Abstract
Depolymerization and de-N-acetylation of chitin by chitinases and deacetylases generates a series of derivatives including chitosans and chitooligosaccharides (COS), which are involved in molecular recognition events such as modulation of cell signaling and morphogenesis, immune responses, and host-pathogen interactions. Chitosans and COS are also attractive scaffolds for the development of bionanomaterials for drug/gene delivery and tissue engineering applications. Most of the biological activities associated with COS seem to be largely dependent not only on the degree of polymerization but also on the acetylation pattern, which defines the charge density and distribution of GlcNAc and GlcNH2 moieties in chitosans and COS. Chitin de-N-acetylases (CDAs) catalyze the hydrolysis of the acetamido group in GlcNAc residues of chitin, chitosan, and COS. The deacetylation patterns are diverse, some CDAs being specific for single positions, others showing multiple attack, processivity or random actions. This review summarizes the current knowledge on substrate specificity of bacterial and fungal CDAs, focusing on the structural and molecular aspects of their modes of action. Understanding the structural determinants of specificity will not only contribute to unravelling structure-function relationships, but also to use and engineer CDAs as biocatalysts for the production of tailor-made chitosans and COS for a growing number of applications.
1. Introduction
Chitin is a linear polysaccharide of β(1→4)-linked N-acetylglucosamine monomers. It was first isolated from fungi in 1811 [1] and its structure was determined in 1929 [2]. Chitin is a major structural component of the exoskeletons of arthropods (insects and crustaceans), of the endoskeletons of mollusks, and it is also found in the cell walls of fungi and diatoms [1,3,4]. It is one of the most abundant organic molecules after cellulose, and the most abundant natural amino polysaccharide. Chitin is present as ordered macrofibrils, mainly in two allomorphs: α-chitin, with antiparallel chains [5], is the most abundant and it is isolated from the exoskeleton of crustaceans, particularly from shrimps and crabs; and β-chitin, with parallel chains [6], is present in the cell walls of diatoms and in the skeletal structures of cephalopods, and commonly extracted from squid pens. β-Chitin is easily converted to α-chitin by alkaline treatment followed by flushing in water [5]. Chitin is also found as γ-chitin in fungi and yeast, which is a combination of the α and β allomorphs [7,8].
Depolymerization (hydrolysis) of chitin by chitinases results in chitooligosaccharides (COS), and de-N-acetylation of chitin and COS yields chitosan and partially acetylated COS (paCOS) or fully deacetylated glucosamine oligomers (Figure 1). In nature, the deacetylation of chitin is almost never complete, and chitosan refers to a family of heteropolysaccharides composed of N-acetylglucosamine and glucosamine, characterized by their degree of polymerization (DP), degree of acetylation (DA), and pattern of acetylation (PA). Only some fungi of the Zygomycota, Basidiomycota and Ascomycota phyla have been reported to be capable of naturally producing chitosans [9]. The free amino groups of the deacetylated units in the polymer are protonated at slightly acidic pH, thus making chitosans the only known natural polycationic polysaccharides [1,3,9]. They interact with polyanionic biomolecules such as polyanionic phospholipidic membranes, glycosaminoglycans at cell surfaces, proteins, and nucleic acids. Depolymerization by hydrolysis of the β-1,4-linkages of chitin and chitosan yields their respective oligosaccharides (COS, paCOS) [10]. Whereas chitin and chitosans act as structural polymers, their oligomers are involved in molecular recognition events, such as cell signaling and morphogenesis, and act as immune response elicitors and host-pathogen mediators [11,12,13,14,15]. The catabolism of chitin and chitosan is summarized in Figure 1. Chitin and chitosan oligosaccharides are further degraded following different organism-dependent pathways to end up in the central energy metabolism.
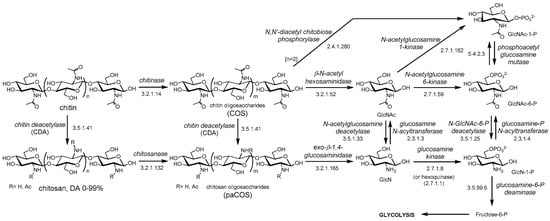
Figure 1.
Chitin catabolism. GlcNAc: N-acetylglucosamine; GlcN: glucosamine; DA: degree of acetylation. COS: chitooligosaccharides (or chitin oligosaccharides), paCOS: partially acetylated chitooligosaccharides (or chitosan oligosaccharides).
The de-N-acetylation of chitin and chitooligosaccharides is catalyzed by chitin deacetylases (CDAs), which exhibit different substrate specificities leading to fully or partially deacetylated products with diverse degrees of acetylation (DA) and patterns of acetylation (PA). In addition to the role CDAs play in the biology of their natural organisms, there is a growing interest in the biochemical characterization of CDAs in order to use them as biocatalysts for the production of partially deacetylated chitooligosaccharides (paCOS) as bioactive molecules in different application fields, or to inhibit them since they are potential targets against pathogenic microorganisms. The aim of this review is to analyze the current knowledge on the biochemistry of chitin deacetylases with regard to their substrate specificity. Although a large number of enzymes have been experimentally identified as chitin deacetylases able to deacetylate chitin either in vivo or in vitro, few of them have been analyzed at the protein level. Here we will focus on those CDAs with characterized activity on chitooligosaccharides (COS) to address the issue of substrate specificity and the deacetylation pattern.
2. Chitin Deacetylases and the Carbohydrate Esterase Family 4 (CE4)
Chitin deacetylases (CDAs, EC 3.5.1.41) and chitooligosaccharides deacetylases (CODs, EC 3.5.1.105) are classified in the carbohydrate esterase family 4 (CE4) in the CAZY database (Carbohydrate Active Enzymes, www.cazy.org) [16]. The CE4 family also contains peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.104), peptidoglycan N-acetylmuramic acid deacetylases (EC 3.5.1.-), poly-β-1,6-N-acetylglucosamine deacetylase (EC 3.5.1.-), and some acetyl xylan esterases (EC 3.1.1.72) [17]. These enzymes share a conserved region known as the NodB homology domain due to its similarity to the NodB oligosaccharide deacetylase, one of the first CE4 enzymes to be characterized [18]. Most currently reported and characterized CDAs and CODs are CE4 enzymes, with the exception of diacetylchitobiose deacetylases (Dacs) from archaea and a COD from Bacillus cereus (BcZBP) that belong to the CE14 family [19,20]. Few other enzymes, such as insect CDAs and a COD from E. coli (ChbG) [21] are in the group of “non-classified” in the CAZY database since they do not share sequence similarities to the other CDA families.
The deacetylase activity from extracts of the fungus Mucor rouxii was the first active CDA identified and partially purified in the mid 1970s [22,23]. Later on, the NodB from a rhizobium species was the first biochemically characterized COD in 1993 [18]. Many other CDAs and CODs were later identified and purified from very diverse organisms, including archaea, marine bacteria, fungi, and insects [24]. These enzymes are diverse in their biochemical properties: molecular masses in the range from 12 to 150 kDa, acidic isoelectric points (pI from 2.7 to 4.8), optimum pH for activity from 4.5 to 12, and significant thermal stability, with optimum temperatures for activity in the range from 30 to 60 °C. Most CDAs are highly inactive on crystalline chitin due to the inaccessibility of the acetyl groups in the tightly packed chitin structure, and have a preference for soluble forms of chitins such as glycol-chitin or chitin oligomers, as well as partially deacetylated chitin (chitosans). It has recently been shown that CDA activity on crystalline chitin is greatly enhanced by oxidative cleavage of the surface polymer chains by lytic polysaccharide monooxygenases (LPMO) [25]. Some CDAs contain carbohydrate binding modules (CBM) fused to the catalytic domain that seem to enhance the deacetylase activity by increasing the accessibility of the substrate to the catalytic domain [26].
CDAs are localized in different cellular compartments, in the periplasm, in the cytosol, or secreted as extracellular enzymes. Periplasmic fungal CDAs are generally tightly coupled to a chitin synthase to rapidly deacetylate newly synthesized chitin before their maturation and crystallization. Extracellular fungal CDAs are secreted to alter the physicochemical properties of the cell wall, which results in protection against exogenous chitinases, or initiates sporulation or autolysis. In bacteria, CDAs are either intracellular, as those involved in Nod factors biosynthesis in Rhizobium species, or extracellular, as those involved in the catabolism of chitin in marine bacteria [24,27,28].
Some CE4 enzymes classified in a specific subfamily also show activity on typical substrates from other subfamilies. Peptidoglycan GlcNAc deacetylases, involved in the de-N-acetylation of the bacterial cell wall peptidoglycan with critical functions in the maturation and turnover of peptidoglycan and in bacterial pathogenicity, are also active on COS. Some CDAs have activity on acetylxylan, as well as some acetylxylan esterases are active on COS, which makes their classification doubtful in some cases.
3. Function and Specificity of CE4 Chitin Deacetylases
3.1. Deacetylation Patterns
Chitin deacetylases exhibit diverse deacetylation patterns, reflecting different substrate specificities and pattern recognition on their linear substrates. The mechanisms of action of enzymes that modify in-chain units on a linear polysaccharide are commonly classified as multiple-attack, multiple-chain, and single-chain mechanisms [29]. In the multiple-attack mechanism, binding of the enzyme to the polysaccharide chain is followed by a number of sequential deacetylations, after which the enzyme binds to another region of the polymeric chain. (i.e., M. rouxii [30,31]). On a polymeric substrate, this mechanism will result in a block-copolymer structure with blocks of GlcNH2 units within the GlcNAc chain. On COS, it will usually result in full deacetylation of the oligomer. In the multiple-chain mechanism, the enzyme forms an active enzyme-polymer complex and catalyzes the hydrolysis of only one acetyl group before it dissociates and forms a new active complex (i.e., C. lindemuthianum CDA [32,33]). It will result in a random distribution of the GlcNH2 and GlcNAc units along the polymeric chain or, in the case of COS substrates, it will render a number of partially deacetylated oligosaccharide intermediates ending in a specific deacetylation pattern or in full deacetylation, depending on the enzyme and the substrate. Finally, a single-chain mechanism includes processive enzymes in which a number of catalytic events occur on a single substrate molecule leading to sequential deacetylation. Some bacterial chitooligosaccharides deacetylases (CODs), which are specific for a single position leading to mono-deacetylated products (i.e., Rhizobium NodB or Vibrio CDA or COD) are also included in the last group.
A fundamental question is how these enzymes define their action pattern. This is relevant not only to understand their biological functions but also to use CDAs (native and engineered variants) as biocatalysts for the production of chitosans with non-random deacetylation patterns, and partially deacetylated COS with tailored patterns of acetylation (see Section 6). A structural model on the determinants of substrate specificity is currently emerging from studies on substrate specificity, determination of 3D structures of enzyme-substrate complexes, and multiple sequence alignments. Many CDAs, particularly from fungal origin, have been identified as involved in chitin deacetylation in vivo, but only few of them have been characterized with regard to substrate specificity and mode of action. To the best of our knowledge, Table 1 compiles the CDA enzymes in family CE4 that have been biochemically characterized and have reported activity on COS substrates, some of them with solved three-dimensional structure by X-ray crystallography. Relevant information on their biological function and substrate specificity is summarized below.

Table 1.
CDAs and CODs with characterized activity on COS.
CDAs participate in diverse biological processes, which include cell wall morphogenesis and host-pathogen interaction in fungi, generation of signaling molecules in bacteria, and participation in the catabolism of chitin as carbon, nitrogen, and energy sources in marine bacteria and fungi. CDAs were thought to be restricted to fungi and bacteria until a first report in 1986 on their presence in arthropods [34]. CDAs seem to be widely present in insects, in cuticles and the perithrophic midgut matrix, but little is known on the function and properties of insect CDAs [35,36], and they are not included here because scarce information on substrate specificities has been reported.
3.2. Fungal CDAs
Fungal CDAs are involved in fungal nutrition, morphogenesis and development [27,29], participating in cell wall formation and integrity [37], in spore formation [38], germling adhesion [39], fungal autolysis [40], and in defense mechanisms for host infection [41].
Fungi that have chitosan (in addition to chitin) as a structural component of the cell wall, secrete CDAs to the periplasmic space that contribute to chitosan biosynthesis from nascent chitin synthesized by chitin synthases. It occurs during exclusive periods corresponding to their particular biological role in the cell cycle of the fungal species: during cell wall formation (i.e., M. rouxi [42] and A. coerulea [43]), during sporulation (i.e., S. cerevisiae [38,44]), or during vegetative growth (i.e., C. neoformas [37]).
Pathogenic fungi secrete CDAs during fungal hyphae penetration to evade plant defense mechanisms and gain access to host tissues. Plants secrete chitinases to break the fungal cell-wall chitin down to chitooligosaccharides (COS), and the released COS are recognized by plant chitin-specific receptors, triggering resistance responses [41]. COS elicitation of resistance mechanisms involve activation of host defense genes [45,46]. There is cumulative evidence that fungi evade plant defense mechanisms by partially deacetylating either their exposed cell wall chitin or the chitooligosaccharides produced by the action of plant chitinases. In both cases, the resulting partially deacetylated oligomers are not well recognized by the specific plant receptors reducing or preventing the elicitation of the defense responses [41,47,48,49].
Filamentous fungi undergo autolysis by self-digestion of aged hyphal cultures due to carbon starvation [40,50]. During this event there is an increased presence of hydrolytic enzymes, especially those involved in cell wall degradation, and CDAs are secreted to the extracellular medium to deacetylate the chitin oligomers produced by chitinases (i.e., A. nidulans [51,52]).
