaluation of the crystallinity enhancement of 13C-labeled bacterial cellulose biosynthesized by Komagataeibacter xylinus under different stimuli: A comparative strategy of analyses. Carbohydr. Res. 2018, 461, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Horn, F. In situ crystallization of bacterial cellulose I. Influences of polymeric additives, stirring and temperature on the formation celluloses Iα and Iβ as revealed by cross polarization/magic angle spinning (CP/MAS)13C NMR spectroscopy. Cellulose 1994, 1, 57–66. [Google Scholar] [CrossRef]
- Yamamoto, H.; Horii, F. CP/MAS 13C NMR analysis of the crystal transformation induced for Valonia cellulose by annealing at high temperatures. Macromolecules 1993, 26, 1313–1317. [Google Scholar] [CrossRef]
- Colvin, J.R.; Leppard, G.G. The biosynthesis of cellulose by Acetobacter xylinum and Acetobacter acetigenus. Can. J. Microbiol. 1977, 23, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Zhang, G.-C.; Liu, M.; Zheng, X.-T.; Han, P.-P.; Jia, S.-R. Metabolic flux analysis of Gluconacetobacter xylinus for bacterial cellulose production. Appl. Microbiol. Biotechnol. 2013, 97, 6189–6199. [Google Scholar] [CrossRef] [PubMed]
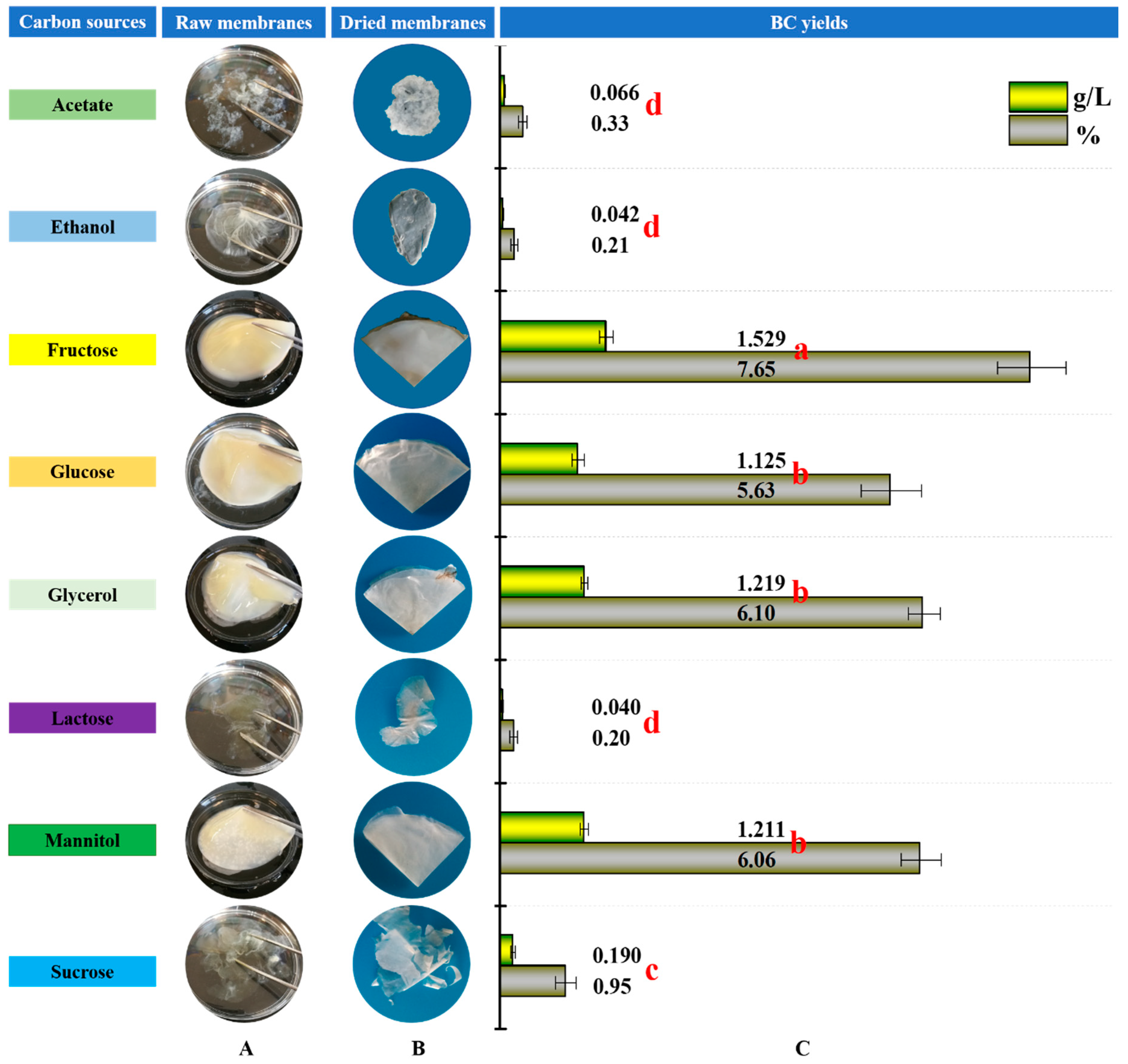
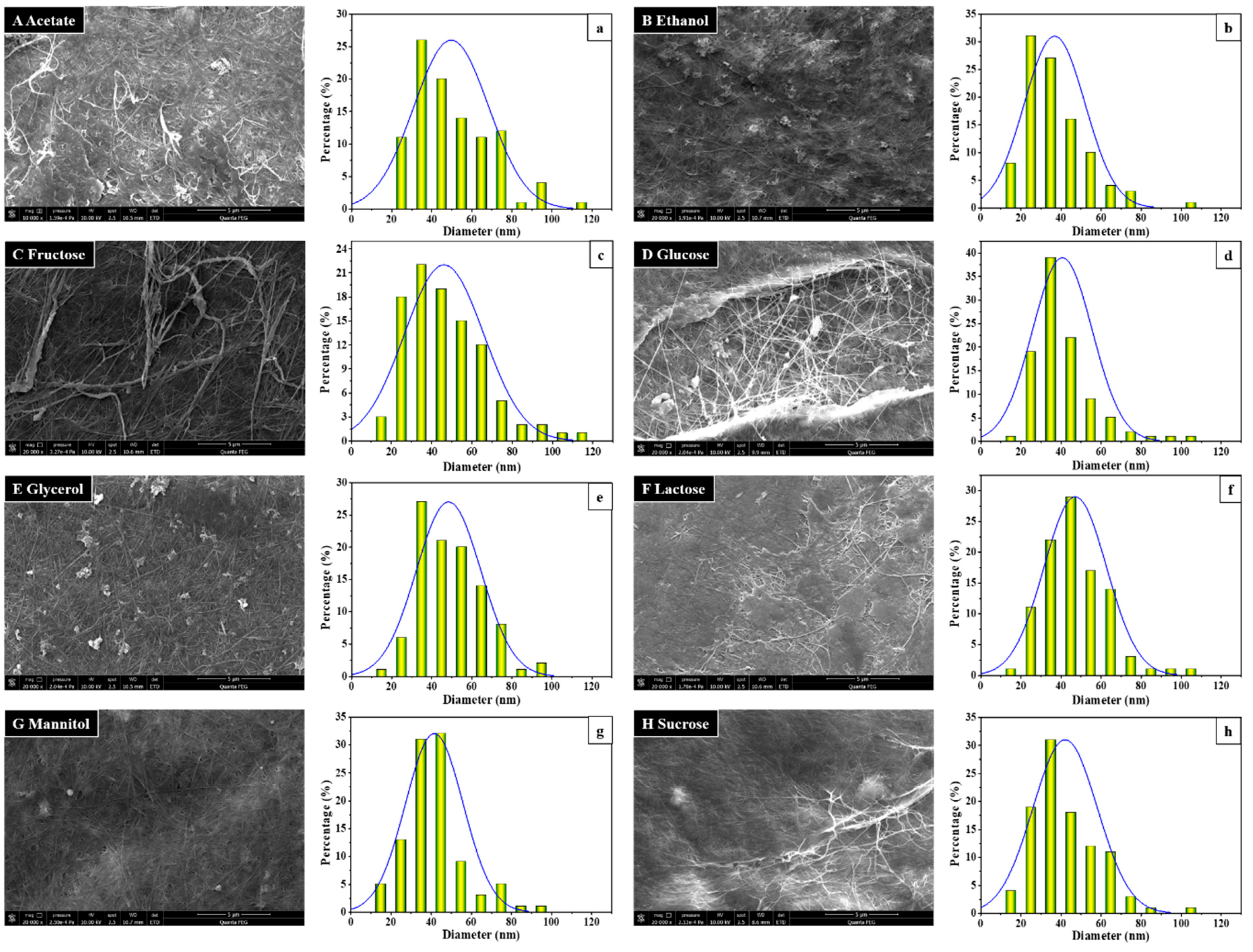

| Bacteria Names | Cultivation Conditions | Carbon Sources | Concentration (%, w/v) | Relative Yields (% Day−1) a | References |
|---|---|---|---|---|---|
| Acetobacter sp. S-35 | Static, 28 °C, 3 days | Sucrose, lactate, glucose, mannitol b, gluconate, glycerol, dulcitol, maltose, lactose, sorbose, ribose, arabinose, xylose, fructose, galactose | 4% except for dulcitol and sorbose with 2% | up to 9.58 | Kojima et al. [40] |
| Acetobacter hansenii ATCC 10821 | Static, 30 °C, 30 days | d-glucose b, d-xylose, d-xylulose, d-xylose/d-xylulose | 2% | 0.037–1.087 | Ishihara et al. [42] |
