Comparison of the Quality of Protein Crystals Grown by CLPC Seeds Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Crystal Images
2.3. Crystal Diffraction
3. Results
3.1. Morphology of Protein Crystal
3.2. Diffraction Pattern of Protein Crystals
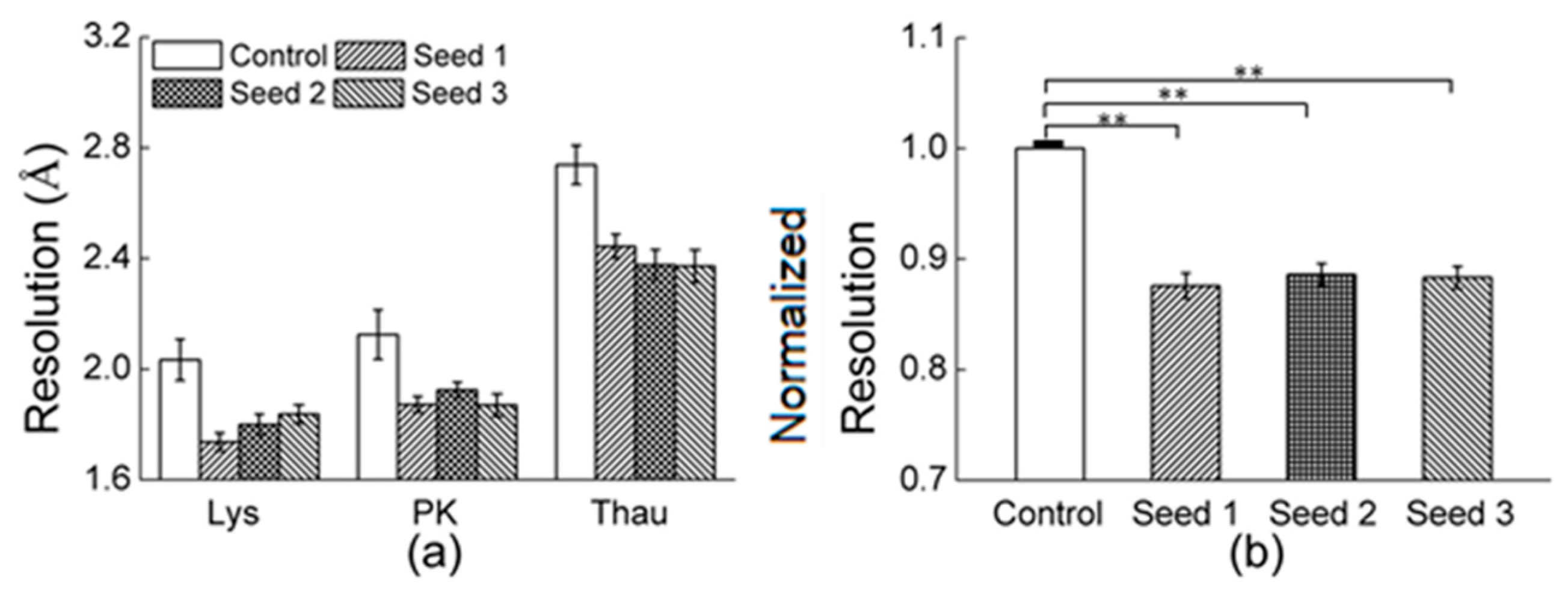
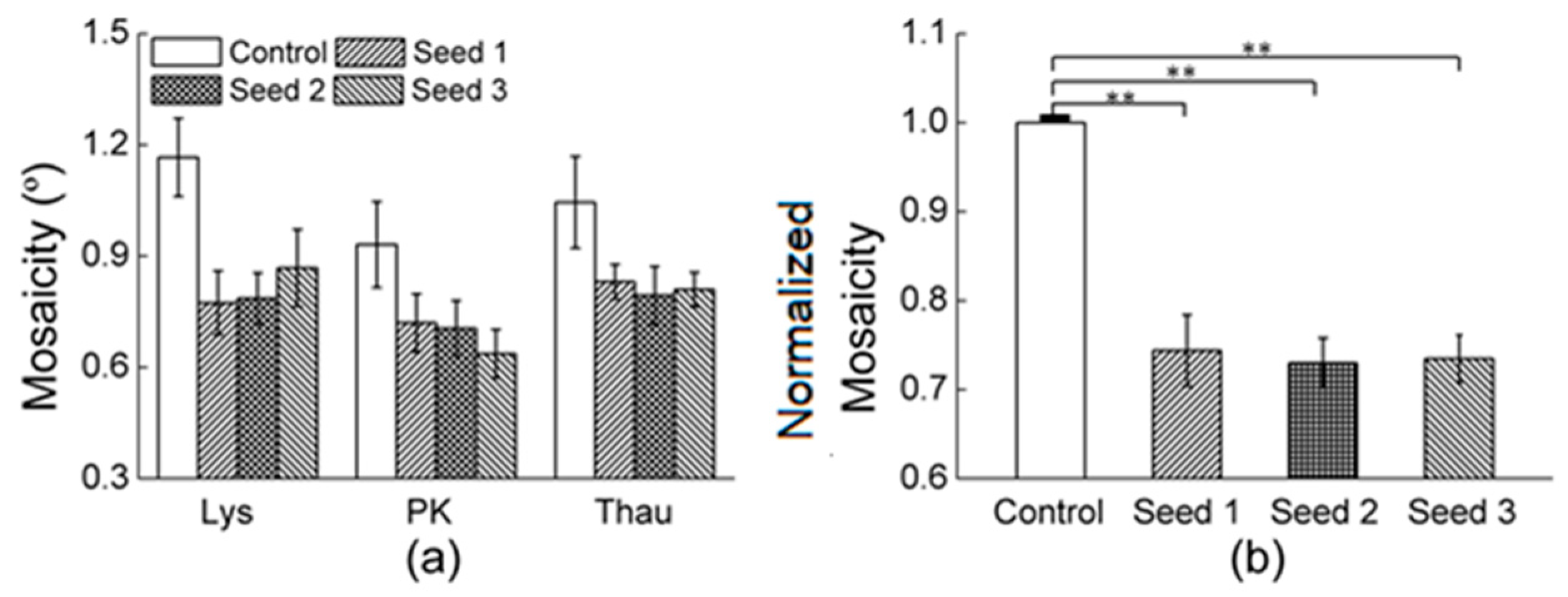
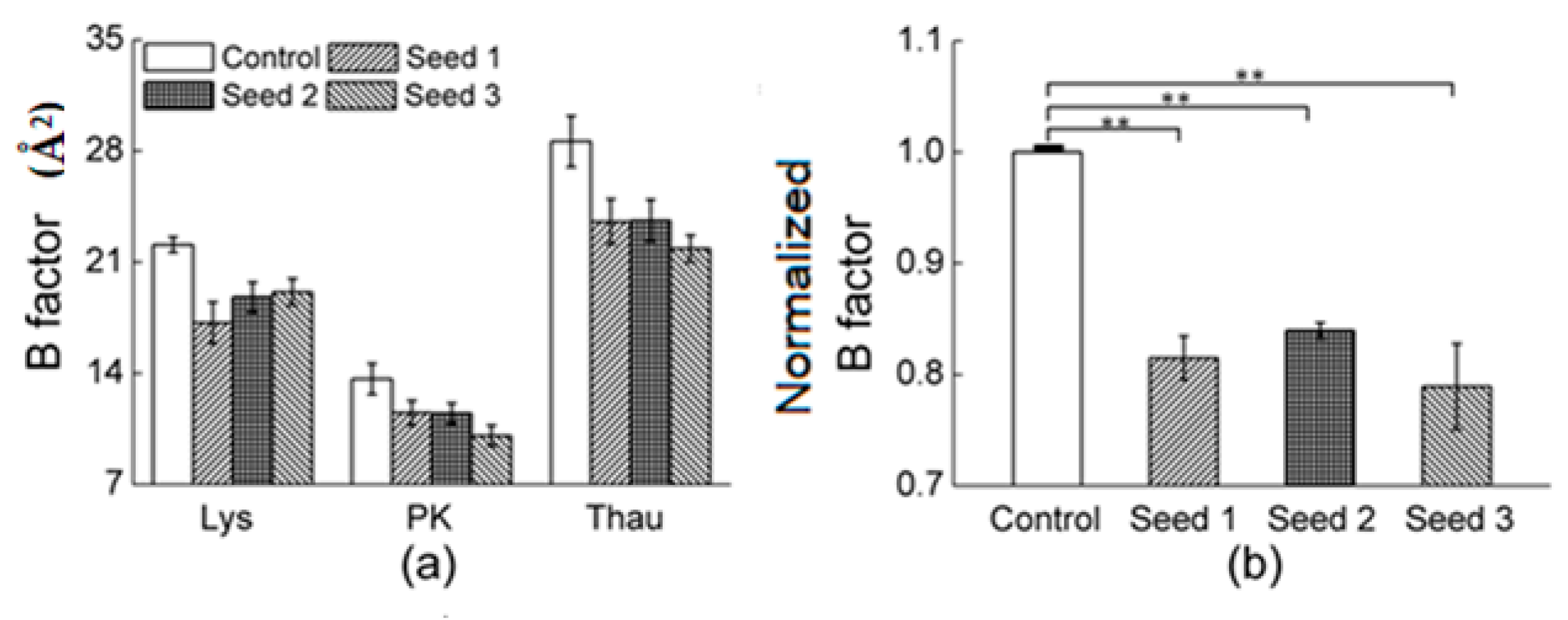
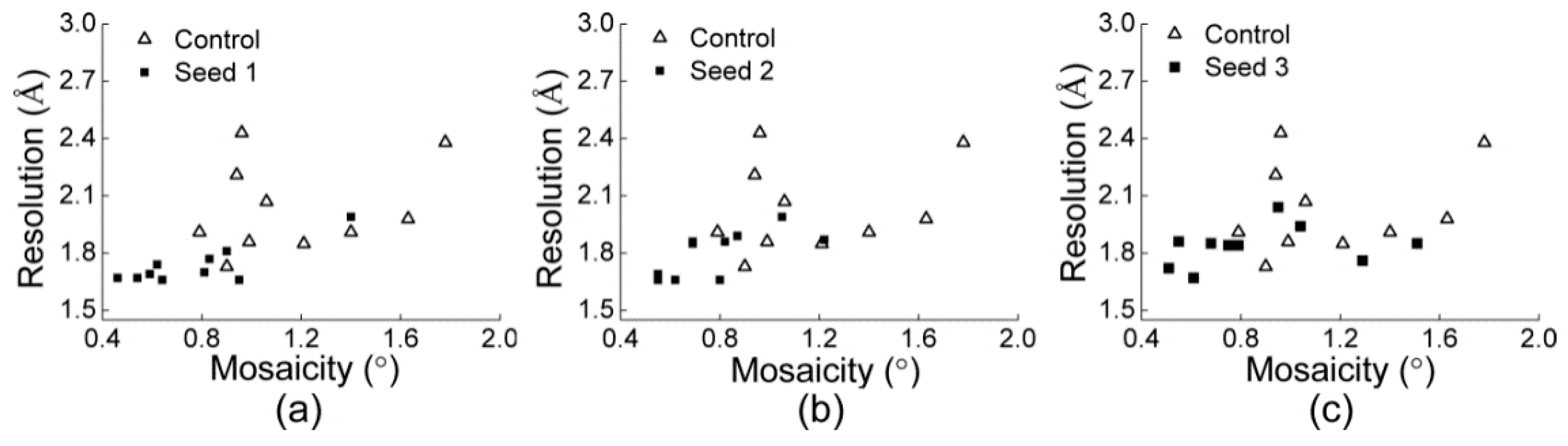
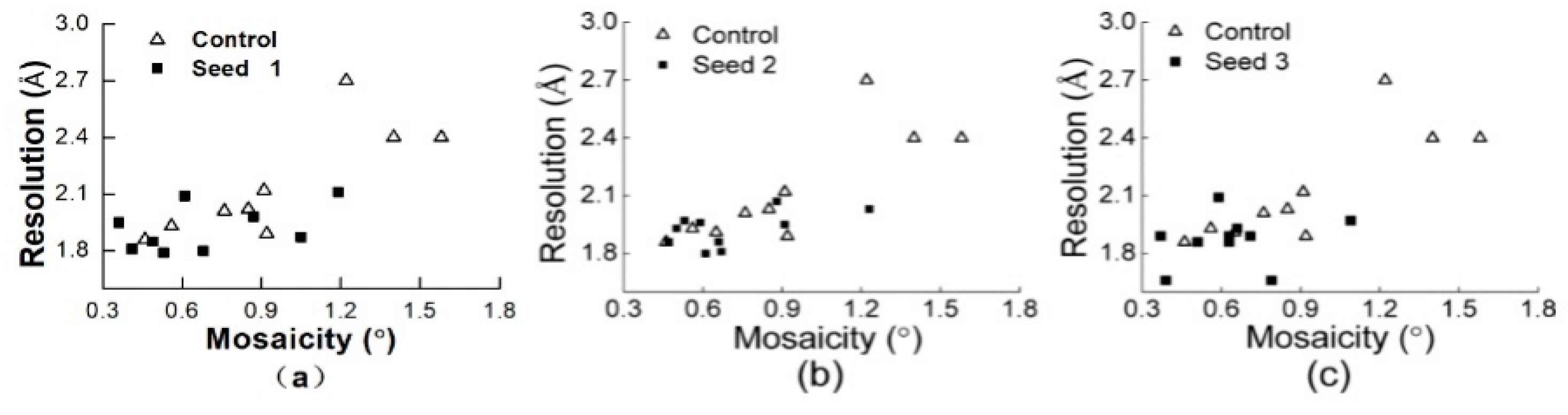
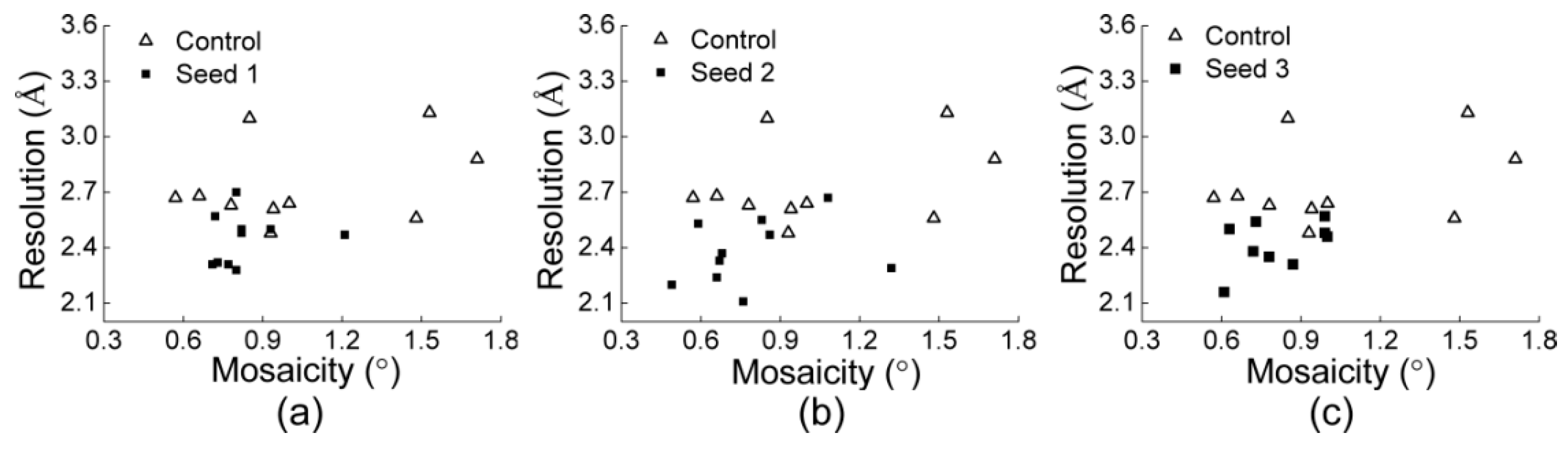
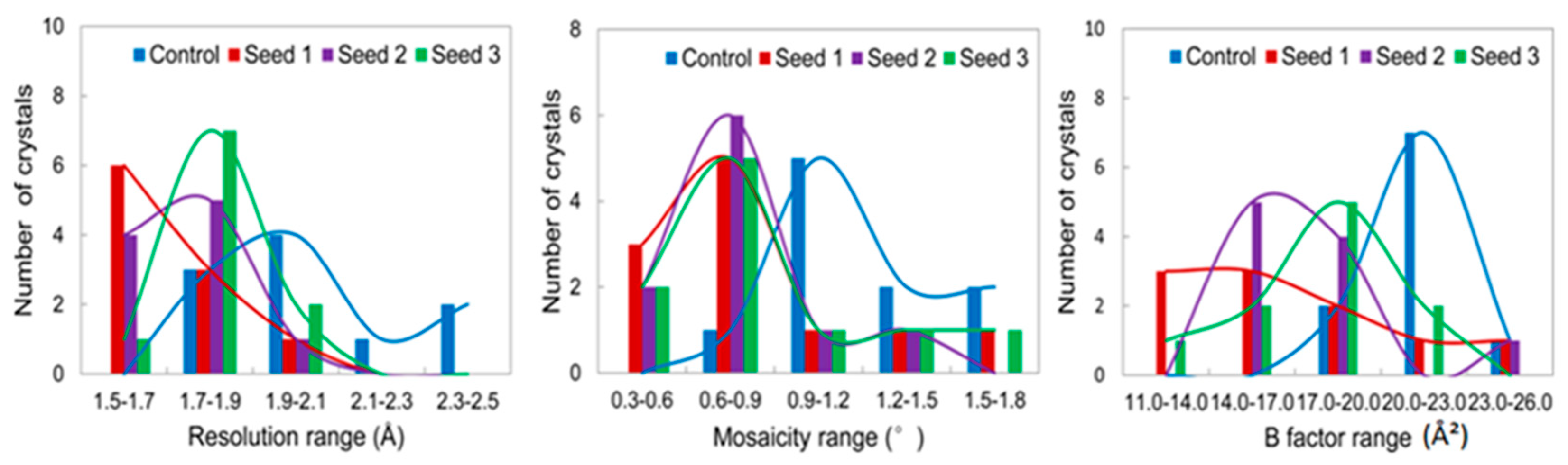
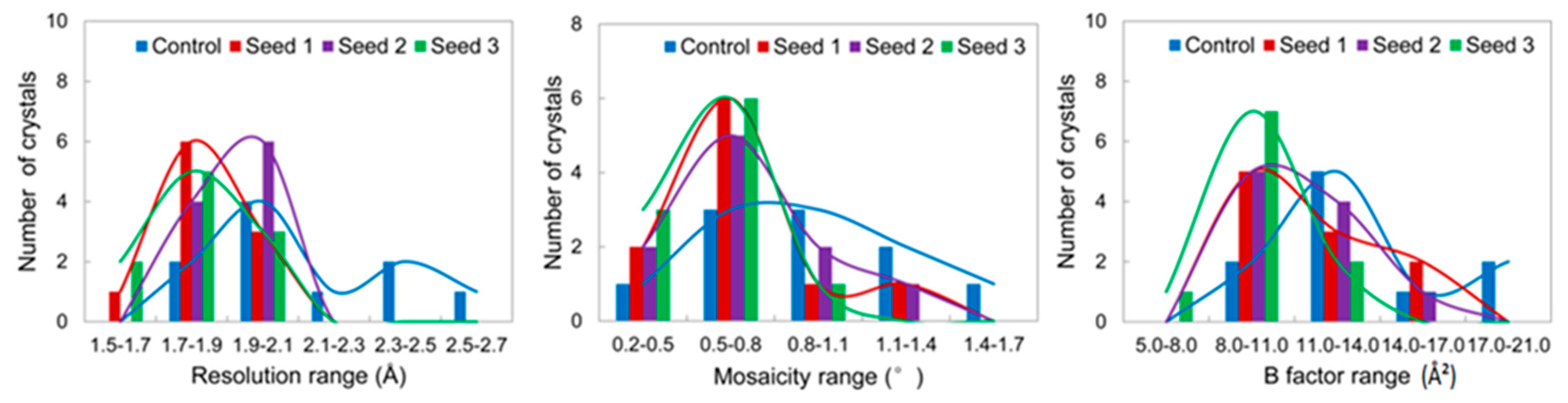
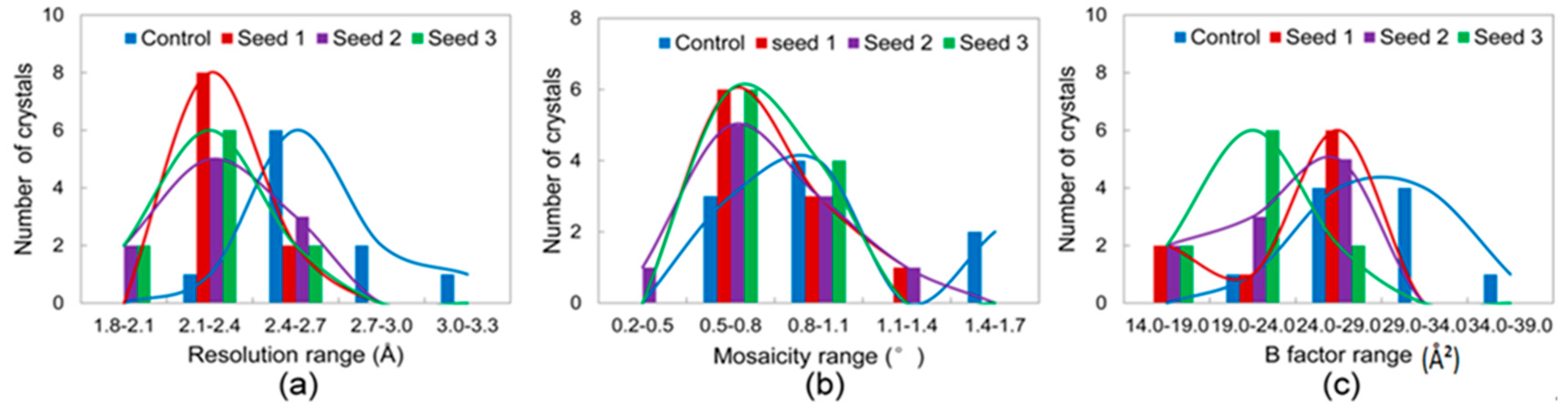
3.2.1. Comparisons of the Diffraction Data
3.2.2. Comparisons of the Diffraction Data for the 5 Degree Range
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kessel, A.; Ben-Tal, N. Introduction to Proteins; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon-on-Thames, UK, 2018; pp. 209–253. [Google Scholar]
- Chayen, N.E. Turning protein crystallisation from an art into a science. Curr. Opin. Struct. Biol. 2004, 14, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Chayen, N.E. Tackling the bottleneck of protein crystallization in the post-genomic. Trends Biotechnol. 2002, 20, 98. [Google Scholar] [CrossRef]
- Koizumi, H.; Uda, S.; Fujiwara, K.; Tachibana, M.; Kojima, K.; Nozawa, J. Crystallization of high-quality protein crystals using an external electric field. J. Appl. Crystallogr. 2015, 48, 1507–1513. [Google Scholar] [CrossRef]
