Abstract
Knowledge of hydrogen locations and protonation states is critical for a fundamental understanding of biological macromolecular function/interactions, and neutron macromolecular crystallography (NMX) is uniquely suited among the experimental structural-determination methods to provide this information. However, despite its potential, NMX remains a relatively niche technique, due to substantial limitations. This review explores NMX’s role amongst the evolving landscape of structural biology, comparing and contrasting it to the historical gold standard of X-ray macromolecular crystallography (X-ray MX) and the increasingly prevalent electron-based methods—i.e., electron microscopy (EM) and electron diffraction (ED). Forthcoming developments (e.g., the European Spallation Source in Lund, Sweden, coming online) are expected to substantially address current limitations and ensure NMX will remain relevant in the coming decades.
1. Introduction
The ability to determine biological macromolecular structures at atomic resolution is critical for understanding both their normal and pathophysiological function. For example, the ability to precisely determine the atomic coordinates of a protein allows for the elucidation of their catalytic mechanism, an understanding how they may be either activated or inhibited by regulatory molecules, and the rational design of drugs specifically targeting them. The first protein structure to be solved at atomic resolution was myoglobin, by John Kendrew in 1959 [1], a feat achieved by analysing the diffraction patterns which resulted from exposing a crystal of the protein with X-rays. In the following decades, X-ray macromolecular crystallography (X-ray MX) became the gold-standard method for determining protein structures at atomic resolutions, with more than one-hundred thousand structures solved to date [2].
Despite its success, X-ray MX can be challenging for some proteins which are difficult to crystalize—e.g., membrane proteins or proteins with flexible domains [3,4]. Historically, as there were no viable alternatives to the method, no crystal meant no structure. However, a series of recent technological advances in electron microscopy (EM) and electron diffraction (ED) has allowed atomic resolution structures to be determined either from samples in solution or crystals of microscopic size (typically a few µm in length/width and several hundred nm in thickness [5]). Consequently, these electron-based techniques have exploded in popularity over the past few years. Furthermore, rapid progress in artificial intelligence has resulted in the emergence of AlphaFold [6], a program which predicts the 3D structure of proteins from their amino acid sequence alone. Though an in silico method, AlphaFold has produced structures for a large number of proteins, and is often able to provide valuable insight into proteins which, for one reason or another, are unable to have their structures solved experimentally. However, it is still important to experimentally validate predicted structures where possible, as even very high-confidence predictions can differ from empirical data at both the global and local scale [7]. Thus, rather than competing with, predictive methods will greatly complement the experimental techniques, and serve as valuable high-quality starting models to be refined by experimentally derived maps (a workflow already implemented in the PHENIX software suite [8]).
Thus, given the current trend within the field of structural biology, where the crystallization of samples appears to be losing relevance as alternate methods gain traction, one naturally wonders about the future prospects of neutron macromolecular crystallography (NMX). In short, despite significant competition and challenges, NMX may be yet to reach its heyday. Indeed, the continued relevance of NMX relies upon one key issue: hydrogens. Hydrogen atoms (H) are key constituents of biological macromolecules—where H represents approximately half of all atoms—and macromolecular function depends heavily upon the hydrogen bonding networks, protonation states, and proton (H+) transfers at the relevant site(s) [9,10]. The ability to detect individual hydrogens is therefore critical for a complete understanding of protein activities and their interactions. Of all the structure determination methods, NMX is (and can be expected to remain) the most powerful for experimentally locating hydrogens.
This concise review will provide the following: (a) a brief overview of NMX and how it compares to other methods; (b) a detailed account of the practicalities of implementing NMX for biological macromolecular targets; (c) a presentation of two cases studies where NMX was able to provide essential insights; and (d) a discussion of the future prospects of NMX within the field of structural biology.
2. Biological Macromolecular Structure Determination
The following is a short summary of the main experimental methods used to determine the atomic structure of biological macromolecules (nuclear magnetic resonance, or NMR, will not be presented here; please refer to [11,12] for a comprehensive review of NMR for biological applications).