| Acetobacter lovaniensis HBB5 | Static, 30 °C, 7 days | Glucose b, sucrose, fructose, ethanol | 2% | 0.008–0.029 | Çoban and Biyik [16] |
| Acetobacter pasteurianus IFO 14814 | Static, 30 °C, 30 days | d-glucose b, d-xylose, d-xylulose, d-xylose/d-xylulose | 2% | 0.130–0.167 | Ishihara et al. [42] |
| Acetobacter xylinum ATCC 10245 | Static, 30 °C, 14 days | Glucose | 1.5% | 5.29 | Hassan et al. [39] |
| Acetobacter xylinumc | NM d, 35 °C, 14 days | Sucrose b, glucose, mannitol b, sorbitol, galactose, lactose, acetic acid, maltose | 5–7% | ≥0.057 e | Ramana et al. [36] |
| Acetobacter xylinus IFO 15606 | Static, 30 °C, 30 days | d-glucose, d-xylose, d-xylulose, D-xylose/d-xylulose b | 2% | 0.147–0.347 | Ishihara et al. [42] |
| Chromobacterium violaceum ATCC 12472 | Static, 32 °C, 3 days | Glucose | 2% | NM | Recouvreux et al. [43] |
| Enterobacter amnigenus GH-1 | Static, 30 °C, 14 days | d-Glucose, d-fructose b, lactose, mannitol, inositol, sucrose, maltose, glycerol | 2% | 1.0 | Hungund and Gupta [44] |
| Enterobacter sp. | Agitated, 30 °C, 24 days | Glucose | 2% | 0.5 | Ago et al. [37] |
| Gluconacetobacter intermedius NEDO-01 | Static, 30 °C, 3 days Agitated, 30 °C, 4 days | Glycerol | 2% | NM 4.25 | Kose et al. [15] |
| Gluconacetobacter sacchari | Static, 30 °C, 4 days | Glucose | 2% | 3.375 | Trovatti et al. [45] |
| Gluconacetobacter xylinus ATCC 53524 | Static, 30 °C, 2 or 4 days | Mannitol b, glucose, glycerol, fructose, sucrose b, galactose | 2% | 5.10 or 4.79 | Mikkelsen et al. [13] |
| Gluconacetobacter xylinus CH001 | Static, 28 °C, 14 days | Xylose | 1–3% | up to 0.482 | Yang et al. [19] |
| Gluconacetobacter xylinus PTCC 1734 | Agitated, 28 °C, 7 days | Mannitol, sucrose b, glucose | 2–5% | ~0.947 | Mohammadkazemi et al. [28] |
| Gluconacetobacter xylinus PTCC 1734 | Static, 28 °C, 20 days | Glucose, fructose, mannitol b, sucrose, glycerol | 2% | up to 2.5 | Tabaii and Emtiazi [41] |