- Koizumi, H.; Uda, S.; Fujiwara, K.; Tachibana, M.; Kojima, K.; Nozawa, J. Technique for high-quality protein crystal growth by control of subgrain formation under an external electric field. Crystals 2016, 6, 95. [Google Scholar] [CrossRef]
- Lu, Q.Q.; Zhang, B.; Tao, L.; Xu, L.; Chen, D.; Zhu, J.; Yin, D.C. Improving protein crystal quality via mechanical vibration. Cryst. Growth Des. 2016, 16, 4869–4876. [Google Scholar] [CrossRef]
- McPherson, A.; Delucas, L.J. Microgravity protein crystallization. NPJ Microgravity 2015, 1, 15010. [Google Scholar] [CrossRef] [PubMed]
- Boyko, K.M.; Timofeev, V.I.; Samygina, V.R.; Kuranova, I.P.; Popov, V.O.; Koval’Chuk, M.V. Protein crystallization under microgravity conditions. Analysis of the results of Russian experiments performed on the International Space Station in 2005–2015. Crystallogr. Rep. 2016, 61, 718–729. [Google Scholar] [CrossRef]
- Guo, Y.Z.; Sun, L.H.; Oberthuer, D.; Zhang, C.Y.; Shi, J.Y.; Di, J.L.; Zhang, B.L.; Cao, H.L.; Liu, Y.M.; Li, J.; et al. Utilisation of adsorption and desorption for simultaneously improving protein crystallisation success rate and crystal quality. Sci. Rep. 2014, 4, 7308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quiocho, F.A.; Richards, F.M. Intermolecular Cross Linking of a Protein in the Crystalline State: Carboxypeptidase-A. Proc. Natl. Acad. Sci. USA 1964, 52, 833–839. [Google Scholar] [CrossRef]
- Lusty, C.J. A gentle vapor-diffusion technique for cross-linking of protein crystals for cryocrystallography. J. Appl. Crystallogr. 1999, 32, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Weichsel, U.; Segets, D.; Thajudeen, T.; Maier, E.M.; Peukert, W. Enhanced Crystallization of Lysozyme Mediated by the Aggregaion of Inorganic Seed Particle. Cryst. Growth Des. 2016, 17, 967–981. [Google Scholar] [CrossRef]