In general, fungal CDAs deacetylate soluble forms of chitin such as glycol-chitin and chitosans of variable DA, but they are inactive, or show low activity, towards insoluble chitins such as crystalline α- and β-chitin and colloidal chitin. Pretreatment of chitin to make surface fibrils more accessible may result in increased deacetylase activity. It has been recently shown that the activity of an A. nidulans CDA on crystalline chitin was enhanced by a lytic polysaccharide monooxygenase (LPMO) that increases substrate accessibility by oxidative cleavage of the chitin chains [25]. Some CDAs also appear to be active on acetylxylan (i.e., AnCDA), but any of them act on peptidoglycans, typical substrates of other CE4 family members. In addition to polymeric substrate, not many CDA have been analyzed with COS as substrates (Table 1). The analysis of the products from enzymatic deacetylation with regards to the extent and pattern of deacetylated provides information about the specificity and mode of action of these enzymes. The first seven entries in Table 1 correspond to CDAs for which the deacetylation pattern on COS has been reported, whereas the rest of the entries are CDAs active on COS but, to the best of our knowledge, the structure of the deacetylated products has not been analyzed.
Mucor rouxii (Amylomyces rouxii) (MrCDA). The dimorphic fungus M. rouxii has a cell wall mainly composed of chitin, chitosan, and mucoric acid. While chitin accounts for 10% of the total dry weight of the cell wall, chitosan reaches 30% [77]. The M. rouxii CDA enzyme was initially found in the cytosol [23], but it is also secreted into the periplasm where it participates in a tandem synthetic mechanism that involves a chitin synthase and a chitin deacetylase working consecutively and synchronously for synthesis and deposition of chitosan polymers at the outer membrane [78]. Decoupling this mechanism prevents the formation of chitosan [42,79]. M. rouxii, like other fungi, has been identified as a suitable microorganism for chitosan production by means of biofermentation processes [80,81]. MrCDA is a monomeric high-mannose-type glycosylated protein with an apparent molecular mass of 75–80 kDa [82]. Kinetic studies indicate that the preferred catalytic metal is Zn2+ like many other CDAs. In terms of activity, its optimal pH and temperature values are between 4.5–5.5 and 50 °C, respectively [23,33]. MrCDA deacetylates chitinous polymers such as glycol-chitin, colloidal chitin, chitosan, and chitin, but also deacetylates acetylxylan [17,23]. On chitooligosaccharides, triacetylchitotriose is the smallest substrate and the activity increases with the degree of polymerization (DP) [23,31,42,78]. The enzyme follows a multiple-attack mechanism [30] but the resulting pattern of acetylation (PA) depends on the DP of the substrate: whereas DP3, DP6 and DP7 substrates are not fully deacetylated, leaving the reducing GlcNAc unmodified [Dn−1A], DP4 and DP5 substrates are fully deacetylated [Dn]. In all cases, deacetylation starts at the non-reducing end residue and then proceeds to the neighboring monomer towards the reducing end [31].
Colletotrichum lindemuthianum (ClCDA). The deuteromycete C. lindemuthianum is a plant pathogen that causes anthracnose, a disease which affects economically important crop species [83]. ClCDA is a heavily glycosylated secreted enzyme allegedly playing a role in the host-pathogen interaction, deacetylating the chitin oligomers resulting from the activity of plant chitinases on the fungal cell wall [83,84]. Less likely is its function in deacetylating the fungal cell wall chitin to evade degradation by plant chitinases, since no chitosan has been observed in the cell wall ultra-structure [85]. Since its discovery in the 1980s it has been purified from its natural host [84,86] as well as expressed in several eukaryotic and bacterial hosts such as Pichia pastoris [87,88] and E. coli [89,90]. The enzyme has a preference for Co2+ and Zn2+ as the catalytic metal cation and its activity is substantially inhibited by Cu2+ or Ni2+, but not inhibited by EDTA or acetate [53,86]. It is a quite thermostable enzyme with an optimum temperature of 60 °C, and a pH optimum of 8.0. ClCDA is active on both chitin polymers (glycol-chitin) and COS. It fully deacetylates COS with a DP equal to or greater than 3, while it only deacetylates the non-reducing GlcNAc of diacetylchitobiose [32,91]. ClCDA acts by a multiple-chain mechanism following a pathway in which the first residue to be deacetylated is the second from the reducing end [32,33]. The initial mono-deacetylation reaction shows no dependency of kcat on DP and a decrease of KM with increasing DP [33,53]. However, kinetics of fully deacetylated products formation show an increase in kcat and reduction in KM that correlate with the increase of DP [86]. It has been reported that this enzyme is reversible, as it is also able to catalyze the acetylation of chitosan oligomers [92,93,94].
Aspergillus nidulans (AnCDA). During cell autolysis, AnCDA is secreted into the extracellular medium to deacetylate the chitin oligomers produced by chitinases [40,50,52,95]. The enzyme has been purified from A. nidulans cultures as a glycosylated enzyme [51]. The recombinant protein has been expressed in E. coli and purified by refolding from inclusion bodies [96] and recently it has been obtained in soluble form [25]. Like ClCDA, AnCDA is a thermostable protein with an optimal temperature of 50 °C and retaining 68% activity after 1 h at 80 °C. Its optimum pH is 7–8 [51,96]. The enzyme is active on soluble chitins (CM-chitin, glycol-chitin), colloidal chitin, chitosan, acetylxylan, and acetylated glucuronoxylan, but not on peptidoglycan [25,51]. AnCDA is active on COS with a DP from 2 to 6 [25]. The enzyme catalyzes mono-deacetylation of (GlcNAc)2 and it is inactive on GlcNAc monosaccharide. Longer substrates than DP2 are fully deacetylated. However, the deacetylation rate exhibits a counter-intuitive relationship with the DP of the substrate: odd-numbered COS (DP5, DP3) have higher apparent rate constants than even-numbered oligomers (DP4, DP2). For the DP6 substrate, time-course monitoring of products formation reveals that the first deacetylation event occurs at random positions except for the reducing end, which reacts much slower to yield the fully deacetylated end product [Dn].
Podospora anserina (PaCDA). The filamentous ascomycete Podospora anserina lives as a saprophyte on herbivore dung [97]. It has a limited lifespan and it is a model organism in cell aging studies [98]. PaCDA was identified in a search for CDAs containing chitin binding domains. The enzyme has been recombinantly expressed in Hansenula polymorpha as a full length protein composed of the CE4 domain flanked by two CBM18 domains [26]. The low activity of the enzyme on colloidal chitin is significantly reduced by deletion of the CBM domains, which supports the hypothesis that the presence of the CBMs helps the enzyme to act on insoluble substrates. PaCDA is active on soluble glycol-chitin, chitosans with a high DA, and COS, with optimum pH and temperature values of 8.0 and 55 °C, respectively. It fully deacetylates COS with a DP ≥ 2 and follows a multiple-chain mechanism. With the DP3 substrate, the first deacetylation event has a clear preference for the reducing end, but all possible isomers are found for both mono- and di-deacetylated intermediate products. With DP4 and DP5 substrates, the residue next to the reducing end is preferentially deacetylated first, with the second deacetylation occurring mainly next to the existing GlcNH2 unit on either side. Deacetylation is faster for longer substrates, with deacetylation of the reducing end occurring as a late event [26].
Puccinia graminis f. sp. Tritici (PgtCDA). The biotrophic basidiomycete Puccinia graminis f. sp. Tritici is the causative agent of the stem rust [99]. The appearance of resistant races of P. graminis affecting wheat cultivars has been recognized as a serious threat to food security [100,101], boosting the interest in understanding the virulence and defense mechanism of this fungal pathogen. Rust fungi promote the formation of complex structures in order to invade the plant cells but at the same time they must prevent the triggering of immune responses [102]. A main transition during infection is from the ectophytically growing appressorium to the endophytically growing substomatal vesicle; while the former exposes chitin on its surface, the latter exposes chitosan [47,103]. PgtCDA may not only participate in the chitin to chitosan transition, making the cell wall less susceptible to host chitinases [104], but also could deacetylate the chitooligosaccharide products, reducing its elicitor properties [105]. PgtCDA has been recombinantly expressed in E. coli as a fusion protein with the maltose binding protein (MBP) [54]. Its optimal pH for activity is between 8 and 9 and its optimal temperature is 50 °C. It is not active on insoluble polymers such as α- or β-chitin, but efficiently deacetylates colloidal chitin, glycol-chitin and chitosans, on which activity increases with the degree of acetylation. With COS substrates, the minimal substrate is tetraacetylchitotetraose (DP4). The structure of the products from enzymatic deacetylation of DP4 to DP6 substrates reveals that the enzyme acts by a multiple-chain mechanism and specifically deacetylates all but the last two GlcNAc units on the non-reducing end [AA(D)n−2] [54].
Pestalotiopsis sp. (PesCDA). The endophytic fungus Pestalotiopsis sp. lives inside the tissues of its plant hosts in tropical areas [106]. To successfully survive in their hosts, endophytes also need to avoid being detected by the plant immune system. A secreted Pestalotiopsis CDA has been identified when chitosan was present in the culture medium [49]. The recombinantly expressed PesCDA is active on colloidal chitin as substrate, chitosans with a DA of 10–60% (higher activity with a higher DA), and COS, but inactive on crystalline α- or β-chitin. When analyzing the activity on COS, tetraacetylchitotetraose is the minimal substrate. With a DP5 substrate, the optimum pH and temperature values are 8.0 and 55 °C, respectively. Through a multiple-chain mechanism, the enzyme deacetylates all residues of the substrates except the reducing end and the last two GlcNAc residues from the non-reducing end, with a pattern of deacetylation [AA(D)n−3A] [49]. The chitosan oligomers obtained from deacetylation of a DP6 substrate by PesCDA have shown that, as opposed to the fully acetylated oligomer, they are no longer elicitors of the plant immune system in rice cells [49].
Pochonia chlamydosporia (PcCDA). The ascomycete Pochonia chlamydosporia infects females and eggs of cyst or root-knot nematodes. It is used as a biocontrol agent against a number of plant parasitic nematodes in food-security crops [107,108,109]. P. chlamydosporia expresses chitosanases and chitin deacetylases during egg infection. Since chitosan is associated with the sites of fungal penetration, it has been suggested that secreted CDAs are involved in nematode infection [110]. A PcCDA has been recently characterized [55]. The full-length protein contains the CE4 catalytic domain flanked by two CBM18 chitin binding domains. The recombinantly expressed PcCDA catalytic domain deacetylates COS with a DP ≥ 4, with preference for longer substrates. It starts deacetylating the penultimate residue from the non-reducing end and continues deacetylating the next residue towards the reducing end, with a pattern of acetylation [ADDAn−3] [55].
The above described CDAs are well characterized in terms of their deacetylation mode of action on COS and the structure of their deacetylated products. A number of other fungal CDAs (Table 1, and below) have also been assayed on COS substrates but, to the best of our knowledge, the deacetylation pattern of the products has not been reported.
Saccharomyces cerevisiae (ScCDA1 and 2). The S. cerevisiae ascospore walls are well ordered structures with two outer layers that confer spore resistance, one made of 95% chitosan and the outermost proteinaceous layer rich in dityrosine [44]. Two CDAs are expressed exclusively during sporulation and are required for spore wall rigidity [38]. Both CDAs have been cloned and expressed in yeast as glycosylated proteins active on glycol-chitin [38,56], and in E. coli [57] as soluble proteins with deacetylase activity on glycol-chitin, chitosan (DA 50%) and COS. More detailed characterization of ScCDA2 expressed in E. coli revealed that at least two GlcNAc residues are required for activity on COS, with maximum activity on DP6 [57]. When glycol chitin is used as substrate the optimum temperature for enzyme activity is 50 °C and the pH optimum is 8.0. It has also been shown that the ScCDAs may act on nascent chitin chains in an in vitro assay system with chitin synthase [56].
Mortierella sp. (MoCDA). Some Mortierella species live as saprotrophs in soil and other organic materials such as decaying plant leafs, fecal pellets or on the exoskeleton of arthropods, whereas other species are endophytes [111]. An extracellular CDA was identified [58] and purified from a Mortierella sp. as a highly glycosylated protein with maximum activity at pH 5.5–6 and 60 °C [112]. MoCDA is active on soluble substrates as chitosans and glycol-chitin but with no detectable activity on β-chitin, colloidal chitin, and CM-chitin. It is active on COS with a DP ≥ 2, with higher activity with increasing DP of the substrate. With diacetylchitobiose, only monodeacetylation was observed. The structure of the deacetylated products from larger oligomers has not been reported.
Absidia sp. (AcoeCDA, AcoryCDA). Absidia strains of Zygomycetes produce chitosan in their cell wall through the tandem action of chitin synthases and deacetylases. In A. coerulea, chitosan accounts for 10% of the vegetative cells and the DA reaches 95%. AcoeCDA was purified and proven to be active on glycol-chitin with a pH optimum of 5 at 50 °C. When the purified enzyme was incubated with a chitin synthase, it converted 90% of the nascent chitin from UDP-GlcNAc into chitosan. It deacetylates COS with more than two GlcNAc units, with increasing activity with longer substrates [43]. Similarly, Absidia corymbifera secretes a CDA active on glycol-chitin and chitosans with optimum pH and temperature of 6.5 and 55 °C, respectively, and active on COS with DP ≥ 2 [59].