Currently, the field of structural biology as a whole is in a state of rapid development, as evidenced by the fact the all-time total of protein structures deposited in the Protein Data Bank (PDB) [2] has doubled in less than ten years, going from 106,569 structures by end-of-year 2015 to 200,775 by end-of-year 2023. To gain insight into the relative prevalence of the different methods, one can look at the total PDB protein structure deposits (as of 9 April 2024) for each: X-ray diffraction (180,967); electron microscopy (19,357); electron diffraction (233); and neutron diffraction (218).
This data highlights the dominance of X-ray MX—which has evolved considerably over the past few decades to encompass a variety of techniques—and has solved an order-of-magnitude more structures than EM, the second most prolific technique, and nearly three orders-of-magnitude more than both ED and NMX. However, these all-time totals do not capture current trends, which can be revealed by the number of protein structures added to the PDB each year for each method over the past decade (Figure 1). In this context, while X-ray MX remains dominant, the increasing prominence of EM (and very recently, ED) is clear. Meanwhile, though it is relatively mature, NMX has remained a niche technique.
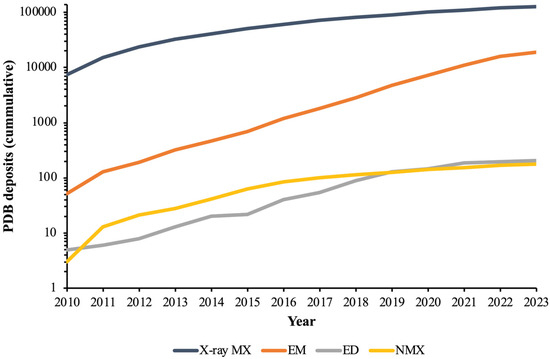
Figure 1.
Cumulative number of protein structures added to the PBD database between the years 2010 and 2023 for the following experimental methods: X-ray MX, EM, ED, and NMX.
To understand this trend, a brief overview of each technique and their relative advantages/disadvantages follow.
2.1. X-ray Macromolecular Crystallography
For a long time, the standard way to determine biological macromolecular structures at atomic resolution was to analyse X-ray diffraction patterns from a crystalline sample. A crystalline sample is required to overcome the weak diffraction provided by individual molecules (the ordered lattice of crystals allows scattered waves to add up in phase, amplifying the signal to observable intensities), while X-rays are required as the resolution able to be achieved is limited to approximately half the wavelength of the scattered particle (the wavelength of X-rays used on standard synchrotron beamlines typically ranges from 0.7 to 2 Å, with the most commonly used wavelength being ~1 Å [13]; for a comprehensive examination of the theory behind crystallography, please refer to [14]).
Another factor to consider is the nature by which the probing particles interact and scatter off the atoms constituting a sample. As photons, X-rays possess a fluctuating electric field, accelerating charged particles which then emit a photon in response (i.e., the scattered particle). Since electrons have a much higher charge-to-mass ratio than atomic nuclei (~2000 times that of a proton), X-rays scatter mainly from electron density and are thus disproportionately sensitive towards heavy elements [14].
Today, the X-ray sources required to analysis one’s sample are readily available. Many have access to laboratory-scale devices—typically utilizing rotating anode sources and charged-coupled device (CCD) detectors—which are capable of analysing crystals a few hundred µm in size over several hours. However, current-generation synchrotron facilities, which offer beam fluxes of ~1013 ph/sec, allow data to be collected from µm-sized crystals in under a minute. This high flux, combined with the ability for remote operation, has made synchrotron facilities the go-to for biological macromolecular determination. Finally, in addition to synchrotron sources, X-ray free-electron laser (XFEL) facilities provide extraordinary bright X-ray pulses on femtosecond timescales, opening up new possibilities for time-resolved studies of fundamental biological processes.