- Koizumi, H.; Uda, S.; Tachibana, M.; Tsukamoto, K.; Kojima, K.; Nozawa, J. Crystallization. Cryst. Growth Des. 2016, 16, 6089–6094. [Google Scholar] [CrossRef]
- Luo, Z.; Morey, J.R.; McDevitt, C.A.; Kobe, B. Heterogeneous nucleation is required for crystallizaion of the ZnuA domain of pneumococcal AdcA. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.B.; Cao, H.L.; Zhang, C.Y.; Yin, D.C. A review on recent advances for nucleants and nucleation in protein crystallization. CrystEngComm 2017, 19, 1143–1155. [Google Scholar] [CrossRef]
- O’dell, W.B.; Swartz, P.D.; Weiss, K.L.; Meilleur, F. Crystallization of fungal lytic polysaccharide monooxygenase exprssed from glycoengineered Pichia pastoris for X-ray and neutron diffraction. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2017, 73, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Narni-Mancinelli, E.; Cantoni, C.; Li, Y.; Guia, S.; Gauthier, L.; Chen, Q.; Moretta, A.; Vély, F.; Eisenstein, E.; et al. Structural Insights into the Inhibitory Mechanism of an Antibody against B7-H6,a Stress-Induced Cellular Ligand for the Natural Killer Cell Receptor NKp30. J. Mol. Biol. 2016, 428, 4457–4466. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, Y.; Gauthier, L.; Chen, Q.; Vivier, E.; Mariuzza, R.A. Expression, crystallization and X-ray diffraction analysis of a complex between B7-H6, a tumor cell ligand for the natural cytotoxicity receptor Nkp30, and an inhibitory antibody. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Kolek, S.A.; Brauning, B.; Stewart, P.D. A novel microseeding method for the crystallization of membrane proteins in lipidic cubic phase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2016, 72, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Abuhammad, A.; McDonough, M.A.; Brem, J.; Makena, A.; Johnson, S.; Schofield, C.J.; Garman, E.F. “To cross-seed or not to cross-seed”: A pilot study using metallo-β-lactamases. Crys. Growth Des. 2017, 17, 913–924. [Google Scholar] [CrossRef]
- D’Arcy, A.; Villard, F.; Marsh, M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. 2007, 63, 550–554. [Google Scholar]
- Rumpf, T.; Gerhardt, S.; Einsle, O.; Jung, M. Seeding for sirtuins: Microseed matrix seeding to obtain crystals of human Sirt 3 and Sirt 2 suitable for soaking. Acta Crystallogr. Sect. F Struct. Boil. Commun. 2015, 71, 1498–1510. [Google Scholar]
- Yan, E.K.; Cao, H.L.; Zhang, C.Y.; Lu, Q.Q.; Ye, Y.J.; He, J.; Huang, L.J.; Yin, D.C. Cross-linked protein crystals by glutaraldehyde and their applications. RSC Adv. 2015, 5, 26163–26174. [Google Scholar] [CrossRef]
- Lee, T.S.; Turner, M.K.; Lye, G.J. Mechanical stability of immobilized biocatalysts (CLECs) in dilute agitated suspensions. Biotechnol. Prog. 2002, 18, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Noritomi, H.; Sasanuma, A.; Kato, S.; Nagahama, K. Catalytic properties of cross-linked enzyme crystals in organic media. Biochem. Eng. J. 2007, 33, 228–231. [Google Scholar] [CrossRef]