Flammulina velutipes (FvCDA). The basidiomycete Flammulina velutipes (called Enokitake in Japan) is commercially cultivated and fruited to produce foods with high nutritional value. A CDA that is expressed at the early stages of fruity body development was recombinantly expressed in Pichia pastoris [60]. FvCDA, active on glycol-chitin and colloidal chitin, deacetylates COS from dimer to pentamer, with activity increasing with the DP of the substrate. The enzyme exhibits the maximum activity at 60 °C and pH 7.
Penicilium oxilicum (PoCDA). An extracellular CDA from Penicilium oxilicum, purified from culture supernatants, exhibits deacetylase activity on glycol-chitin at pH 9, a common value for extracellularly secreted CDAs as opposed to intracellular CDAs with typical pH optima in the range of 5 to 7. PoCDA is active on COS with activity increasing from DP2 to DP5 [61].
Aspergillus flavus (AfCDA). In the search for extracellularly secreted CDAs for industrial applications, optimization of solid substrate fermentation and submerged fermentation of Aspergillus flavus has been reported [62,113]. The AfCDA enzyme purified from the extracellular medium has optimal activity on glycol-chitin and colloidal chitin at pH 8 and 50 °C. When assayed with COS as substrates, AfCDA is active on DP4 but has no activity on shorter substrates [62].
Scopulariopsis brevicaulis (SbCDA). Scopulariopsis spp. are common soil saprophytes. Few species have been associated with human diseases, including S. brevicaulis. They are dermatomycotic molds and mainly have been associated with onychomycosis [114,115]. SbCDA is an extracellular enzyme that is active on chitin and chitosans. The purified native enzyme is also active on COS with at least two GlcNAc units, and the activity increases with the DP of the substrate. With DP6, optimum conditions for deacetylation are pH 7.5 and 55 °C [63].
Rhizopus sp. (RcCDA, RsCDA). Rhizopus species have been screened as CDA producers. A R. circicans CDA has been cloned and recombinantly expressed in Pichia pastoris [64]. RcCDA has maximum activity on glycol-chitin at pH 5–6 and 37 °C. On COS, only activity on a DP6 substrate has been reported. A CDA from Rhizopus stolonifer (or nigricans) has also been isolated as an active enzyme on glycol-chitin but no activity on COS has been reported [64,116]. Fermentation conditions of other Rhizopus species as CDA producers are being studied for the bioconversion of chitin to chitosan [117].
Other chitosan producers have been identified and studied as a source of chitosans, with many reports on screening and fermentation optimization, but the corresponding chitin deacetylases have not been characterized yet. Some examples include Gongronella butleri [65,118], Phycomyces blakesleeanus [66,119], and Schizophyllum commune [67].
Cryptococcus neoformans (CnCDA). Cryptococcus neoformans is a dimorphic basidiomycetous human fungal pathogen that causes cryptococcal meningoencephalitis, particularly in immunocompromised patients [37]. C. neoformans has substantial chitosan in its cell wall during vegetative growth that is necessary for virulence and persistence in the mammalian host [120,121]. Three CDAs are predicted to be GPI-anchored to the cell wall, suggesting that they transverse the plasma membrane or attach to the cell wall to deacetylate the chitin generated by a chitin synthase as it is extruded through the plasma membrane [37]. The GPI-anchor of CnCDA2 has proven to be required for membrane association but dispensable for cell wall association [122]. Activity of C. neoformans CDAs on COS substrates has not been reported. Interestingly, screening studies to identify cryptococcal antigens that stimulate an immune response on murine T cell hybridomas reactive with cryptococcal proteins, have shown that two of the CDAs are immunogenic [123,124].
3.3. Protozoan CDAs
Entamoeba histolytica (EhCDA). Entamoeba histolytica is an anaerobic parasitic amoebozoan that predominantly infects humans and other primates causing amoebiasis [125]. The genome contains two putative CDAs, one of which has been cloned and recombinantly expressed in Saccharomyces cerevisiae [68]. EhCDA deacetylates COS, being active on DP5 and DP6, but with no detected activity on DP4 [68].
3.4. Bacterial CDAs
The predominant CE4 deacetylases in bacteria are chitin oligosaccharide deacetylases (CODs), active on low molecular mass COS and essentially inactive on polymeric chitin and chitosans. These include rhizobial NodB deacetylases and CODs from marine bacteria. But bacterial CDAs other than CODs are being discovered from screening programs and data mining of sequenced genomes and metagenomes, as in the recent case of an Arthobacter CDA.
Sinorhizobium meliloti (NodB). Rhizobial NodB is part of the Nod operon involved in the biosynthesis of Nod factors, the morphogenic signal molecules produced by rhizobia, which initiate the development of root nodules in leguminous plants [126]. NodB is active on chitooligosaccharides from DP2 to DP5 with no differences in kcat, but KM decreases with increasing DP [18,127,128,129]. Specifically, kcat/KM is 5-fold higher for DP5 than for DP2 substrates. DP4 or DP5 substrates are the natural substrates depending on the Rhizobial strain. SmNodB optimum activity between pH 7 and 8 at 30 °C [18]. NodB is highly specific deacetylating only the non-reducing end residue [DAn−1] although traces of a second deacetylation event have been observed upon long incubations [18,130,131].
Vibrio species (VcCDA, VpCDA, VaCDA). Chitin oligosaccharide deacetylases (COD) from the Vibrionaceae family are involved in the chitin degradation cascades occurring in sea water [132,133,134,135]. They have been identified in many Vibrio species, such as V. algynolyticus [73,136], V. parahaemolyticus [72,137], V. cholera [70], V. harveyi [138] and others. The V. parahaemolyticus and Vibrio sp. SN184 CDAs only deacetylate DP2 and DP3 substrates, whereas the Vibrio cholera chitin deacetylase (VcCDA) has a broader specificity, accepting substrate from DP2 to DP6 [69,70]. VcCDA has a 10-fold higher activity on DP2 than on DP4 [69], and specifically deacetylates the penultimate residue from the non-reducing end, generating monodeacetylated products with the pattern [ADAn−2] [69,70,130].
Shewanella species (SwCOD, SbCOD). In addition to the Vibrio genus, CODs have been recently identified and characterized from the Shewanella genus, marine bacteria found in extreme aquatic habitats (low temperature and high pressure). Shewanella sp. CODs share high sequence identity (50–60%) with Vibrio CODs, and have essentially the same biochemical properties. The S. woodyi enzyme (SwCOD) contains two CBM12 chitin binding domains at the C-terminus, deacetylates the reducing end on diacetylchitobiose [AD], and the activity drastically decreases from DP2 to DP4 substrates, with no activity detected on a DP5 substrate [74]. The S. baltica enzyme (SbCOD) contains a single CBM12 at the C-terminus, it is active of diacetylchitobiose with the same deacetylation pattern [AD] but it is less active on a DP3 than on a DP4 substrate [75].
Arthrobacter sp. (ArCE4). A bioinformatics search for monodomain and extracellular CDAs in annotated genomes and metagenomes identified ArCE4 as a CDA from an Arthrobacter species [76], a Gram-positive bacteria known to grow on chitin and secrete chitinases [139,140,141]. ArCE4 is active on chitosan (DA 64%), acetylxylan, and insoluble chitin. It also deacetylates COS substrates with DP ≥ 2. The activity increases with increasing DP, with higher activity against DP5 compared to DP6. As shown with the DP5 substrate, the enzyme follows a multiple-chain mechanism where different mono- and di-deacetylated products are obtained. Whereas the first deacetylation occurs at all three internal positions, di-deacetylation mainly takes place at the GlcNAc unit next to the reducing end and at either of the two other internal units (ADDAA and ADADA). The final products have a pattern of acetylation [Dn−1A], where the reducing end unit is not deacetylated [76].
4. Structural Determinants of Activity and Specificity
Structural analysis of CE4 enzymes with solved 3D structure have been recently reviewed [142], comparing and highlighting the differences between the different subfamilies based on substrate preferences. Here we focus and summarize the current knowledge on the structure and specificity of CDAs as a subfamily of CE4 enzymes. The closer similarity and activity on the same substrates provides a framework to analyze the structural determinants responsible for the different modes of action that lead to different patterns of deacetylation in their products.
4.1. 3D Structures
Some CDAs are mono-domain proteins and some others have a multi-domain architecture composed of the CE4 catalytic domain (or NodB homology domain), and several other domains, such as carbohydrate binding modules (CBMs [143]) and domains with unknown function. The function of the CBMs is not clear and might be diverse depending on the biological role of each enzyme in its organism. In extracellular CDAs acting on the cell wall chitin, they may facilitate solubilization and access to the substrate (i.e., PaCDA with two CBMs, where deletion of one or both confirmed their proposed function in supporting the enzymatic conversion of insoluble chitin [26]). In CDAs acting on low molecular weight COS, the CBMs may be involved in enzyme localization. This is the case of COD enzymes from marine bacteria (Vibrio and Shewanella species), where the small substrate does not span out of the active site, and the CBMs might bind to chitinous material in order to keep the COD activity close to the site where COS are generated by the action of chitinases.
The first CE4 enzymes with 3D structure determined by X-ray crystallography were the peptidoglycan deacetylases BsPdaA [144] and SpPgdA [145], and the first CDA was that from Colletotrichum lindemuthianum (ClCDA) [53]. Currently, only five CDAs in the CE4 family have known 3D structure (Figure 2). The CE4 catalytic domain is characterized by a distorted (β/α)8 barrel fold. The distorted barrel, which often lacks one of the αβ repeats of regular TIM barrels, creates a groove into which the extended polymer substrate binds [144,146,147]. Seven or eight parallel β-strands form the β-barrel surrounded by α-helices. In addition, a series of loops decorate the β-barrel and make up the majority of the carbohydrate binding pocket as discussed below.
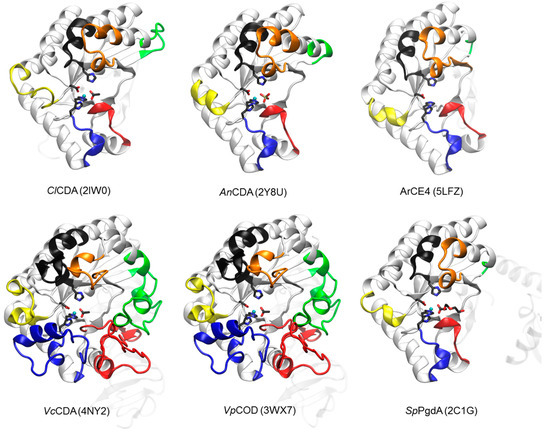
Figure 2.
3D structures of CDAs determined by X-ray crystallography. Loops 1 to 6 colored as in Figure 3. The peptidoglycan deacetylase SpPgdA is also included for comparison (see text). In parenthesis, PDB accession codes.
4.2. The NodB Homology Domain and Conserved Active Site Motifs
The multiple sequence alignment of the CE4 domain for the CDAs listed in Table 1 was guided by the structural superimposition of the available X-ray structures (Figure 2) and is presented in Figure 3. Compared to most of the CDA members, the VcCDA enzyme has substantially longer insertions, and it was key to defining the loops that differentiate CDAs and shape the binding site cleft of these enzymes. Sequences of enzymes without structural date were incorporated into the alignment by means of Hidden Markov Model comparisons. As seen in Figure 3, the conserved motifs and non-conserved insertions are evenly distributed along the sequences of CDAs.
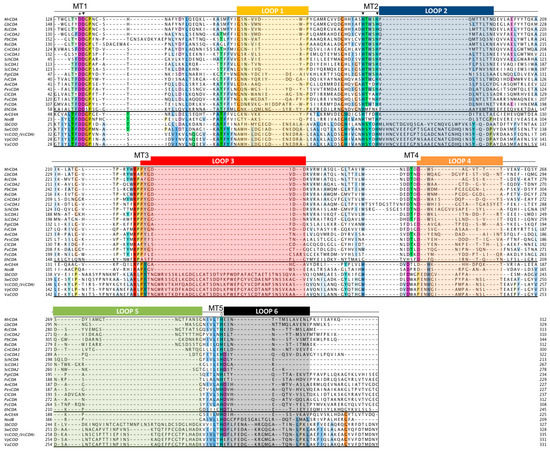
Figure 3.
Multiple sequence alignment of the CDA enzymes listed in Table 1. Loops are highlighted with colored boxes according to [69]. Conserved catalytic motifs are labelled MT1-5. The ‘His-His-Asp’ metal binding triad (▼), catalytic base (*), and catalytic acid (◊) are labelled.
The conserved motifs are related to enzymatic activity (Motifs 1 to 5) and are typically located at the center of the active site structure. The non-conserved insertions correspond to both un-structured and structured loops of variable length, sequence, and geometry that surround the active site. These loops are numbered from Loop 1 to Loop 6 in the sequence alignment (Figure 3). As discussed in Section 4.5, they are key elements in determining the substrate specificity of different CDAs.
As members of the CE4 family, CDAs share the ≈150 aa-long NodB homology domain (CE4 domain). This region is defined by five conserved motifs that, according to the order they appear in the sequence, are named Motif 1 to Motif 5. These consensus motifs were first proposed in 2005 by sequence alignment of representative enzyme members of the CE4 family when the 3D structure of the peptidoglycan deacetylase SpPgdA was solved [144]. Motif 1 (TFDD) is highly conserved in CDAs and contains the general base aspartate (first D) and the metal-binding aspartate (second D). Motif 2 (H(S/T)xxH) is a zinc-binding motif, where the two His residues bind the metal cation and the Ser or Thr residue forms a hydrogen bond with the second His, stabilizing the local conformation of the loop-shaped motif. These two His from Motif 2 plus the metal-binding Asp from Motif 1 are often designated the His-His-Asp metal-binding triad of CE4 enzymes. Motif 3 (RxPY) forms one of the sides of the active site groove and establishes stabilizing interactions with other active site residues. Motif 4 (DxxD(W/Y)) forms the other side of the active site groove, including a hydrophobic residue exposed to the solvent and a buried Asp. Motif 5 (I(V/I)LxHD) contains the catalytic general acid His residues and a Leu, which is part of a hydrophobic pocket that accommodates the acetate methyl group of the substrate.