The substantial resources directed towards X-ray MX over the past several decades has resulted in the development of advanced techniques which are available to users in addition to the canonical procedure. Standard X-ray MX experiments involve the analysis of single crystals (either loop- or capillary-mounted for cryogenic or room-temperature (RT) data collection, respectively) which are rotated within the incident beam in order to capture all possible Bragg peaks. However, the high-flux beams of modern synchrotron and XFEL facilities have allowed for other possibilities. These include ‘serial crystallography’, in which individual exposures from thousands of randomly orientated microcrystals are collected in succession. As each crystal is exposed only once in serial crystallography, radiation damage is minimized, allowing structures to be more easily solved at physiologically relevant temperatures [15]. Critically, this technique allows for time-resolved experiments aimed at linking structure to function—e.g., how structural changes over time allow for the light-driven transport of sodium ions [16]—by first initiating processes within sample microcrystals (typically using either light or diffusion activation), then probing the samples with X-rays after defined time intervals [17]. Other advanced X-ray MX methods have been developed, including automated handling, in situ data collection, precise control over the crystallization process, and their combinations [18,19,20,21,22,23].
Because of this half-century of development—the readily available high-flux sources, almost fully automated sample handling and data processing pipelines, and the gathering of vast amounts of knowledge and experience—X-ray MX has become an extremely accessible and high-throughput method. If one’s sample can be crystallized, X-ray diffraction provides the most reliable and efficient method for determining biological macromolecular structures at atomic resolution. However, as mentioned above, X-rays are inherently insensitive to lighter elements, and even the highest-resolution X-ray structures have difficulty resolving hydrogens. Thus, X-ray MX is often unsuitable for addressing biological questions that require detailed knowledge of hydrogen positions.
2.2. Electron Microscopy
In contrast to X-ray and NMX, in which structural information is obtained by analysing diffraction patterns, the EM method is akin to traditional microscopy. However, as resolution in microscopy is limited to approximately half the wavelength of the illuminating beam, to enable atomic resolutions, electrons are used in place of visible-light photons (typically, electrons of 100–300 keV are used in EM, corresponding to wavelengths of 0.020–0.037 Å).
Rather than undergoing crystallization, biological macromolecules are instead stored in a thin film of vitrified ice under cryogenic conditions, in order to keep them hydrated within the high-vacuum environment of an EM sample chamber and to protect them from radiation damage following exposure to the electron beam. This is achieved by dispensing the solubilized sample on a holey carbon or gold grid, blotting away excess liquid, then rapidly freezing the sample by plunging it into liquid ethane. Though this process requires substantial optimization in its own right, it enables structural determination of specific conformations, macromolecular types, and complexes that are unable to be crystallized and thus unsuitable for MX studies. Furthermore, the quantities of sample required are minimal compared to MX, with only a few micrograms of sample needed. Prepared grids, containing hundreds-of-thousands to millions of single macromolecules, are imaged. These images contain 2D projections at various random orientations, which may then be combined to obtain a 3D volume of the sample [24].
Historically, the resolutions able to be achieved with EM precluded the ability to visualize individual atoms. However, a ‘resolution revolution’ over the past few years has allowed structures at near-atomic (~1 Å) resolutions to be obtained [25,26]. Indeed, high-resolution macromolecular structures are now routinely achieved, thanks to several recent technology advancements in data collection and processing which have significantly improved signal-to-noise. For their 1.7 Å structure of the β3 GABAA receptor, Nakane et al. [27] employed a new electron source (a cold field-emission gun producing an electron beam with a narrow energy spread), an improved energy filter (to remove inelastically scattered electrons), and a direct electron detector (offering increased sensitivity). By contrast, for their 1.25 Å structure of apoferritin, Yip et al. [28] employed an improved monochromator (to reduce the energy spread of the electron beam) combined with a novel spherical aberration corrector (to minimize optical aberrations).
Such developments have greatly expanded the number of proteins able to be solved to high-resolution (2–4 Å) by cryo-EM, including small soluble proteins and membrane proteins [26].
2.3. Electron Diffraction
Electron diffraction (ED) is a method in which a structure is determined by rotating frozen crystalline samples in an electron beam while diffraction is recorded. Though similar to its X-ray and neutron counterparts, ED is only now just entering the mainstream, due to recent technological advancements needed for the method to be suitable for radiation-sensitive biological macromolecules, namely sufficiently fast and sensitive detectors [29].