- Roy, J.J.; Abraham, T. Continuous biotransformation of pyrogallol to purpurogallin using cross-linked enzyme crystals of laccase as catalyst in a packed-bed reactor. J. Chem. Technol. Biotechnol. 2006, 81, 1836–1839. [Google Scholar] [CrossRef]
- Khalaf, N.; Govardhan, C.P.; Lalonde, J.J.; Persichetti, R.A.; Wang, Y.F.; Margolin, A.L. Cross-linked enzyme crystals as highly active catalysts in organic solvents. J. Am. Chem. Soc. 1996, 118, 5494–5495. [Google Scholar] [CrossRef]
- Iimura, Y.; Yoshizaki, I.; Rong, L.; Adachi, S.; Yoda, S.; Komatsu, H. Development of a reusable protein seed crystal processed by chemical cross-linking. J. Cryst. Growth 2005, 275, 554–560. [Google Scholar] [CrossRef]
- Yan, E.K.; Zhao, F.Z.; Zhang, C.Y.; Yang, X.Z.; Shi, M.; He, J.; Liu, Y.L.; Liu, Y.; Hou, H.; Yin, D.C. Seeding Protein Crystallization with Cross-Linked Protein Crystals. Cryst. Growth Des. 2018, 18, 1090–1100. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.F.; Zhang, C.Y.; Guo, Y.Z.; Xie, S.X.; Zhou, R.B.; Cheng, Q.D.; Yan, E.K.; Liu, Y.L.; Lu, X.L.; et al. A protein crystallisation screening kit designed using polyethylene glycol as major precipitant. CrystEngComm 2015, 17, 5488–5495. [Google Scholar] [CrossRef]
- Hou, H.; Liu, Y.; Wang, B.; Jiang, F.; Tao, H.R.; Hu, S.Y.; Yin, D.C. Recrystallization: A method to improve the quality of protein crystals. J. Appl. Cryst. 2015, 48, 758–762. [Google Scholar] [CrossRef]