4.3. Phylogeny of CE4 Chitin Deacetylases
Based on the above multiple sequence alignment, a clustering of the CE4 domain sequences of characterized CDAs based on phylogenetic analysis is presented in Figure 4. This is a reduced phylogenic analysis limited to the CDAs with reported activity on COS as listed in Table 1. Fungal and bacterial CDAs are clearly segregated in two clades, with a protist CDA (EhCDA) located between both groups.
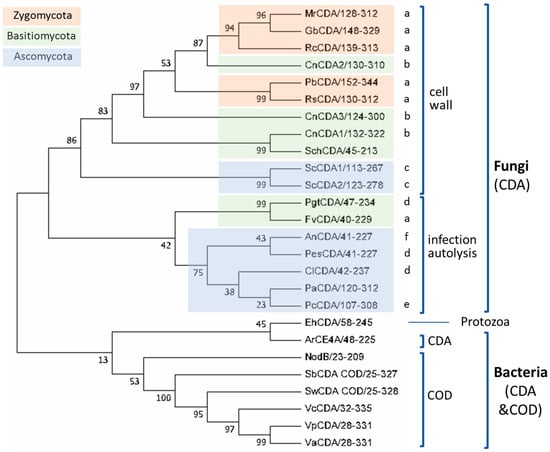
Figure 4.
Phylogenetic analysis of CDAs from the multiple sequence alignment presented in Figure 3. A bootstrap analysis with 500 replicates was carried out on the trees inferred from the neighbor joining method. The consensus tree is shown with bootstrap values at each node of the tree. Biological functions: cell wall biosynthesis: (a) cell wall, (b) vegetative growth, (c) sporulation; host infection, (d) defense, (e) interaction/infection, (f) autolysis (see text).
Fungal enzymes from organisms belonging to different phyla (Zygomycota, Basidiomycota, and Ascomycota) are distributed throughout the fungal clade. Within the clade, CDAs appear grouped in two clusters related with their biological function. The first cluster contains orthologous CDAs of different phyla known to have a role in cell wall chitosan biosynthesis at different stages of the fungal cell cycle, such as MrCDA during cell wall formation [42], CnCDAs during vegetative growth [37], or ScCDAs during sporulation [38,44]. Although there is no experimental proof of their biological function, the CDAs from Gonglonella, Phycomyces, as well as those from Rhizopus are likely to be also involved in cell wall formation due to their location in the same cluster of the phylogenetic tree and their taxonomic classification (mucorales inside the Zygomycota phylum). Regarding the cellular location of the enzymes in this cluster, most of them are secreted to the periplasm or are GPI-anchored to the cell wall, where they are coupled with chitin synthases for chitosan biosynthesis. The second cluster is mainly composed of extracellular CDAs that participate in host infection, either as a defense mechanism to prevent the elicitation of host defense mechanisms (PgtCDA, PesCDA), or involved in the interaction with the host or as a virulent factor (ClCDA, PcCDA). Extracellular CDAs involved in cell autolysis (AnCDA) also fall in this group.
Most of the bacterial enzymes included in the alignment are chitin oligosaccharide deacetylases (COD) and are more distantly related to the fungal CDAs. These form a different clade in the phylogenetic tree (Figure 4). The enzymes from marine bacteria (Vibrio and Shewanella species) are clustered together with high sequence similarity and have similar biological functions and biochemical properties. NodB has a more distant relationship with the other CODs.
4.4. Catalytic Mechanism
CDA enzymes operate by metal-assisted acid/base catalysis. The general mechanism was first proposed for the peptidoglycan GlcNAC deacetylase SpPgdA when solving its X-ray structure [144] and short after supported by the 3D structures of the acetylxylan esterases SlAxeA and CtAxeA [148]. The catalytic machinery involves the conserved active site motifs containing the metal-binding triad and the general acid and base residues. Only the structures of four different CDA have been solved up to date (Colletotrichum, Aspergillus, Vibrio, and Arthrobacter CDAs, Table 1), all consistent with the proposed metal-assisted mechanism. VcCDA was the first CE4 enzyme for which the 3D structure of enzyme-substrate complexes were solved by X-ray crystallography [69]. The structure of complexes of an inactive mutant (at the general base Asp residue) with diacetylchitobiose (DP2) and triacetylchitotriose (DP3) in productive binding for catalysis showed that a sugar hydroxyl group of the substrate also participates in metal coordination. Specifically (Figure 5), the Zn2+ cation is coordinated by the imidazole nitrogens of His97 and His101, the carboxylate group of Asp40, and the O7 atom of the N-acetyl group and O3 hydroxyl of the GlcNAc ring. The distorted octahedral coordination is completed by a water molecule. Upon activation, this water molecule is proposed to be the nucleophile responsible for removal of the N-acetyl group. Just recently, a second structure of an enzyme-substrate complex has been reported for the Arthrobacter sp. CDA (ArCE4) [76]. The diacetylchitobiose ligand bound into the active site also shows the same type of interactions with the conserved active site residues.
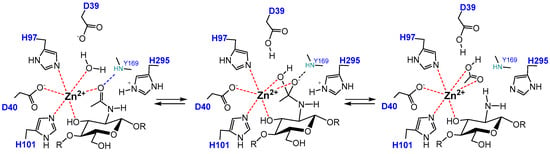
Figure 5.
Metal-assisted general acid/base mechanism proposed for CE4 deacetylases. Scheme based on the 3D structure of the enzyme·substrate complex VcCDAD39S·DP2 [61]. D39 is the general base and His295 is the general acid.
The proposed mechanism of CDAs and related CE4 enzymes is shown in Figure 5. In the first step, metal coordination polarizes the carbonyl amide of the substrate which reacts with the nucleophilic water molecule activated by the general base (Asp), leading to a tetrahedral oxyanion intermediate. Next, protonation of the nitrogen group of the intermediate by the general acid (His) facilitates C-N bond breaking with release of acetate and the generation of a free amine in product. Kinetic evidence for an oxyanion tetrahedral intermediate and significant charge development at the first transition state was provided by Hammett linear free energy correlations using the ClCDA enzyme with α-haloacetamido substrate analogues [53]. In most of the enzymes, the catalytic acid and base residues are part of two conserved “charge relay” side chain pairs that may contribute to modulate the pKa of the catalytic residues [53,144]: the catalytic base (Asp) is tethered by a conserved Arg from MT3 (RxxPY) and the catalytic acid (His) is tethered by a conserved Asp from MT4, DxxD(W/Y).
4.5. Determinants of Substrate Specificity
The series of crystal structures of the Vibrio cholerae chitin oligosaccharide deacetylase (VcCDA or VcCOD) reported in 2014 [69] were the first 3D structures of a CE4 enzyme in complex with substrates. These data provided a first insight into structure-function relationships for this family of enzymes and highlighted the role of the loops that shape the binding site cleft in substrate binding. Recently by the end of 2017, the 3D structure of an Arthrobacter sp. CDA (ArCE4) in complex with substrate has also been reported [76]. Albeit sharing the same molecular function, both enzymes represent two different scenarios regarding the binding site topology and, hence, substrate specificity. VcCDA has a rather closed binding cleft and is highly specific for monodeacetylation of COS, whereas ArCE4 has a more open binding cleft and is able to fully deacetylate their COS substrates (Figure 6). A comparative structural analysis of both enzyme structures has been recently reviewed [142].
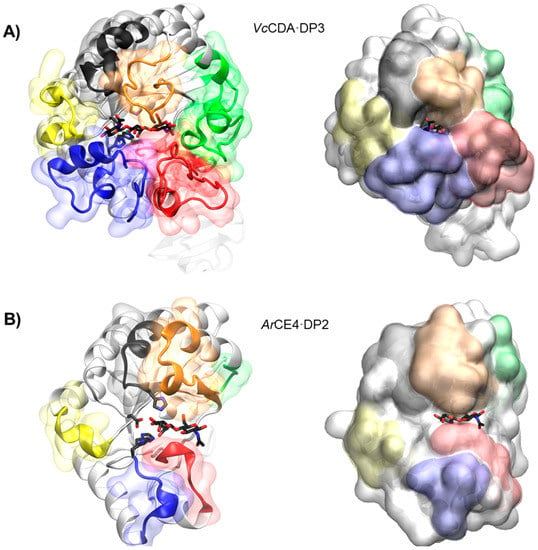
Figure 6.
3D structures of enzyme·substrate complex. (A) VcCDA with DP3 substrate and (B) ArCE with DP2 substrate. Loops 1 to 6 are colored as in Figure 3.
4.5.1. VcCDA. Long Loops and High Specificity
Currently available structures of VcCDA include the unliganded form of the enzyme and the binary complexes with N-acetylglucosamine (DP1), diacetylchitobiose (DP2), and triacetylchitotriose (DP3). These structures revealed two significant observations: a series of non-conserved loops (labeled Loop 1 to 6 in Figure 3 and Figure 6) that shape the binding cleft, and the dynamics of the loops that assemble the active site for catalysis [69].
In all structures, the substrate is confined in a small binding cleft that is shaped by a series of long loops surrounding the active site (see Figure 6A for the VcCDA·DP3 complex). Given the topology of VcCDA protein surface, the binding of longer COS is prevented because these loops cap both the reducing and non-reducing ends of the substrate. Indeed, the catalytic efficiency of VcCDA drops substantially on oligomers longer than DP2 [69] and attempts to solve the structure of VcCDA bound to substrates longer than DP3 in a catalytically competent mode have been unsuccessful. Another consequence of this constricting topology is the high specificity of VcCDA to exclusively deacetylate the penultimate residue from the non-reducing end of the substrates. There is no room for the ligand to slide along the binding cleft, thus it can only accommodate one GlcNAc unit of the oligomeric chain at the catalytic site where deacetylation takes place. The binding of substrates induces a conformational change of Loop 4 from an open conformation in the unliganded enzyme to a closed conformation in the enzyme·DP2 complex or a semi-closed conformation in the enzyme·DP3 complex. It is triggered by a staking interaction between a Trp residue located in apical site of Loop 4 and the GlcNAc unit at the catalytic center, locking the substrate in the active site in the proper orientation for catalysis.
4.5.2. ArCE4. Short Loops and Broad Specificity
In contrast to VcCDA, the crystal structure of ArCE4 in complex with diacetylchitobiose [76] reveals a flatter protein surface with the substrate bound to a more open binding cleft (Figure 6B). Even though the enzyme was co-crystalized with tetraacetylchitotetraose (DP4), only two GlcNAc units are observed in the structure. This indicates a weak binding of part of the COS substrate on this flat topology of the protein surface. The catalytic center in both enzymes is in the same position with respect to the protein core, being the main difference, the size and shape of the loops surrounding the active site. Since the binding cleft is more open, the enzyme can accommodate longer COS. Indeed, the enzymatic activity of ArCE4 increases as the length of the chitin oligomer chain increases. The lack of protein caps at either the reducing and non-reducing ends of the substrate can also explain the multiple-chain mechanism proposed for this enzyme. Deacetylation takes place at all GlcNAc units of the substrate (except the reducing end) because it can freely bind to ArCE4 in different binding modes exposing different GlcNAc units of the oligomeric chain at the catalytic site.
4.5.3. The Subsite Capping Model
The diversity of deacetylation patterns exhibited by chitin deacetylases and related CE4 enzymes can be attributed to the differential accessibility of the linear chitin oligosaccharide chain to the separate subsites along the substrate binding cleft of their structures. Considering all CE4 enzymes with reported activity on polymeric chitin or COS, these can be classified into two groups. One group is represented by general chitin deacetylases (CDA), and a second group is formed by chitin oligosaccharide deacetylases (COD). The two structures of the enzymes-substrate complexes described above are reference models for the protein surface topologies and substrate binding mechanisms of these two groups: CDAs (ArCE4) and CODs (VcCDA). These two structures provide a unified view of the determinants of substrate specificity in chitin deacetylases in terms of the “subsite capping model” proposed in [69]. According to this model, substrate accessibility is affected by the length, shape, and dynamics of a series of loops surrounding the active site of CE4 enzymes. These loops are numbered from 1 to 6 and their location in the sequences and structures of CDAs and CODs is highlighted in Figure 2 and Figure 3.
The group of CDAs bears short loops, and their structures exhibit a flat and open binding cleft. The substrate binding mechanism in this group of enzymes may be similar to that described for the reference structure of ArCE4. According to the model, the substrate may be able to slide along the binding cleft or to bind in different modes resulting in processive or multiple-chain attack mechanisms of deacetylation. This can already be anticipated for CDA enzymes of known structure (Figure 2) because the flat protein surface is already evident, but also for CDA enzymes of unknown structure given the similar sequence lengths of the loops evidenced in the alignment (Figure 3). This could be an explanation of why CDA enzymes in general are not specific for the deacetylation at a single N-acetylglucosamine unit. However, the patterns of deacetylation differ among the different CDAs. The surface charge distribution along the binding cleft and other structural features yet to be disclosed may also participate in defining the mode of action and deacetylation pattern by each particular enzyme.