| Komagataeibacter medellinensis | Static, 28 °C, 8 days | Fructose, glucose b, sucrose | 2% | 0.238–1.75 | Molina-Ramírez et al. [2] |
| Komagataeibacter rhaeticus PG2 | Static, 28 °C, 15 days | Fructose, lactose, xylose, sucrose, galactose, mannitol, sorbitol, and glycerol | 2% | up to 2.3 | Thorat and Dastager [46] |
| Komagataeibacter saccharivorans PE 5 | Static, 30 °C, 14 days | Mannitol | 1.5% | 6.00 | Hassan et al. [39] |
| Komagataeibacter xylinus KX, TISTR 086, 428, 975 and 1011 | Static, 30 °C, 7 days Agitated, 30 °C, 7 days | Glucose b, fructose, lactose b, maltitol, sucralose, xylitol | 5% | 0.326–0.526 up to 1.34 | Singhsa et al. [12] |
| Komagataeibacter xylinus B-12068 | Static, 30 °C, 7 days | Glucose b, sucrose, galactose, maltose, mannitol | 2% | 0.071–1.571 | Volova et al. [47] |
| Rhodococcus sp. MI 2 | Static, 25 °C, 14 days | Glucose, fructose b, sucrose, lactose, sorbitol, and mannitol | 2% | ~2.25 | Tanskul et al. [38] |
| Saccharomyces cerevisiae CGMCC1670 | Static, 30 °C, 22 days | Glucose | 5% | ~0.118 | Tan et al. [48] |
| Komagataeibacter sp. W1 | Static, 30 °C, 14 days | Acetate, ethanol, fructose, glucose, glycerol, lactose, mannitol b, sucrose | 2% f | 0.015–0.547 | This study |
| Carbon Sources | Peak 1 (100Iα or 10Iβ) | Peak 2 (010Iα or 110Iβ) | Peak 3 (110Iα or 200Iβ) | At 2θ Scale | C.I. | % Crystalline | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| d-Spacing (nm) | ACS (nm) | d-Spacing (nm) | ACS (nm) | d-Spacing (nm) | ACS (nm) | Iam | Ima | |||
| Acetate | 0.60 | 6.7 | 0.53 | 15.5 | 0.39 | 7.8 | 133 | 636 | 0.79 | 83 |
| Ethanol | 0.60 | 6.9 | – a | – | 0.39 | 7.2 | 126 | 602 | 0.79 | 83 |
| Fructose | 0.60 | 8.2 | 0.53 | 9.4 | 0.39 | 8.9 | 165 | 1323 | 0.88 | 89 |
| Glucose | 0.60 | 8.8 | 0.53 | 6.8 | 0.39 | 9.2 | 132 | 844 | 0.84 | 86 |