The main advantage of ED is that it allows complete datasets to be collected from a low number of extremely small micro- and nanocrystals, a feat extremely difficult to achieve by other means. For example, while the high brilliance of XFELs allows analysis of crystals of ~1 µm size, as samples are destroyed immediately following exposure, thousands of randomly orientated crystals must be serially exposed in order to yield a complete dataset [29]. For ED, once crystalline material has been identified and vitrified on grids (as per the EM method), crystal samples are ion-beam milled to an optimal thickness of ~100–300 nm (thinner samples possess reduced scattering power and are susceptible to physical deformations, and thicker samples suffer from increased diffuse/inelastic scattering) [30].
Prepared samples are then loaded into an appropriately set up cryogenic transmission electron microscope (cryo-TEM), i.e., microscopes equipped with a fast readout detector (such as CMOS-based cameras), a field-emission gun (FEG) as an electron source (for increased beam coherence), and an energy filter (to reduce noise and help identify faint reflections) [29].
As the collected diffraction data are essentially analogous to those from X-ray diffraction experiments, standard crystallographic software is used for data processing (while phases can be determined using direct ab initio methods for small molecules/peptides, in general, molecular replacement is used for proteins).
2.4. Neutron Macromolecular Crystallography
NMX operates on the same general principle as X-ray MX, except that a crystalline sample amplifies diffraction events from an incident beam of neutrons. This use of neutrons differentiates NMX in two key respects. First, as electrically neutral particles (and lacking a fluctuating electric field, as with photons), neutrons interact almost exclusive with atomic nuclei. As a result, neutrons are relatively sensitive towards the lighter elements, making NMX ideal for detecting hydrogens (a topic discussed in detail in Section 3). Second, as a nucleon, the ability to generate an incident beam with sufficient intensity to analyse biological macromolecules remains a significant challenge. This is the main disadvantage of NMX: the nature of currently available neutron sources, and the relatively large size of biological macromolecules themselves, combine to severely limit its application, despite the method having its beginnings in the early 1980s [31].
Current neutron sources possess incident-beam flux orders of magnitudes below that of their X-ray- and electron-source counterparts: e.g., the Quasi-Laue Diffractometer (LADI-III) at the Institut Laue-Langevin (ILL; Grenoble, France) provides a flux of ~1.1 × 108 n cm−2 s−1 [32]. As diffracted intensity is proportional to the incident-beam intensity and inversely proportional to the unit-cell volume squared (Equation (1)), the weak fluxes of neutron sources and the large unit cells of protein crystals result in low Bragg peak intensities.
where ⟨Ι⟩ is the diffracted intensity, Ι0 is the incident beam intensity, |F| is the structure factor magnitude, V is crystal volume, λ is the incident wavelength, and ν0 is the unit cell volume. Equation from O’Dell et al. [11]. However, as diffracted intensity is also proportional to crystal volume, a large crystal size is able to compensate for an otherwise weak signal. The typical sample size required for current generation of neutron facilities is ~1 mm3, with a lower limit of ~0.1–0.2 mm3 for fully deuterated samples (the growth of large crystals and the deuteration of samples is covered in Section 4).
Once a suitable sample is obtained, neutron diffraction experiments are able to be performed only at a handful of facilities around the world, of which there are two main types: nuclear reactors and pulsed spallation sources. Reactors use the nuclear fission of uranium atoms to generate a neutron beam, while spallation sources use a particle accelerator to fire high-intensity proton pulses at a heavy metal target (e.g., tungsten), resulting in the ejection of a large number of neutrons.
NMX instruments employ one of three main data collection methods, depending on the nature of the neutron source: monochromatic or quasi-Laue for reactor-based facilities, and time-of-flight (TOF) for spallation-based facilities [33] (note that while reactor sources are able to implement TOF methods, typically by employing a mechanical ‘chopper’ to time-slice the neutron beam, low fluxes limit reactor-based TOF instruments to spectroscopy or small-molecule powder-diffraction experiments [34,35,36,37,38]). Currently available NMX instruments and the data collection method are shown in Table 1.