On the contrary, the group of COD enzymes bears longer loops and their structures have narrower binding pockets and buried active sites. According to the subsite capping model, the substrate is constrained to bind in very specific binding modes resulting in single-site deacetylations. This is the case for the reference structure of VcCDA in complex with substrates, but it can also be anticipated for other COD enzymes for which the 3D structure is still unknown. For instance, Loop 6 in RmNodB is longer than in other CDAs. This loop is located on the non-reducing end site of the binding cleft and may cap the accessibility of the substrate after subsite 0 (the catalytic site) thus defining the deacetylation specificity for the non-reducing end of the substrate. Likewise, the Shewanella CODs have a Loop 6 with the same length than the Vibrio CODs, but shorter than NodB, and both exhibit the same mono-deacetylation specificity for the penultimate GlcNAc residue from the reducing end of the substrate.
For most CDAs, the reducing end of the substrate is not deacetylated, or it is the least reactive GlcNAc unit. As seen in the ArCE4·DP2 complex 3D structure [64], binding to the +1 subsite seems to be dominated by the stacking interaction of the GlcNAc unit of the substrate with a Trp in Motif 4 at the beginning of Loop 4. This aromatic residue is highly conserved (MT4, DxxD(W/Y), Figure 3). CDA enzymes having this aromatic residue prefer a sugar bound in the +1 subsite; they do not deacetylate the reducing end of their substrates, as it is the case for ArCE4, PesCDA [89] and PcCDA [95], or the reducing end is the slowest position to be deacetylated, as shown for ClCDA [49] and AnCDA [12]. On the contrary, PgtCDA, which deacetylates the reducing end GlcNAc unit of all substrates from DP4 to DP6, lacks the +1 aromatic residue [89]. Different is the case of Vibrio and Sewanella CODs that have the equivalent aromatic residue in a slightly different position after a two-amino acid insertion in the MT4 motif, and it is located farther in Loop 4 (Figure 3). In the VcCDA enzyme this loop moves from an open to a closed conformation upon substrate binding, and the same is expected for the other closely related CODs that have the same Loop 4 size. As a consequence of the induced fit, the Trp residue now establishes a stacking interaction with the GlcNAc unit in subsite 0. DP2 is the preferred substrate for this group of COD enzymes, and it is deacetylated at the reducing end [62].
Gaining further structural information of protein-ligand complexes of CDA and COD enzymes, and other CE4 in general, will contribute further to decipher the structural and sequential determinants of substrate specificity in this family of enzymes. This will pave the way to the rational design or discovery of novel CDA with controlled specificities on the deacetylation of oligomeric and polymeric chitin for the biotechnological production of chitosans and paCOS with defined patterns of acetylation.
5. Application of Chitin Deacetylases
5.1. Targets for Antifungals
Fungal infections have an enormous impact on human health. Fungi are generally opportunistic pathogens affecting immunocompromised individuals including those with AIDS, receiving immunosuppressive drugs or undergoing cancer treatments. The cell wall is a meaningful target for antifungal therapies. Current major classes of antifungal drugs target cell membrane ergosterol biosynthesis (azoles), ergosterol function by disrupting membrane integrity (polyenes), or 1,3-β-glucan synthase preventing the formation of the cell-wall structural polysaccharide 1,3-β-glucan (echinocandins) [149]. New targets to overcome the emerging drug resistance by pathogenic fungi are becoming critical to treat life-threatening fungal infections. Other promising targets are the so called cell wall proteins (CWP) which mediate important cellular processes, including adhesion, invasion, biofilm formation and flocculation [122]. In fungal chitosan producers, chitin deacetylases are a class of CWP and potential targets for drug design. Cryptococcus neoformans, one of the most deadly pathogens, requires chitosan for virulence. Lack of chitosan in the cell wall has detrimental consequences in fungal growth and results in the complete loss of sporulation [120,121]. Thus, CDAs represent a promising target for anticryptococcal therapeutics [37,120], but no CDA inhibitors have been reported yet.
In pathogenic plants, major strategies to prevent fungal pathogenesis are related to the inhibition of fungal chitinases, which are required for chitin remodeling in the cell wall [150,151,152]. Different types of chitinase inhibitors have been reported, including potent natural inhibitors such as allosamidin [153] and the cyclic pentapeptides argifin and argadin [154]. However, inhibition must be selective so as not to interfere with the plant chitinases involved in triggering the plant defense mechanisms. Another potential and promising strategy is the inhibition of extracellularly secreted fungal CDAs since they constitute a defense mechanism to evade the plant immune system, as discussed in Section 3.2. As in the case of human fungal pathogens, no inhibitors have been yet reported against CDAs from plant pathogenic fungi.
5.2. Biocatalysts for the Enzymatic Production of Chitosans and paCOS
Chitosans can be found in a large number of applications in such distant areas as agriculture, cosmetics, water treatment, medicine and the food industry [155,156,157,158,159]. In addition to chitosan polymers, their oligomers (paCOS) have also proven to have relevant potential applications in agriculture and pharmaceutical industries [160]. The physicochemical and biological properties of chitosans and paCOS have been shown to be strongly dependent on their degree of polymerization and their degree of acetylation [161,162]. Many of the identified CDAs arose from screening programs addressed to find efficient biocatalysts to overcome the current industrial chitosan production by highly concentrated alkali treatment of chitin. Some examples include CDAs from Mortiriella sp. [58], Rhizopus sp. [117], or Gongronella sp. [118].
Chemical methods for the production of COS and paCOS are based on chemical depolymerization of chitosan [163,164], total synthesis of chitosan oligomers [165,166], partial chemical deacetylation of fully acetylated COS, or chemical re-N-acetylation of glucosamine oligomers based in two-step procedures or one-pot synthesis [167,168,169]. The drawbacks of chemical strategies are the unwanted side reactions and the randomness of the chemical reactions. Current efforts are addressed to develop enzymatic routes for COS and paCOS production with defined DP, DA, and PA.
Enzymatic approaches include depolymerization of chitin or chitosan polymers using hydrolytic enzymes, chitinases and chitosanases, enzymatic polymerization by transglycosidation using transglycosylating hydrolases, and enzymatic de-acetylation and re-acetylation of chitin oligomers using chitin deacetylases, strategies recently reviewed in [170]. Based on the current knowledge on the specificity of a number of fungal and bacterial CDAs, recent reports have combined enzymes with different specificities to have access to a large family of paCOS with defined structures. The first proof of concept was to show that two specific CODs, NodB and VcCDA, each accept the monodeacetylated product from the other, leading to specific di-deacetylation, and that both enzymes can work in a one-pot process [130]. Recently, the use of different recombinant CDAs from bacterial and fungal origin to produce all of fourteen possible partially acetylated chitosan tetramers combining different enzymatic deacetylations and enzymatic N-acetylations has been reported [171] (Figure 7).
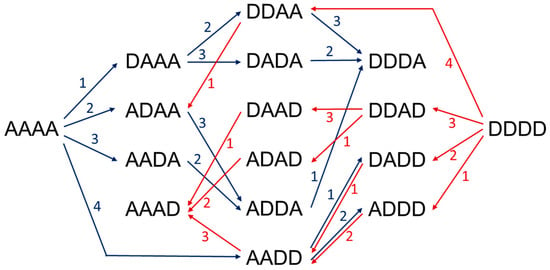
Figure 7.
Production routes of all possible chitin and chitosan tetramers using 4 different CDAs to specifically deacetylate or N-acetylate paCOS. A: GlcNAc, D: GlcNH2. Blue arrows, deacetylation reactions, red arrows, N-acetylation reactions in the presence of excess acetate.
The production of paCOS using in vivo strategies is an alternative to increase the scalability of the process. The first example towards a more general cell factory approach for the in vivo synthesis of paCOS was based on NodB deacetylase. By in vivo studies with Escherichia coli expressing different combinations of the nodABCS genes of Azorhizobium caulinodans, Nod factor intermediates were identified, as well as the sequence of the biosynthetic steps [172]. The nod gene cluster encodes a series of enzymes, which include the NodC chitin oligosaccharide synthase that produces fully acetylated chitin oligomers, the NodB chitin oligosaccharide deacetylase that deacetylates the non-reducing end unit, the NodA N-acyl transferase that transfer a fatty acid chain to the free amine group, and the NodS N-methyl transferase. Further transformations by other nod proteins elaborate the final Nod signaling factors [173]. In a first cell factory approach, high density cells of E. coli expressing nodC or nodBC genes produced in high yield (up to 2.5 g/L) penta-N-acetyl-chitopentaose and its deacetylated derivative tetra-N-acetyl-chitopentaose, which were easily purified by charcoal adsorption and ion-exchange chromatography [174]. The strategy was further extended to the production of sulfated and O-acetylated derivatives of these two compounds by coexpressing nodC or nodBC with nodH and/or nodL that encode chitooligosaccharide sulfotransferase and chitooligosaccharide O-acetyltransferase, respectively [175]. Other Nod analogues have also been generated with further modifications [176,177,178]. The cell factory approach, currently limited to one deacelylation pattern based on the use of NodB, is a promising technology to be developed by incorporating the diversity of CDAs with different deacetylation patterns in order to access a large family of paCOS and derivatives.
6. Conclusions
We have here summarized the current knowledge on substrate specificity of fungal and bacterial chitin deacetylases, their modes of action, and their use as biocatalysts for the production of chitosans and chitosan oligosaccharides with defined pattern of acetylation. By combining multiple sequence alignments and 3D structures of enzyme·substrate complexes of representative enzymes, a unified view of the determinants of substrate specificity is proposed in terms of the “subsite capping model.” According to this model, substrate accessibility is affected by the length, shape, and dynamics of a series of loops surrounding the active site of CE4 enzymes. The group of CDAs active on polymeric substrates and COS bear short loops, and their structures exhibit a flat and open binding cleft. The substrate may be able to slide along the binding cleft or to bind in different modes, resulting in processive or multiple-chain attack mechanisms of deacetylation. Other structural features not yet disclosed, such as the charge distribution along the binding cleft may also participate in defining the mode of action and deacetylation pattern by each particular enzyme. The group of COD enzymes active on low molecular mass COS bear longer loops and their structures have narrower binding pockets and buried active sites. The substrate is constrained to bind in very specific binding modes resulting in single-site deacetylations.
But a deeper knowledge on substrate specificity requires further structural information of protein-ligand complexes of CDA and COD enzymes in order to decipher the structural and sequential determinants of substrate specificity in this family of enzymes aimed at the rational design or discovery of novel CDAs with controlled specificities on the deacetylation of oligomeric and polymeric chitin for biotechnological applications.
Although CDAs have been proposed as targets for antifungal drugs, no specific inhibitors have been yet reported. This is an open field that deserves attention not only for drug design but also to probe the signaling function of CDAs and CODs through their specific deacetylation of COS substrates.
Applications of CDAs and CODs as biocatalyst are currently being developed as a novel methodology to produce partially acetylated COS with tailored patterns of acetylation. Since not all patterns for COS of different sizes are yet available, enzyme discovery and protein engineering offer new opportunities for the biotechnological production of chitosans and paCOS with defined patterns of acetylation.
Acknowledgments
Work supported by the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement n°613931, and grant BFU2016-77427-C2-1-R from MINECO, Spain. Laia Grifoll and Sergi Pascual acknowledge a predoctoral contract from the NANO3BIO project. Hugo Aragunde acknowledges a predoctoral fellowship from Generalitat de Catalunya.