- Hou, H.; Shi, M.; Chen, Z.H.; Ahmad, F.; Liu, Y.; Deng, X.D.; Yin, D.C. A high-performance protein crystallization plate pre-embedded with crosslinked protein microcrystals as seeds. CrystEngComm. 2018, 20, 4713–4718. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillaion mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Bourenkov, G.P.; Popov, A.N. Optimization of data collection taking radiation damage into account. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 409–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carugo, O. Atomic displacement parameters in structural biology. Amino Acids. 2018, 50, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Liu, Q.; Qu, G.; Feng, Y.; Reetz, M.T. Utility of B-Factors in Protein Science: Interpreting Rigidity, Flexibility, and Internal Motion and Engineering Thermostability. Chem. Rev. 2019, 3, 1626–1665. [Google Scholar] [CrossRef]
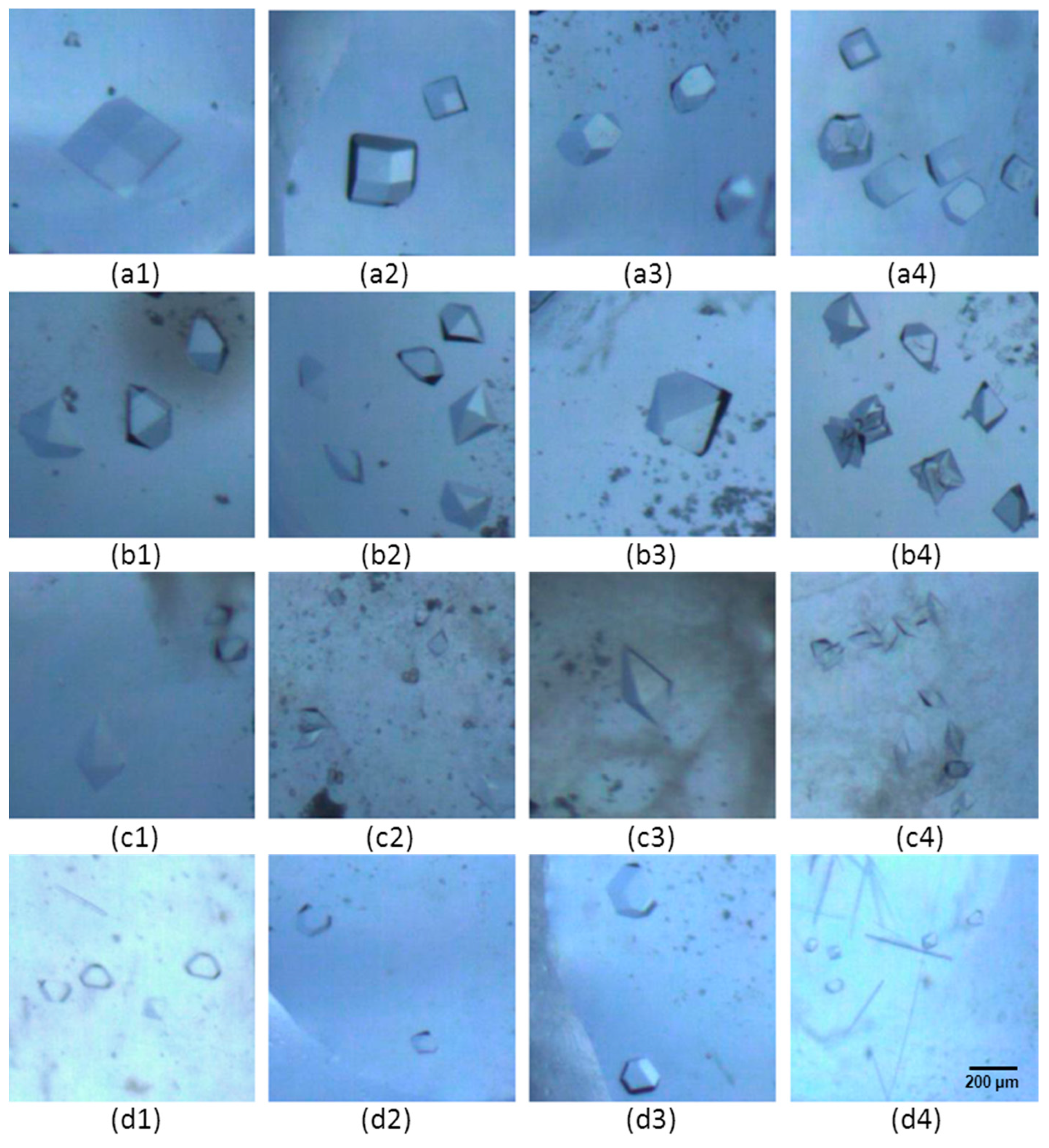
| Protein | Initial C (mg mL−1) | T (K) | Crystallization Time (days) | Buffer | Precipitant |
|---|---|---|---|---|---|
| lysozyme | 70 | 293 | 2 | 0.1 M sodium acetate pH 4.60 | 80 mg mL−1 NaCl |
| Proteinase K | 30 | 293 | 2 | 0.05 M sodium cacodylate, 0.08 M magnesium acetate pH 6.50 | 20% (w/v) PEG 8000 |
| Thaumatin | 20 | 293 | 5 | 0.1 M HEPES-Na pH 7.00 | 0.2 M Potassium sodium tartrate tetrahydrate, 20% (w/v) PEG 3350 |
| Glucose isomerase | 7 | 293 | 3 | 0.1 M HEPES-Na pH 7.00 | 0.02 M magnesium chloride hexahydrate, 22% (w/v) poly (acrylic acid sodium salt) 5100 0.1 M HEPES, pH 7.5 |
| Crystallographic Data | Lysozyme | Proteinase K | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | Seeds 1 | Seeds 2 | Seeds 3 | Control | Seeds 1 | Seeds 2 | Seeds 3 | |
| Space group | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 |