Table 1.
Summary of currently available NMX facilities.
For monochromatic diffraction analysis, the incident neutron beam is filtered to a single wavelength by a graphite or silicon monochromator. Such a beam produces well-defined Bragg peaks that are relatively easy and convenient to process (as it is analogous to data processing in standard X-ray MX). On the other hand, the low number of reflections observed per image means hundreds of exposures are required to obtain a complete dataset, necessitating collection times in the order of weeks. For quasi-Laue diffraction analysis, source neutrons within a wavelength range (a typical wavelength range spans ~1 Å) allow more Bragg peaks to be captured per exposure, substantially reducing collection time (tens instead of hundreds of exposures). For samples with large unit cells, however, the use of multiple wavelengths can result in overlapping reflections and increased background from incoherent scattering [33]. Finally, TOF diffraction analysis combines the low-background/low-reflection overlap of a monochromatic beam with the high measurement efficiency of Laue diffraction [39]. This is achieved by exploiting the fact that a particle’s wavelength is inversely proportional to its momentum [40]. For massive particles such as neutrons, variation in wavelength corresponds to a significant change in velocity. Thus, given the relatively low velocities of the cold neutrons used in NMX experiments (at λ = 2 Å, neutrons have a velocity of 3956 m/s) [41], neutrons of different wavelengths are able to be filtered by their TOF between the source and detector.
For monochromatic or quasi-Laue NMX experiments, generalized MX programs may be used to process the neutron diffraction data. Meanwhile, for TOF experiments, beamline specific programs are required for data processing, e.g., STARGazer [42] and Mantid [43]. For structural refinement, joint X-ray and neutron refinement is often performed (ideally, from X-ray and neutron data collected from the same sample), using MX software such as Phenix [44]. The addition of X-ray diffraction data is valuable, as they provide high-resolution information for heavy elements (e.g., carbon, oxygen, and nitrogen), resulting in better-quality maps/improved R factors. Finally, OMIT maps are used to unambiguously locate hydrogens throughout the obtained structure. OMIT maps are widely used to verify whether particular atoms are present in the studied molecule, and are calculated by removing the atoms-of-interest from the model, updating the corresponding structure factors, and computing a residual map [45].
3. Hydrogen Determination in Biological Macromolecules
H atoms are key constituents of biological macromolecules, whose function depends heavily on the hydrogen bonding networks, protonation states, and proton transfers that occur at vital catalytic and/or binding sites. The ability to detect individual hydrogens is therefore critical for a complete understanding of biological macromolecule activities and their interactions [11]. It is in this context that NMX demonstrates significant advantage over the other methods, due to the nature of how they interact with matter vs. X-rays and electrons (Figure 2).
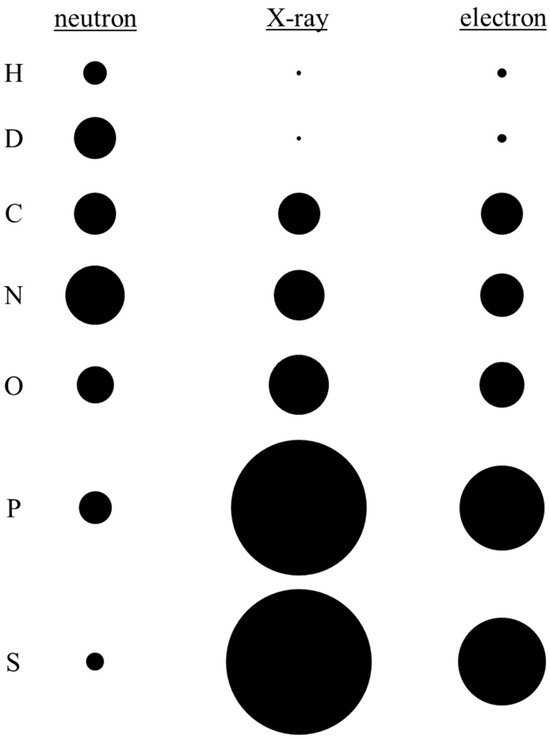
Figure 2.