Author Contributions
Antoni Planas conceived and designed the review; Laia Grifoll, Sergi Pascual, Hugo Aragunde, Xevi Biarnés, and Antoni Planas compiled the references and prepared the figures. Xevi Biarnés and Antoni Planas wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AXE | Acetylxylan esterase |
| An | (GlcNAc)n |
| CDA | Chitin deacetylase |
| COS | Chitooligosaccharides |
| Dn | (GlcNH2)n |
| DA | Degree of acetylation |
| DP | Degree of polymerization |
| GlcNAc | N-acetylglucosamine |
| PA | Pattern of acetylation |
| paCOS | Partially acetylated chiton oligosaccharides |
| PDB | Protein data bank |
References
- Peniche Covas, C.A.; Argüelles-Monal, W.; Goycoolea, F.M. Chitin and chitosan: Major sources, properties and applications. In Monomers, Polymers and Composites from Renewable Resources; Belgacem, M.N., Gandini, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; Volume 1, pp. 517–542. ISBN 9780080453163. [Google Scholar]
- Karrer, P.; Hofmann, A. Über den enzymatischen Abbau von Chitin und Chitosan I. Helv. Chim. Acta 1929, 12, 616–637. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Dutta, P.K.; Duta, J.; Tripathi, V.S. Chitin and Chitosan: Chemistry, properties and applications. J. Sci. Ind. Res. (India) 2004, 63, 20–31. [Google Scholar] [CrossRef]
- Noishiki, Y.; Takami, H.; Nishiyama, Y.; Wada, M.; Okada, S.; Kuga, S. Alkali-induced conversion of β-chitin to α-chitin. Biomacromolecules 2003, 4, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Kong, B.G.; Jeong, Y.I.; Lee, C.H.; Nah, J.W. Physicochemical characterization of α-chitin, β-chitin, and γ-chitin separated from natural resources. J. Polym. Sci. Part A Polym. Chem. 2004, 42, 3423–3432. [Google Scholar] [CrossRef]
- Kumirska, J.; Czerwicka, M.; Kaczyński, Z.; Bychowska, A.; Brzozowski, K.; Thöming, J.; Stepnowski, P. Application of spectroscopic methods for structural analysis of chitin and chitosan. Mar. Drugs 2010, 8, 1567–1636. [Google Scholar] [CrossRef] [PubMed]
- Kaya, M.; Mujtaba, M.; Ehrlich, H.; Salaberria, A.M.; Baran, T.; Amemiya, C.T.; Galli, R.; Akyuz, L.; Sargin, I.; Labidi, J. On chemistry of γ-chitin. Carbohydr. Polym. 2017, 176, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, G.S.; Kaur, S.; Brar, S.K.; Verma, M. Green synthesis approach: Extraction of chitosan from fungus mycelia. Crit. Rev. Biotechnol. 2013, 33, 379–403. [Google Scholar] [CrossRef] [PubMed]
- Hoell, I.A.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structure and function of enzymes acting on chitin and chitosan. Biotechnol. Genet. Eng. Rev. 2010, 27, 331–366. [Google Scholar] [CrossRef]
- Xia, W.; Liu, P.; Zhang, J.; Chen, J. Biological activities of chitosan and chitooligosaccharides. Food Hydrocoll. 2011, 25, 170–179. [Google Scholar] [CrossRef]
- Yu, R.; Liu, W.; Li, D.; Zhao, X.; Ding, G.; Zhang, M.; Ma, E.; Zhu, K.Y.; Li, S.; Moussian, B.; et al. Helicoidal organization of chitin in the cuticle of the migratory locust requires the function of the chitin deacetylase2 enzyme (LmCDA2). J. Biol. Chem. 2016, 291, 24352–24363. [Google Scholar] [CrossRef] [PubMed]
- Winkler, A.J.; Dominguez-Nuñez, J.A.; Aranaz, I.; Poza-Carrión, C.; Ramonell, K.; Somerville, S.; Berrocal-Lobo, M. Short-chain chitin oligomers: Promoters of plant growth. Mar. Drugs 2017, 15, 40. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Min, M.; Du, N.; Gu, Y.; Hode, T.; Naylor, M.; Chen, D.; Nordquist, R.E.; Chen, W.R. Chitin, chitosan, and glycated chitosan regulate immune responses: The novel adjuvants for cancer vaccine. Clin. Dev. Immunol. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Sharon, N. Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R., Esko, J., Freeze, H., Stanley, P., Bertozzi, C.R., Hart, G., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Caufrier, F.; Martinou, A.; Dupont, C.; Bouriotis, V. Carbohydrate esterase family 4 enzymes: Substrate specificity. Carbohydr. Res. 2003, 338, 687–692. [Google Scholar] [CrossRef]
- John, M.; Rohrig, H.; Schmidt, J.; Wieneke, U.; Schell, J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 1993, 90, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Mine, S.; Niiyama, M.; Hashimoto, W.; Ikegami, T.; Koma, D.; Ohmoto, T.; Fukuda, Y.; Inoue, T.; Abe, Y.; Ueda, T.; et al. Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N-diacetylchitobiose deacetylase. FEBS J. 2014, 281, 2584–2596. [Google Scholar] [CrossRef] [PubMed]
- Fadouloglou, V.E.; Deli, A.; Glykos, N.M.; Psylinakis, E.; Bouriotis, V.; Kokkinidis, M. Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. FEBS J. 2007, 274, 3044–3054. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Mahadevan, S. The ChbG gene of the chitobiose (chb) operon of Escherichia coli encodes a chitooligosaccharide deacetylase. J. Bacteriol. 2012, 194, 4959–4971. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Ito, E. A pathway of chitosan formation in Mucor rouxii: Enzymatic deacetlation of chitin. Biochem. Biophys. Res. Commun. 1974, 56, 669–675. [Google Scholar] [CrossRef]
- Araki, Y.; Ito, E. A Pathway of Chitosan Formation in Mucor rouxii Enzymatic Deacetylation of Chitin. Eur. J. Biochem. 1975, 55, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ju, W.; Jo, G.; Jung, W.; Park, R. Perspectives of Chitin Deacetylase Research. In Biotechnology of Biopolymers; InTech: London, UK, 2011; pp. 131–145. [Google Scholar] [CrossRef]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Horn, S.J.; Vaaje-Kolstad, G.; van Aalten, D.M.F.; Eijsink, V.G.H. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef] [PubMed]
- Hoßbach, J.; Bußwinkel, F.; Kranz, A.; Wattjes, J.; Cord-Landwehr, S.; Moerschbacher, B.M. A chitin deacetylase of Podospora anserina has two functional chitin binding domains and a unique mode of action. Carbohydr. Polym. 2018, 183, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ghormade, V.; Kulkarni, S.; Doiphode, N.; Rajamohanan, P.R.; Deshpande, M.V. Chitin deacetylase: A comprehensive account on its role in nature and its biotechnological applications. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2010; pp. 1054–1066. [Google Scholar]
- Tsigos, I.; Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 2000, 18, 305–312. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, R.D.; Muzzarelli, R.A.A. Chitin deacetylases: Properties and applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Martinou, A.; Bouriotis, V.; Stokke, B.T.; Vårum, K.M. Mode of action of chitin deacetylase from Mucor rouxii on partially N-acetylated chitosans. Carbohydr. Res. 1998, 311, 71–78. [Google Scholar] [CrossRef]
- Tsigos, I.; Zydowicz, N.; Martinou, A.; Domard, A.; Bouriotis, V. Mode of action of chitin deacetylase from Mucor rouxii on N-acetylchitooligosaccharides. Eur. J. Biochem. 1999, 261, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Tokuyasu, K.; Mitsutomi, M.; Yamaguchi, I.; Hayashi, K.; Mori, Y. Recognition of chitooligosaccharides and their N-acetyl groups by putative subsites of chitin deacetylase from a Deuteromycete, Colletotrichum lindemuthianum. Biochemistry 2000, 39, 8837–8843. [Google Scholar] [CrossRef] [PubMed]
- Hekmat, O.; Tokuyasu, K.; Withers, S.G. Subsite structure of the endo-type chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum: An investigation using steady-state kinetic analysis and MS. Biochem. J. 2003, 374, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Gooday, G.W. Chitin deacetylases in invertebrates. In Chitin in Nature and Technology; Springer: Boston, MA, USA, 1986; pp. 263–267. [Google Scholar]
- Muthukrishnan, S.; Merzendorfer, H.; Arakane, Y.; Yang, Q. Chitin Metabolic Pathways in Insects and Their Regulation. In Extracellular Composite Matrices in Arthropods; Cohen, E., Moussian, B., Eds.; Springer: Berlin, Germany, 2016; pp. 31–65. ISBN 9783319407401. [Google Scholar]
- Dixit, R.; Arakane, Y.; Specht, C.A.; Richard, C.; Kramer, K.J.; Beeman, R.W.; Muthukrishnan, S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008, 38, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Christodoulidou, A.; Bouriotis, V.; Thireos, G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 31420–31425. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, I.A.; Gurr, S.J. Chitosan Mediates Germling Adhesion in Magnaporthe oryzae and Is Required for Surface Sensing and Germling Morphogenesis. PLoS Pathog. 2016, 12, 1–34. [Google Scholar] [CrossRef] [PubMed]
- White, S.; McIntyre, M.; Berry, D.R.; McNeil, B. The autolysis of industrial filamentous fungi. Crit. Rev. Biotechnol. 2002, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.L.; Bartnicki-Garcia, S. Chitosan Synthesis by the Tandem Action of Chitin Synthetase and Chitin Deacetylase from Mucor rouxii. Biochemistry 1984, 23, 1065–1073. [Google Scholar] [CrossRef]
- Gao, X.-D.; Katsumoto, T.; Onodera, K. Purification and characterization of chitin deacetylase from Absidia coerulea. J. Biochem. 1995, 117, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Christodoulidou, A.; Briza, P.; Ellinger, A.; Bouriotis, V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 1999, 460, 275–279. [Google Scholar] [CrossRef]
- Hadwiger, L.A. Anatomy of a nonhost disease resistance response of pea to Fusarium solani: PR gene elicitation via DNase, chitosan and chromatin alterations. Front. Plant Sci. 2015, 6, 373. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, L.A. Pea-Fusarium solani interactions contributions of a system toward understanding disease resistance. Phytopathology 2008, 98, 372–379. [Google Scholar] [CrossRef] [PubMed]
- El Gueddari, N.E.; Rauchhaus, U.; Moerschbacher, B.M.; Deising, H.B. Developmentally regulated conversion of surface-exposed chitin to chi-tosan in cell walls of plant pathogenic fungi. New Phytol. 2002, 156, 103–112. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-Induced Dimerization Activates a Plant Immune Receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Cord-Landwehr, S.; Melcher, R.L.J.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef] [PubMed]
- Emri, T.; Molnár, Z.; Szilágyi, M.; Pócsi, I. Regulation of autolysis in Aspergillus nidulans. Appl. Biochem. Biotechnol. 2008, 151, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, C.; Nuero, O.M.; Santamaría, F.; Reyes, F. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr. Microbiol. 1995, 30, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Reyes, F.; Calatayud, J.; Martinez, M.J. Endochitinase from Aspergillus nidulans implicated in the autolysis of its cell wall. FEMS Microbiol. Lett. 1989, 51, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M. Structure and Mechanism of Chitin Deacetylase from the Fungal Pathogen Colletotrichium lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Cord-Landwehr, S.; Singh, R.; Bernard, F.; Kolkenbrock, S.; El Gueddari, N.E.; Moerschbacher, B.M. A recombinant fungal chitin deacetylase produces fully defined chitosan oligomers with novel patterns of acetylation. Appl. Environ. Microbiol. 2016, 82, 6645–6655. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Martinez, A.; Grifoll-Romero, L.; Aragunde Pazos, H.; Enea Sancho-Vaello; Biarnés, X.; Lopez-Llorca, L.V.; Planas, A. Expression and specificity of a chitin deacetylase catalytic doain from the nematophagous fungus Pochonia chlamydosporia potentially involved in pathogenicity. Sci. Rep. 2018, 8, 2170. [Google Scholar] [CrossRef] [PubMed]
- Mishra, C.; Mishra, C.; Semino, C.; Semino, C.; Mccreath, K.J.; Mccreath, K.J.; de la Vega, H.; de la Vega, H.; Jones, B.J.; Jones, B.J.; et al. Cloning and expression of two chitin deacetylase gens of Saccharomyces cerevisiae. Yeast 1997, 13, 327–336. [Google Scholar] [CrossRef]
- Martinou, A.; Koutsioulis, D.; Bouriotis, V. Cloning and expression of a chitin deacetylase gene (CDA2) from Saccharomyces cerevisiae in Escherichia coli: Purification and characterization of the cobalt-dependent recombinant enzyme. Enzyme Microb. Technol. 2003, 32, 757–763. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhao, Y.; Oh, K.T.; Nguyen, V.N.; Park, R.D. Enzymatic deacetylation of chitin by extracellular chitin deacetylase from a newly screened Mortierella sp. DY-52. J. Microbiol. Biotechnol. 2008, 18, 759–766. [Google Scholar] [PubMed]
- Zhao, Y.; Kim, Y.J.; Oh, K.T.; Nguyen, V.N.; Park, R.D. Production and characterization of extracellular chitin deacetylase from Absidia corymbifera DY-9. J. Appl. Biol. Chem. 2010, 53, 119–126. [Google Scholar] [CrossRef]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 289, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Pareek, N.; Vivekanand, V.; Saroj, S.; Sharma, A.K.; Singh, R.P. Purification and characterization of chitin deacetylase from Penicillium oxalicum SAEM-51. Carbohydr. Polym. 2012, 87, 1091–1097. [Google Scholar] [CrossRef]
- Karthik, N.; Binod, P.; Pandey, A. SSF production, purification and characterization of chitin deacetylase from Aspergillus flavus. Biocatal. Biotransform. 2017. [Google Scholar] [CrossRef]
- Cai, J.; Yang, J.; Du, Y.; Fan, L.; Qiu, Y.; Li, J.; Kennedy, J.F. Purification and characterization of chitin deacetylase from Scopulariopsis brevicaulis. Carbohydr. Polym. 2006, 65, 211–217. [Google Scholar] [CrossRef]