| Cell dimensions | ||||||||
| a (Å) | 76.75 | 76.75 | 78.75 | 76.65 | 68 | 68.14 | 67.94 | 68.04 |
| b (Å) | 76.75 | 76.75 | 78.75 | 76.65 | 68 | 68.14 | 67.94 | 68.04 |
| c (Å) | 38.45 | 37.15 | 37.13 | 37.55 | 101.83 | 101.09 | 101.65 | 101.93 |
| α (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| β (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| γ (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Resolution (Å) | 50–1.87 | 50–1.67 | 50–1.67 | 50–1.78 | 50–1.91 | 50–1.71 | 50–1.89 | 50–1.76 |
| I/σ I | 25.53 (4.12) | 48.91 (4.96) | 44.24 (2.88) | 24.98 (2.19) | 14.65(2.09) | 22.54 (2.25) | 17.67 (2.07) | 18.07 (2.03) |
| Mosaicity | 0.71 | 0.34 | 0.33 | 0.5 | 0.44 | 0.38 | 0.42 | 0.34 |
| Redundancy | 3.3 (2.1) | 7.3 (6.1) | 7.9 (5.6) | 5.0 (2.2) | 5.9 (3.2) | 6.2 (3.5) | 6.4 (3.4) | 5.7 (3.4) |
| Completeness (%) | 90.1 (71.2) | 99.6 (99.1) | 99.6 (98.1) | 96.1 (72.2) | 98.6 (85.8) | 99.4 (92.6) | 94.9 (85.7) | 99.4(92.2) |
| No. reflections | 30030 (9041) | 102123 (14047) | 109611 (13960) | 53896 (10792) | 112066 (19009) | 163657 (26610) | 119827 (18797) | 141447 (24371) |
| Crystallographic Data | Thaumatin | Glucose Isomerase | |||
|---|---|---|---|---|---|
| Control | Seeds 1 | Seeds 2 | Seeds 3 | Seeds 3 | |
| Space group | P43212 | P21212 | P43212 | P43212 | P21212 |
| Cell dimensions | |||||
| a (Å) | 58.24 | 58.15 | 57.72 | 57.6 | 84.59 |
| b (Å) | 58.24 | 58.15 | 57.72 | 57.6 | 92.57 |
| c (Å) | 150.38 | 150.33 | 149.75 | 150.01 | 98.95 |
| α (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| β (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| γ (°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