Neutron, electron, and X-ray scattering cross-sections for the major elemental constituents of biological macromolecules: hydrogen (H); deuterium (D); carbon (C); nitrogen (N); oxygen (O); phosphorus (P); and sulfur (S). The scattering cross-sections are normalized at element C for direct comparison.
3.1. X-rays
As previously mentioned, X-rays scatter according to electron density, and are thus disproportionately insensitive towards lighter elements—the limited photon-scattering power provided by a single electron makes hydrogens particularly difficult to detect. Consequently, even at ultra-high resolutions (≤1 Å, where a significant portion of hydrogens may be located within a protein), highly polarized H atoms and H+ ions (often the most relevant) remain essentially invisible [46]. Consequently, it is widely recognized within the field of structural biology that X-ray MX is generally inappropriate for answering scientific questions pertaining to hydrogens.
3.2. Electrons
However, this situation improves for the electron-based techniques, where recent developments in both EM and ED have suggested they may potentially compete with NMX in terms in locating hydrogens. As charged particles, electrons interact both with other electrons and atomic nuclei. Accordingly, in comparison to X-rays, hydrogens have a relatively large electron-scattering cross-section [47,48]. Thus, for a given resolution, hydrogens should be more visible in structures solved using electrons than for those solved with X-rays. Regarding EM, two recent high-resolution structures—a 1.2 Å resolution structure of apoferritin (PDB ID: 8J5A) [49] and a 1.7 Å structure of the β3 GABAA receptor (PDB ID: 7A5V) [27]—demonstrated that large numbers of individual hydrogens can be revealed by the method. In the apoferritin EM structure, ~70% hydrogen atoms were revealed—i.e., positive densities ≥ 2σ in the hydrogen OMIT map, which corresponded to riding positions. Furthermore, within the apoferritin core, a majority of main-chain and side-chain hydrogens could be identified, including those involved in hydrogen bonding and/or associated with waters (Figure 3). Meanwhile, in the case of the β3 GABAA receptor, hydrogen bonding networks at well-ordered regions and ordered waters at the ligand-binding site were able to be observed.
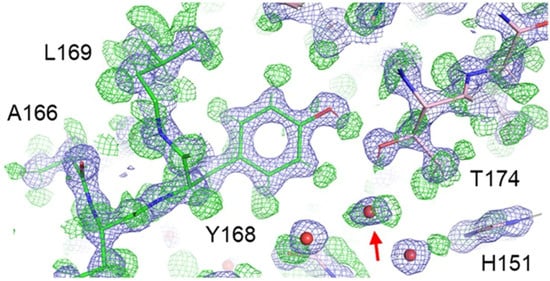
Figure 3.
Local environment at Tyr168 of the 1.2 Å apoferritin structure obtained by EM. Experimental densities (blue mesh) and positive densities in the hydrogen OMIT map (green mesh). A water molecule with both hydrogens resolved is highlighted (red arrow). Figure from Maki-Yonekura et al. [49].
Nevertheless, it is critical to note that these two structures represent the extremes of what is possible to obtain with EM; the majority of structures determined by EM will be at resolutions of ~2 Å (or lower), beyond the limit required to unambiguously detect hydrogens. Indeed, apoferritin represents the absolute best-case scenario—due to a unique combination of high-stability and 24-fold symmetry, it is used to benchmark the high-resolution limit able to be achieved by EM facilities. In the case of the β3 GABAA receptor, its five-fold symmetry allows for more efficient data collection than for most samples, and even with the 1.7 Å structure obtained, hydrogens are unable to be resolved at the critical ligand-binding site. Thus, to reliably visualize biological macromolecular hydrogens with EM, resolutions of ~1 Å will need to be approached (a substantially challenging endeavour). Even then, the vast majority of hydrogens identified will be limited to those of the unlabile variety (i.e., those bonded to carbon atoms) in the most well-defined, relatively rigid regions.