- Gauthier, C.; Clerisse, F.; Dommes, J.; Jaspar-Versali, M.F. Characterization and cloning of chitin deacetylases from Rhizopus circinans. Protein Expr. Purif. 2008, 59, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Maw, T.; Tan, T.K.; Khor, E.; Wong, S.M. Complete cDNA sequence of chitin deacetylase from Gongronella butleri and its phylogenetic analysis revealed clusters corresponding to taxonomic classification of fungi. J. Biosci. Bioeng. 2002, 93, 376–381. [Google Scholar] [CrossRef]
- Mélida, H.; Sain, D.; Stajich, J.E.; Bulone, V. Deciphering the uniqueness of Mucoromycotina cell walls by combining biochemical and phylogenomic approaches. Environ. Microbiol. 2015, 17, 1649–1662. [Google Scholar] [CrossRef] [PubMed]
- Smirnou, D.; Krcmar, M.; Prochazkova, E.V.A. Chitin-Glucan complex production by Schizophyllum commune submerged cultivation. Pol. J. Microbiol. 2011, 60, 223–228. [Google Scholar] [PubMed]
- Das, S.; Van Dellen, K.; Bulik, D.; Magnelli, P.; Cui, J.; Head, J.; Robbins, P.W.; Samuelson, J. The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin). Mol. Biochem. Parasitol. 2006, 148, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Andrés, E.; Albesa-Jové, D.; Biarnés, X.; Moerschbacher, B.M.; Guerin, M.E.; Planas, A. Structural basis of chitin oligosaccharide deacetylation. Angew. Chem. Int. Ed. 2014, 53, 6882–6887. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.X.; Wang, X.; Roseman, S. The chitin catabolic cascade in the marine bacterium Vibrio cholerae: Characterization of a unique chitin oligosaccharide deacetylase. Glycobiology 2007, 17, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Sugiyama, K.; Sakaki, Y.; Hakamata, W.; Park, S.Y.; Nishio, T. Structure-based analysis of domain function of chitin oligosaccharide deacetylase from Vibrio parahaemolyticus. FEBS Lett. 2015, 589, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Kadokura, K.; Rokutani, A.; Yamamoto, M.; Ikegami, T.; Sugita, H.; Itoi, S.; Hakamata, W.; Oku, T.; Nishio, T. Purification and characterization of Vibrio parahaemolyticus extracellular chitinase and chitin oligosaccharide deacetylase involved in the production of heterodisaccharide from chitin. Appl. Microbiol. Biotechnol. 2007, 75, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, K.; Yamagishi, M.; Ohta, T.; Motosugi, M.; Izumida, H.; Sano, H.; Adachi, K.; Miwa, T. Purification and Properties of Two Deacetylases Produced by Vibrio alginolyticus H-8. Biosci. Biotechnol. Biochem. 1997, 61, 1113–1117. [Google Scholar] [CrossRef]
- Hirano, T.; Uehara, R.; Shiraishi, H.; Hakamata, W.; Nishio1, T. Chitin Oligosaccharide Deacetylase from Shewanella woodyi ATCC51908. J. Appl. Glycosci. 2015, 62, 153–157. [Google Scholar] [CrossRef]
- Hirano, T.; Shiraishi, H.; Ikejima, M.; Uehara, R.; Hakamata, W.; Nishio, T. Chitin oligosaccharide deacetylase from Shewanella baltica ATCC BAA-1091. Biosci. Biotechnol. Biochem. 2017, 81, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Tuveng, T.R.; Rothweiler, U.; Udatha, G.; Vaaje-Kolstad, G.; Smalås, A.; Eijsink, V.G.H. Structure and function of a CE4 deacetylase isolated from a marine environment. PLoS ONE 2017, 12, e0187544. [Google Scholar] [CrossRef] [PubMed]
- Bartnicki-Garcia, S.; Nickerson, W.J. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of Mucor rouxii. Biochim. Biophys. Acta 1962, 58, 102–119. [Google Scholar] [CrossRef]
- Kafetzopoulos, D.; Martinou, A.; Bouriotis, V. Bioconversion of chitin to chitosan: Purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. USA 1993, 90, 2564–2568. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.L.; Bartnicki-Garcia, S. The co-ordination of chitosan and chitin synthesis in Mucor rouxii. J. Gen. Microbiol. 1984, 130, 2095–2102. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Adhya, M.; Guha, A.K.; Chatterjee, B.P. Chitosan from Mucor rouxii: Production and physico-chemical characterization. Process Biochem. 2005, 40, 395–400. [Google Scholar] [CrossRef]
- Synowiecki, J.; Al-Khateeb, N.A.A.Q. Mycelia of Mucor rouxii as a source of chitin and chitosan. Food Chem. 1997, 60, 605–610. [Google Scholar] [CrossRef]
- Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Isolation of chitin deacetylase from Mucor rouxii by immunoaffinity chromatography. J. Chromatogr. A 1993, 644, 35–41. [Google Scholar] [CrossRef]
- Kauss, H.; Bauch, B. Chitin deacetylase from Colletotrichum lindemuthianum. Methods Enzymol. 1988, 161, 518–523. [Google Scholar] [CrossRef]
- Tsigos, I.; Bouriotis, V. Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J. Biol. Chem. 1995, 270, 26286–26291. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.J.; Ride, J.P. Chemical detection and ultrastructural localization of chitin in cell walls of Colletotrichum lindemuthianum. Physiol. Mol. Plant Pathol. 1990, 37, 39–53. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ohnishi-Kameyama, M.; Hayashi, K. Purification and characterization of extracellular chitin deacetylase from Colletotrichum lindemuthianum. Biosci. Biotechnol. Biochem. 1996, 60, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Blondeau, K.; Stevens, W.F.; Hegarat, F.L. Expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris: Purification and characterization. Protein Expr. Purif. 2004, 38, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Chen, X.; Zhai, C.; Ma, L. Synthesis and high expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris GS115. J. Microbiol. Biotechnol. 2012, 22, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Tokuyasu, K.; Kaneko, S.; Hayashi, K.; Mori, Y. Production of a recombinant chitin deacetylase in the culture medium of Escherichia coli cells. FEBS Lett. 1999, 458, 23–26. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ohnishi-Kameyama, M.; Hayashi, K.; Mori, Y. Cloning and expression of chitin deacetylase gene from a deuteromycete, Colletotrichum lindemuthianum. J. Biosci. Bioeng. 1999, 87, 418–423. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ono, H.; Ohnishi-Kameyama, M.; Hayashi, K.; Mori, Y. Deacetylation of chitin oligosaccharides of dp 2-4 by chitin deacetylase from Colletotrichum lindemuthianum. Carbohydr. Res. 1997, 303, 353–358. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ono, H.; Hayashi, K.; Mori, Y. Reverse hydrolysis reaction of chitin deacetylase and enzymatic synthesis of β-d-GlcNAc-(1→4)-GlcN from chitobiose. Carbohydr. Res. 1999, 322, 26–31. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ono, H.; Mitsutomi, M.; Hayashi, K.; Mori, Y. Synthesis of a chitosan tetramer derivative, β-d-GlcNAc-(1→4)-β-d-GlcNAc-(1→4)-β-d-GlcNAc-(1→4)-d-GlcN through a partial N-acetylation reaction by chitin deacetylase. Carbohydr. Res. 2000, 325, 211–215. [Google Scholar] [CrossRef]
- Kang, L.X.; Liang, Y.X.; Ma, L.X. Novel characteristics of chitin deacetylase from Colletotrichum lindemuthianum: Production of fully acetylated chitooligomers, and hydrolysis of deacetylated chitooligomers. Process Biochem. 2014, 49, 1936–1940. [Google Scholar] [CrossRef]
- Reyes, F.; Calatayud, J.; Vazquez, C.; Martínez, M.J. β-N-Acetylglucosaminidase from Aspergillus nidulans which degrades chitin oligomers during autolysis. FEMS Microbiol. Lett. 1990, 65, 83–87. [Google Scholar] [CrossRef]
- Wang, Y.; Song, J.Z.; Yang, Q.; Liu, Z.H.; Huang, X.M.; Chen, Y. Cloning of a heat-stable chitin deacetylase gene from Aspergillus nidulans and its functional expression in Escherichia coli. Appl. Biochem. Biotechnol. 2010, 162, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Espagne, E.; Lespinet, O.; Malagnac, F.; Da Silva, C.; Jaillon, O.; Porcel, B.M.; Couloux, A.; Aury, J.-M.; Ségurens, B.; Poulain, J.; Anthouard, V.; et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008, 9, R77. [Google Scholar] [CrossRef] [PubMed]
- Lorin, S.; Dufour, E.; Sainsard-Chanet, A. Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.; Upadhyaya, N.M.; Sperschneider, J.; Park, R.F.; Szabo, L.J.; Steffenson, B.; Ellis, J.G.; Dodds, P.N. Changing the Game: Using Integrative Genomics to Probe Virulence Mechanisms of the Stem Rust Pathogen Puccinia graminis f. sp. tritici. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Hodson, D.P.; Jin, Y.; Lagudah, E.S.; Ayliffe, M.A.; Bhavani, S.; Rouse, M.N.; Pretorius, Z.A.; Szabo, L.J.; Huerta-Espino, J.; et al. Emergence and Spread of New Races of Wheat Stem Rust Fungus: Continued Threat to Food Security and Prospects of Genetic Control. Phytopathology 2015, 105, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Singh, R.P. Resistance in U.S. Wheat to Recent Eastern African Isolates of Puccinia graminis f. sp. tritici with Virulence to Resistance Gene Sr31. Plant Dis. 2006, 90, 476–480. [Google Scholar] [CrossRef]
- Mendgen, K.; Hahn, M. Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 2002, 7, 352–356. [Google Scholar] [CrossRef]
- Broeker, K.; Fehser, S.; Tenberge, K.B.; Moerschbacher, B.M. Two class III chitin synthases specifically localized in appressoria and haustoria of Puccinia graminis f. sp. tritici. Physiol. Mol. Plant Pathol. 2011, 76, 27–33. [Google Scholar] [CrossRef]
- Ride, J.P.; Barber, M.S. Purification and characterization of multiple forms of endochitinase from wheat leaves. Plant Sci. 1990, 71, 185–197. [Google Scholar] [CrossRef]
- Vander, P.; Vårum, K.M.; Domard, A.; El Gueddari, N.E.; Moerschbacher, B.M. Comparison of the Ability of Partially N-Acetylated Chitosans and Chitooligosaccharides to Elicit Resistance Reactions in Wheat Leaves. Plant Physiol. 1998, 118, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Stud. Mycol. 2014, 79, 121–186. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Llorca, L.V.; Olivares-Bernabeu, C.; Salinas, J.; Jansson, H.-B.; Kolattukudy, P.E. Pre-penetration events in fungal parasitism of nematode eggs. Mycol. Res. 2002, 106, 499–506. [Google Scholar] [CrossRef]
- Manzanilla-Lopez, R.H.; Esteves, I.; Finetti-Sialer, M.M.; Hirsch, P.R.; Ward, E.; Devonshire, J.; Hidalgo-Diaz, L. Pochonia chlamydosporia: Advances and Challenges to Improve Its Performance as a Biological Control Agent of Sedentary Endo-parasitic Nematodes. J. Nematol. 2013, 45, 1–7. [Google Scholar] [PubMed]
- Larriba, E.; Jaime, M.D.L.A.; Carbonell-Caballero, J.; Conesa, A.; Dopazo, J.; Nislow, C.; Martín-Nieto, J.; Lopez-Llorca, L.V. Sequencing and functional analysis of the genome of a nematode egg-parasitic fungus, Pochonia chlamydosporia. Fungal Genet. Biol. 2014, 65, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Martinez, A.; Lenfant, N.; Escudero, N.; Zavala-Gonzalez, E.A.; Henrissat, B.; Lopez-Llorca, L.V. CAZyme content of Pochonia chlamydosporia reflects that chitin and chitosan modification are involved in nematode parasitism. Environ. Microbiol. 2016, 18, 4200–4215. [Google Scholar] [CrossRef] [PubMed]
- Wani, Z.A.; Kumar, A.; Sultan, P.; Bindu, K.; Riyaz-Ul-Hassan, S.; Ashraf, N. Mortierella alpina CS10E4, an oleaginous fungal endophyte of Crocus sativus L. enhances apocarotenoid biosynthesis and stress tolerance in the host plant. Sci. Rep. 2017, 7, 8598. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jo, G.-H.; Ju, W.-T.; Jung, W.-J.; Park, R.-D. A Highly N-Glycosylated Chitin Deacetylase Derived from a Novel Strain of Mortierella sp. DY-52. Biosci. Biotechnol. Biochem. 2011, 75, 960–965. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Parameswaran, B.; Pandey, A. Production of chitin deacetylase by Aspergillus flavus in submerged conditions. Prep. Biochem. Biotechnol. 2016, 46, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Estrella, M.; Gomez-Lopez, A.; Mellado, E.; Buitrago, M.J.; Monzón, A.; Rodriguez-Tudela, J.L. Scopulariopsis brevicaulis, a fungal pathogen resistant to broad-spectrum antifungal agents. Antimicrob. Agents Chemother. 2003, 47, 2339–2341. [Google Scholar] [CrossRef] [PubMed]
- Tosti, A.; PiracciniI, B.M.; Stinchi, C.; Lorenzi, S. Onychomycosis due to Scopulariopsis brevicaulis: Clinical features and response to systemic antifungals. Br. J. Dermatol. 1996, 135, 799–802. [Google Scholar] [CrossRef] [PubMed]
- El Ghaouth, A.; Arul, J.; Grenier, J.; Asselin, A. Effect of chitosan and other polyions on chitin deacetylase in Rhizopus stolonifer. Exp. Mycol. 1992, 16, 173–177. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, S.; Fang, J.; Deng, Y.; Wang, D.; Zhao, Y. Optimization of the fermentation conditions of Rhizopus japonicus M193 for the production of chitin deacetylase and chitosan. Carbohydr. Polym. 2014, 101, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Maw, T.; Tan, T.K.; Khor, E.; Wong, S.M. Selection of Gongronella butleri strains for enhanced chitosan yield with UV mutagenesis. J. Biotechnol. 2002, 95, 189–193. [Google Scholar] [CrossRef]
- Yonemura, A.; Nagashima, T.; Murayama, T. Expression of Chitin Deacetylase Gene from Phycomyces blakesleeanus in Aspergillus oryzae and Neurospora crassa; The Society for Bioscience and Bioengineering: Osaka, Japan, 2007; p. 129. Available online: http://dl.ndl.go.jp/view/download/digidepo_10529404_po_ART0009175183.pdf?contentNo=1&alternativeNo= (accessed on 19 February 2018).