| Glycerol | 0.60 | 8.6 | 0.53 | 10.7 | 0.39 | 8.9 | 140 | 978 | 0.86 | 87 |
| Lactose | – | – | – | – | 0.39 | 1.8 | 99 | 273 | 0.64 | 73 |
| Mannitol | 0.60 | 7.9 | 0.53 | 8.0 | 0.39 | 8.8 | 185 | 1674 | 0.89 | 90 |
| Sucrose | 0.60 | 23.6 | – | – | 0.39 | 4.6 | 112 | 322 | 0.65 | 74 |
| Peak Number | Wavenumber (cm−1) | Functional Groups | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Acetate | Ethanol | Fructose | Glucose | Glycerol | Lactose | Mannitol | Sucrose | ||
| 1 | 3344 | 3343 | 3343 | 3343 | 3344 | 3345 | 3341 | 3346 | O–H stretching vibration |
| 2 | 2895 | 2896 | 2895 | 2895 | 2895 | 2897 | 2894 | 2899 | C–H stretching of CH2 and CH3 groups |
| 3 | – a | – | – | – | – | 1730 | – | 1734 | UK c |
| 4 | 1648 | 1645 | – | 1645 | 1645 | 1649 | 1642 | 1647 | H–O–H bending of absorbed water |
| 5 | 1564 | 1573 | 1574 | – | 1574 | – | – | – | UK |
| 6 | 1424 | 1427 | 1423 | 1427 | 1423 | – | 1426 | 1423 | CH2 symmetric bending or O–H in plane bending |
| 7 | 1361 | 1360 | 1361 | 1360 | 1361 | 1355 | 1361 | 1361 | C–H bending |
| 8 | 1336 | 1335 | 1335 | 1335 | 1336 | 1336 | 1335 | 1335 | C–H deformation or O–H in-plane bending |
| 9 | 1315 | 1315 | 1315 | 1315 | 1315 | 1315 | 1315 | 1315 | Out-of-plane wagging of the CH2 groups |
| 10 | 1280 | 1281 | 1280 | 1280 | 1281 | 1281 b | 1280 | 1280 b | C–H bending |
| 11 | 1248 | – | 1249 | 1249 | 1249 b | – | – | 1249 b | UK |
| 12 | 1203 b | 1205 | 1205 | 1205 | 1204 | 1203 b | 1205 | 1202 b | C–H bending |
| 13 | 1160 | 1161 | 1161 | 1161 | 1161 | 1160 | 1160 | 1161 | C–O–C antisymmetric bridge stretching of 1, 4-β-d-glucoside |
| 14 | 1108 | 1108 | 1108 | 1108 | 1108 | 1109 | 1108 | 1108 | C–C bonds of the monomer units of polysaccharide or C–O bending vibration |
| 15 | 1055 | 1055 | 1054 | 1054 | 1055 | 1056 | 1055 | 1054 | The bending of C–O–H bond of carbohydrates or C–O–C pyranose ring skeletal vibration |