It is a similar situation for ED, though this method is perhaps more suitable for identifying hydrogens than EM. In a recent study investigating the potential to detect hydrogens with ED, Clabbers et al. [50] analysed a 0.87 Å structure of hen egg-white lysozyme (PDB ID: 7ULY), obtained using data collected from 16 microcrystals ion-beam milled to an optimal thickness of ~300 nm and using an electron-counting mode with an exposure of 0.64 e− Å−2 per crystal [51]. The use of low electron exposures is critical, as radiation damage negatively affects sample integrity and the ability to localize hydrogens [52]. To identify hydrogens, Clabbers et al. [50] first generated a hydrogen OMIT map, then selected peaks ≥ 2.0 σ and located within 0.5 Å of any idealized riding position, resulting in 367/1067 (~35%) hydrogens being identified. It is important to note that while lower contour levels increased the number of hydrogens identified, this led to substantially increased noise, making unambiguous identification significantly more challenging.
With respect to the main chain, the authors identified 61/141 (~43%) of Cα-H and 76/127 (~60%) of peptide N–H hydrogens. For side chains—generally more difficult to resolve due to their increased flexibility—several hydrogen bonding arrangements/protonation states were identified, including a 2.4 σ peak between His15-NE2 and Thr89-OG1, indicative of a hydrogen bond and the protonation of His15-ND1. For the equivalent X-ray structure solved at 0.93 Å [53], difference maps contoured at 1.8 σ showed 77/127 (~60%) of peptide N–H hydrogens, with a similar number of hydrogen atoms being identified overall.
In general, both EM and ED are expected to better resolve hydrogens atoms at a given resolution than X-ray MX, due to the comparatively large electron-scattering factors for light elements. Indeed, this was noted to be the case in an analysis by Yamashita et al. [54], which compared the number of hydrogens able to be observed for multiple (apo)ferritin structures obtained either by EM or X-ray MX. However, the same study found that as the resolution of the EM structures decreased from 1.25 Å to 1.84 Å to 2.0 Å, the number of hydrogens observed went from ~70% to ~17% to only a few. Thus, while the electron-based methods are capable of locating hydrogens within biological macromolecules, as with X-rays, this will generally require atomic-level resolutions. This is a substantial challenge for most samples—likely only achievable for relatively few cases—and leads to the conclusion that EM and ED are not the most suitable methods for mapping hydrogens in biological macromolecules.
3.3. Neutrons
As opposed to the other structural determination methods, NMX is uniquely suited for detecting hydrogens within one’s sample. As neutrons scatter from atomic nuclei almost exclusively, they are the most sensitive towards light elements. In fact, hydrogen (and its isotope deuterium) possesses a neutron-scattering cross-section comparable in magnitude to that of carbon, nitrogen, and oxygen (Figure 2), thus enabling them to be reliably detected at the resolutions typically achieved by protein crystals (~2.5 Å). The efficacy of NMX in detecting hydrogens at site-of-interests within biological macromolecules—i.e., the functionally relevant hydrogens—will be demonstrated through two case studies: one investigating the catalytic mechanism of urate oxidase (Section 5) and the other detailing inhibitor binding to the SARS-CoV-2 main protease (Section 6).
However, there are unique considerations to be aware of when performing neutron diffraction studies on biological macromolecular samples, chiefly the following: crystallization, deuteration, data collection temperatures, and the lack of radiation damage. These topics will be explored in detail in the following section.
4. NMX: Practical Considerations for Biological Macromolecules
4.1. Sample Crystallization
Producing the large, well-diffracting protein crystals required for NMX is the main limiting factor of the technique. While the other structural determination methods are relatively tolerant towards sample size (X-ray MX and ED are capable of analysing µm- or even nm-sized crystals, and EM does not require crystals at all), as explained in Section 2.4, NMX requires relatively extreme crystal sizes in order to compensate for low source fluxes [55]. The relative size difference between the crystals needed by each diffraction-based structural determination method can be visualized in Figure 4.