- Baker, L.G.; Specht, C.A.; Lodge, J.K. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot. Cell 2011, 10, 1264–1268. [Google Scholar] [CrossRef] [PubMed]
- Doering, T.L. How Sweet it is! Cell Wall Biogenesis and Polysaccharide Capsule Formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 2009, 63, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.M.; Baker, L.G.; Specht, C.A.; Lodge, J.K. A glycosylphosphatidylinositol anchor is required for membrane localization but dispensable for cell wall association of chitin deacetylase 2 in Cryptococcus neoformans. mBio 2012, 3, e00007–e00012. [Google Scholar] [CrossRef] [PubMed]
- Levitz, S.M.; Nong, S.-H.; Mansour, M.K.; Huang, C.; Specht, C.A. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 2001, 98, 10422–10427. [Google Scholar] [CrossRef] [PubMed]
- Biondo, C.; Beninati, C.; Delfino, D.; Oggioni, M.; Mancuso, G.; Midiri, A.; Tomaselli, G.; Teti, G.; Biondo, C.; Beninati, C.; et al. Identification and Cloning of a Cryptococcal Deacetylase That Produces Protective Immune Responses. Infect. Immun. 2002, 70, 2383–2391. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.; Anderson, I.; Davies, R.; Alsmark, U.C.M.; Samuelson, J.; Amedeo, P.; Roncaglia, P.; Berriman, M.; Hirt, R.P.; Mann, B.J.; et al. The genome of the protist parasite Entamoeba histolytica. Nature 2005, 433, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.; Maillet, F.; Plazanet, C.; Debellé, F.; Ferro, M.; Truchet, G.; Promé, J.C.; Dénarié, J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc. Natl. Acad. Sci. USA 1996, 93, 15305–15310. [Google Scholar] [CrossRef] [PubMed]
- Egelhoff, T.T.; Long, S.R. Rhizobium meliloti nodulation genes: Identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J. Bacteriol. 1985, 164, 591–599. [Google Scholar] [PubMed]
- Spaink, H.P.; Wijfjes, A.H.M.; der van Drift, K.M.G.M.; Haverkamp, J.; Thomas-Oates, J.E.; Lugtenberg, B.J.J. Structural identification of metabolites produced by the NodB and NodC proteins of Rhizobium leguminosarum. Mol. Microbiol. 1994, 13, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Chambon, R.; Pradeau, S.; Fort, S.; Cottaz, S.; Armand, S. High yield production of Rhizobium NodB chitin deacetylase and its use for in vitro synthesis of lipo-chitinoligosaccharide precursors. Carbohydr. Res. 2017, 442, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Hamer, S.N.; Cord-Landwehr, S.; Biarnés, X.; Planas, A.; Waegeman, H.; Moerschbacher, B.M.; Kolkenbrock, S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 2015, 5, 8716. [Google Scholar] [CrossRef] [PubMed]
- Röhrig, H.; Schmidt, J.; Wieneke, U.; Kondorosi, E.; Barlier, I.; Schell, J.; John, M. Biosynthesis of lipooligosaccharide nodulation factors: Rhizobium NodA protein is involved in N-acylation of the chitooligosaccharide backbone. Proc. Natl. Acad. Sci. USA 1994, 91, 3122–3126. [Google Scholar] [CrossRef] [PubMed]
- Keyhani, N.O.; Roseman, S. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta Gen. Subj. 1999, 1473, 108–122. [Google Scholar] [CrossRef]
- Zobell, C.; Rittenberg, S. The occurrence and characteristics of chitinoclastic bacteria in the sea. J. Bacteriol. 1937, 35, 275–287. [Google Scholar]
- Meibom, K.L.; Li, X.B.; Nielsen, A.T.; Wu, C.-Y.; Roseman, S.; Schoolnik, G.K. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 2004, 101, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Roseman, S. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, K.; Murase, K.; Ohta, T.; Etoh, H. Cloning and sequencing of the deacetylase gene from Vibrio alginolyticus H-8. J. Biosci. Bioeng. 2000, 90, 561–563. [Google Scholar] [CrossRef]
- Kadokura, K.; Sakamoto, Y.; Saito, K.; Ikegami, T.; Hirano, T.; Hakamata, W.; Oku, T.; Nishio, T. Production of a recombinant chitin oligosaccharide deacetylase from Vibrio parahaemolyticus in the culture medium of Escherichia coli cells. Biotechnol. Lett. 2007, 29, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Maebara, Y.; Uehara, R.; Sakaki, Y.; Shiraishi, H.; Ichimura, S.; Hakamata, W.; Nishio, T. Chitin oligosaccharide deacetylase from Vibrio harveyi ATCC BAA-1116: Gene cloning, overexpression, purification, and characterization. Chitin Chitosan Res. 2012, 19, 321–324. [Google Scholar]
- Jacquiod, S.; Franqueville, L.; Cécillon, S.; Vogel, T.M.; Simonet, P. Soil bacterial community shifts after Chitin enrichment: An integrative metagenomic approach. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Dsouza, M.; Taylor, M.W.; Turner, S.J.; Aislabie, J. Genomic and phenotypic insights into the ecology of Arthrobacter from Antarctic soils. BMC Genom. 2015, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Mavromatis, K.; Vorgias, C.E.; Buchon, L.; Gerday, C.; Bouriotis, V. Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: Isolation and partial characterization of the enzymes. J. Bacteriol. 2001, 183, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Aragunde-pazos, H.; Biarnés, X.; Planas, A. Substrate recognition and specificity of chitin deacetylases and related family 4 carbohydrate esterases. Int. J. Mol. Sci. 2018, 19, 412. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-Binding modules: Fine-Tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Schuttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M.F. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Van Aalten, D.M.F. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Lett. 2004, 570, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.M.; Nascimento, A.S.; Polikarpov, I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017, 1, 35–51. [Google Scholar] [CrossRef]
- Nishiyama, T.; Noguchi, H.; Yoshida, H.; Park, S.Y.; Tame, J.R.H. The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: A circularly permuted member of the carbohydrate esterase 4 family. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Hernick, M.; Fierke, C.A. Zinc hydrolases: The mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 2005, 433, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.L.; Polvi, E.J.; Shekhar-Guturja, T.; Cowen, L.E. Elucidating drug resistance in human fungal pathogens. Future Microbiol. 2014, 9, 523–542. [Google Scholar] [CrossRef] [PubMed]
- Takaya, N.; Yamazaki, D.; Horiuchi, H.; Ohta, A.; Takagi, M. Cloning and characterization of a chitinase-encoding gene (chiA) from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci. Biotechnol. Biochem. 1998, 62, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Hartl, L.; Zach, S.; Seidl-Seiboth, V. Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl. Microbiol. Biotechnol. 2012, 93, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Aoun, M. Host defense mechanisms during fungal pathogenesis and how these are overcome in susceptible plants: A review. Int. J. Bot. 2017, 13, 82–102. [Google Scholar] [CrossRef][Green Version]
- Huang, G. Chitinase Inhibitor Allosamidin and Its Analogues: An Update. Curr. Org. Chem. 2012, 16, 115–120. [Google Scholar] [CrossRef]
- Rao, F.V.; Houston, D.R.; Boot, R.G.; Aerts, J.M.F.G.; Hodkinson, M.; Adams, D.J.; Shiomi, K.; Omura, S.; Van Aalten, D.M.F. Specificity and affinity of natural product cyclopentapeptide inhibitors against A. fumigatus, human, and bacterial chitinases. Chem. Biol. 2005, 12, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Younes, I.; Rinaudo, M. Chitin and chitosan preparation from marine sources. Structure, properties and applications. Mar. Drugs 2015, 13, 1133–1174. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H.; Chan, W.Y. Chitosan: An update on potential biomedical and pharmaceutical applications. Mar. Drugs 2015, 13, 5156–5186. [Google Scholar] [CrossRef] [PubMed]
- Anitha, A.; Sowmya, S.; Kumar, P.T.T.S.; Deepthi, S.; Chennazhi, K.P.; Ehrlich, H.; Tsurkan, M.; Jayakumar, R. Chitin and chitosan in selected biomedical applications. Prog. Polym. Sci. 2014, 39, 1644–1667. [Google Scholar] [CrossRef]
- Abdul Khalil, H.P.S.; Saurabh, C.K.; Adnan, A.S.; Nurul Fazita, M.R.; Syakir, M.I.; Davoudpour, Y.; Rafatullah, M.; Abdullah, C.K.; Haafiz, M.K.M.; Dungani, R. A review on chitosan-cellulose blends and nanocellulose reinforced chitosan biocomposites: Properties and their applications. Carbohydr. Polym. 2016, 150, 216–226. [Google Scholar]
- Pestov, A.; Bratskaya, S. Chitosan and Its Derivatives as Highly Efficient Polymer Ligands. Molecules 2016, 21, 330. [Google Scholar] [CrossRef] [PubMed]
- Das, S.N.; Madhuprakash, J.; Sarma, P.V.S.R.N.; Purushotham, P.; Suma, K.; Manjeet, K.; Rambabu, S.; Gueddari, N.E.E.; Moerschbacher, B.M.; Podile, A.R. Biotechnological approaches for field applications of chitooligosaccharides (COS) to induce innate immunity in plants. Crit. Rev. Biotechnol. 2015, 35, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Sorlier, P.; Denuzière, A.; Viton, C.; Domard, A. Relation between the degree of acetylation and the electrostatic properties of chitin and chitosan. Biomacromolecules 2001, 2, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Omura, Y.; Shigemoto, M.; Akiyama, T.; Saimoto, H.; Shigemasa, Y.; Nakamura, I.; Tsuchido, T. Antimicrobial Activity of Chitosan with Different Degrees of Acetylation and Molecular Weights. Biocontrol Sci. 2003, 8, 25–30. [Google Scholar] [CrossRef]
- Domard, A.; Cartier, N. Glucosamine oligomers: 1. Preparation and characterization. Int. J. Biol. Macromol. 1989, 11, 297–302. [Google Scholar] [CrossRef]
- Einbu, A.; Varum, K.M. Depolymerization and de-N-acetylation of chitin oligomers in hydrochloric acid. Biomacromolecules 2007, 8, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Kuyama, H.; Nakahara, Y.; Nukada, T.; Ito, Y.; Nakahara, Y.; Ogawa, T. Stereocontrolled synthesis of chitosan dodecamer. Carbohydr. Res. 1993, 243, C1–C7. [Google Scholar] [CrossRef]
- Barroca-Aubry, N.; Pernet-Poil-Chevrier, A.; Domard, A.; Trombotto, S. Towards a modular synthesis of well-defined chitooligosaccharides: Synthesis of the four chitodisaccharides. Carbohydr. Res. 2010, 345, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, M.X.; Sauvageau, J.C.M.; Kumirska, J.; Thöming, J. Studies on acetylation patterns of different chitosan preparations. Carbohydr. Polym. 2009, 78, 678–684. [Google Scholar] [CrossRef]
- Abla, M.; Marmuse, L.; Delolme, F.; Vors, J.P.; Ladavière, C.; Trombotto, S. Access to tetra-N-acetyl-chitopentaose by chemical N-acetylation of glucosamine pentamer. Carbohydr. Polym. 2013, 98, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Trombotto, S.; Ladavière, C.; Delolme, F.; Domard, A. Chemical preparation and structural characterization of a homogeneous series of chitin/chitosan oligomers. Biomacromolecules 2008, 9, 1731–1738. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Moerschbacher, B.M. The cell factory approach toward biotechnological production of high-value chitosan oligomers and their derivatives: An update. Crit. Rev. Biotechnol. 2017, 37, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Hembach, L.; Cord-Landwehr, S.; Moerschbacher, B.M. Enzymatic production of all fourteen partially acetylated chitosan tetramers using different chitin deacetylases acting in forward or reverse mode. Sci. Rep. 2017, 7, 17692. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, P.; D’Haeze, W.; Geelen, D.; Promé, D.; Van Montagu, M.; Geremia, R.; Promé, J.C.; Holsters, M. Biosynthesis of Azorhizobium caulinodans Nod factors: Study of the activity of the nodABCS proteins by expression of the genes in Escherichia coli. J. Biol. Chem. 1995, 270, 29217–29223. [Google Scholar] [CrossRef] [PubMed]
- Poinsot, V.; Crook, M.B.; Erdn, S.; Maillet, F.; Bascaules, A.; Ané, J.M. New insights into Nod factor biosynthesis: Analyses of chitooligomers and lipo-chitooligomers of Rhizobium sp. IRBG74 mutants. Carbohydr. Res. 2016, 434, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Samain, E.; Drouillard, S.; Heyraud, A.; Driguez, H.; Geremia, R.A. Gram-scale synthesis of recombiant chitooligosaccharides in Escherichia coli. Carbohydr. Res. 1997, 302, 35–42. [Google Scholar] [CrossRef]
- Samain, E.; Chazalet, V.; Geremia, R.A. Production of O-acetylated and sulfated chitooligosaccharides by recombinant Escherichia coli strains harboring different combinations of nod genes. J. Biotechnol. 1999, 72, 33–47. [Google Scholar] [CrossRef]
- Cottaz, S.; Samain, E. Genetic engineering of Escherichia coli for the production of NI, NII-diacetylchitobiose (chitinbiose) and its utilization as a primer for the synthesis of complex carbohydrates. Metab. Eng. 2005, 7, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Bettler, E.; Samain, E.; Chazalet, V.; Bosso, C.; Heyraud, A.; Joziasse, D.H.; Wakarchuk, W.W.; Imberty, A.; Geremia, R.A. The living factory: In Vivo production of N-acetyllactosamine containing carbohydrates in E. coli. Glycoconj. J. 1999, 16, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Southwick, A.M.; Wang, L.X.; Long, S.R.; Lee, Y.C. Activity of Sinorhizobium meliloti NodAB and nodH enzymes on thiochitooligosaccharides. J. Bacteriol. 2002, 184, 4039–4043. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).