| 16 | 1031 | 1031 | 1031 | 1031 | 1031 | 1032 | 1030 | 1032 | |
| 17 | 1003 b | 1005 b | 1003 b | 1004 | 997 | 1002 b | 1003 | 1002 b | UK |
| 18 | 899 b | 899 | – | 899 | – | – | 895 | – | Antisymmetric out-of-phase ring stretching of β-glucosidic linkages between the glucose units |
| 19 | 847 | 835 | – | – | – | 836 | – | 836 | UK |
| 20 | 660 | 664 | 663 | 664 | 662 | 666 | 663 | 657 | O–H out-of-phase bending vibration |
| 21 | 600 | 609 | 602 | 609 | 597 | 602 | 601 | 597 | |
| 22 | 563 | 559 | 557 | 558 | 563 | 564 | 558 | 562 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.-S.; Han, Y.-H.; Chen, J.-L.; Zhang, D.-C.; Shi, X.-X.; Ye, Y.-X.; Chen, D.-L.; Li, M. Insights into Bacterial Cellulose Biosynthesis from Different Carbon Sources and the Associated Biochemical Transformation Pathways in Komagataeibacter sp. W1. Polymers 2018, 10, 963. https://doi.org/10.3390/polym10090963
Wang S-S, Han Y-H, Chen J-L, Zhang D-C, Shi X-X, Ye Y-X, Chen D-L, Li M. Insights into Bacterial Cellulose Biosynthesis from Different Carbon Sources and the Associated Biochemical Transformation Pathways in Komagataeibacter sp. W1. Polymers. 2018; 10(9):963. https://doi.org/10.3390/polym10090963
Chicago/Turabian StyleWang, Shan-Shan, Yong-He Han, Jia-Lian Chen, Da-Chun Zhang, Xiao-Xia Shi, Yu-Xuan Ye, Deng-Long Chen, and Min Li. 2018. "Insights into Bacterial Cellulose Biosynthesis from Different Carbon Sources and the Associated Biochemical Transformation Pathways in Komagataeibacter sp. W1" Polymers 10, no. 9: 963. https://doi.org/10.3390/polym10090963
APA StyleWang, S.-S., Han, Y.-H., Chen, J.-L., Zhang, D.-C., Shi, X.-X., Ye, Y.-X., Chen, D.-L., & Li, M. (2018). Insights into Bacterial Cellulose Biosynthesis from Different Carbon Sources and the Associated Biochemical Transformation Pathways in Komagataeibacter sp. W1. Polymers, 10(9), 963. https://doi.org/10.3390/polym10090963