| Resolution (Å) | 50–2.24 | 50–2.10 | 50–2.18 | 50–1.96 | 50–2.45 |
| I/σ I | 21.1 (3.24) | 21.31 (2.01) | 16.02 (2.11) | 20.3 (2.1) | 9.73 (2.08) |
| Mosaicity | 0.65 | 0.66 | 0.56 | 0.49 | 0.74 |
| Redundancy | 5.4 (2.8) | 5.1 (2.5) | 6.4 (3.6) | 6.8 (3.7) | 3.4 (2.2) |
| Completeness (%) | 95 (79.6) | 96.5 (70) | 99.2 (88.8) | 98.6 (87) | 94 (77.5) |
| No. reflections | 67058 (12531) | 77875 (15312) | 89329 (13900) | 127704 (18732) | 47421 (13810) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Yan, E.-K.; Liu, Y.; Wu, Z.-Q.; Liu, Y.-L.; Hou, H.; Zhang, C.-Y.; Lu, Q.-Q.; Deng, X.-D.; Yin, D.-C. Comparison of the Quality of Protein Crystals Grown by CLPC Seeds Method. Crystals 2019, 9, 501. https://doi.org/10.3390/cryst9100501
Li J, Yan E-K, Liu Y, Wu Z-Q, Liu Y-L, Hou H, Zhang C-Y, Lu Q-Q, Deng X-D, Yin D-C. Comparison of the Quality of Protein Crystals Grown by CLPC Seeds Method. Crystals. 2019; 9(10):501. https://doi.org/10.3390/cryst9100501
Chicago/Turabian StyleLi, Jin, Er-Kai Yan, Yue Liu, Zi-Qing Wu, Ya-Li Liu, Hai Hou, Chen-Yan Zhang, Qin-Qin Lu, Xu-Dong Deng, and Da-Chuan Yin. 2019. "Comparison of the Quality of Protein Crystals Grown by CLPC Seeds Method" Crystals 9, no. 10: 501. https://doi.org/10.3390/cryst9100501
APA StyleLi, J., Yan, E.-K., Liu, Y., Wu, Z.-Q., Liu, Y.-L., Hou, H., Zhang, C.-Y., Lu, Q.-Q., Deng, X.-D., & Yin, D.-C. (2019). Comparison of the Quality of Protein Crystals Grown by CLPC Seeds Method. Crystals, 9(10), 501. https://doi.org/10.3390/cryst9100501