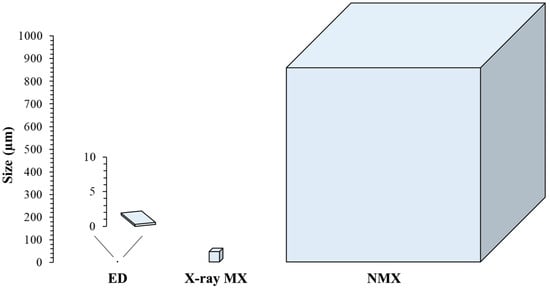
Figure 4.
Approximate relative size of the crystalline sample required for each experimental method utilizing diffraction (ED, X-ray MX, and NMX). Note the extremely small crystal sizes used in ED, which only appear as a few pixels when placed on the same scale as X-ray MX and NMX.
Thus, crystallization experiments aimed at generating samples for neutron diffraction studies often employ the use of advanced techniques and equipment.
Crystallization itself involves a phase transition in which a solid, crystalline state forms out of an aqueous solution once supersaturation is achieved [56]. Supersaturation, which is reached under specific physicochemical conditions, occurs when a solute (i.e., the sample) exists in solution at concentrations greater than the solubility limit [57]. As sample solubility is dependent upon a wide range of parameters—e.g., concentration, temperature, pH, solvent, ionic strength, added organic molecules—this phase space is multidimensional. In the process of crystallization, the kinetic pathway taken through this phase space determines nucleation and growth, and consequently crystal size, number, and morphology (Figure 5).
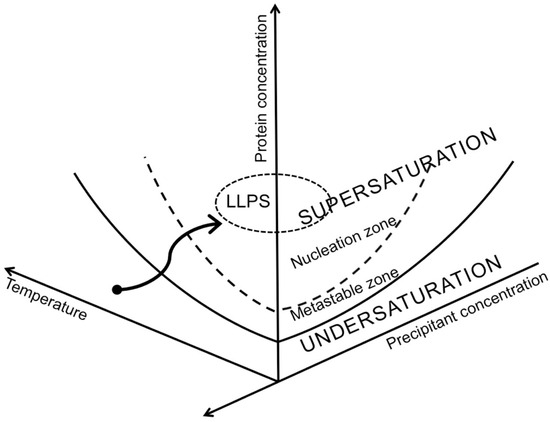
Figure 5.
Schematic view of a multidimensional crystallization-phase diagram. The solid line represents sample solubility as a function of both temperature and precipitant concentration. Once sample concentration falls to this limit, crystal growth can no longer occur. The dashed line represents the boundary above which supersaturation is high enough for nucleation (i.e., crystal formation) to spontaneously occur. The area between the solid line (solubility limit) and the dashed line is the region where supersaturation exists, but nucleation probability is zero and extended crystal growth takes place. For protein systems where short-range attractive interactions dominate, a demixion results—i.e., a metastable liquid−liquid phase separation (LLPS) where protein molecules condense into a dense phase that resembles liquid droplets, and this dense phase coexists with a dilute-protein phase. Illustrated by the finely dashed line, the location of the LLPS coexistence line relative to the solubility line indicates metastability with respect to the crystalline phase. Finally, the black arrow indicates an example kinetic trajectory taken through phase space during a crystallization experiment. Figure from Junius et al. [22].
To produce the large, mm3-scale crystals needed for NMX studies, control over the kinetics and thermodynamics of the crystallization process is required. Such control is able to be achieved by employing specially designed equipment allowing the fine control of phase space, such as the OptiCrys crystallization apparatus developed by Junius et al. [23]. The device consists of a sample-containing dialysis chamber, a flow-cell reservoir, a Peltier device for temperature control, and a microscope for real-time imaging. As a result, the OptiCrys allows the ability to reversibly manipulate the physicochemical parameters of the mother liquor by adjusting both the temperature of the chamber and the chemical composition of the reservoir solution. When combined with the ability to visually monitor samples in real time, crystallization phase space is able to be explored and characterized in order to identify the metastable zone (i.e., area of crystal growth). This knowledge, combined with the ability for fine physicochemical manipulations, enables crystal samples to undergo multiple rounds of growth, dissolution, and re-growth—resulting in high-quality samples of maximum size (Figure 6).
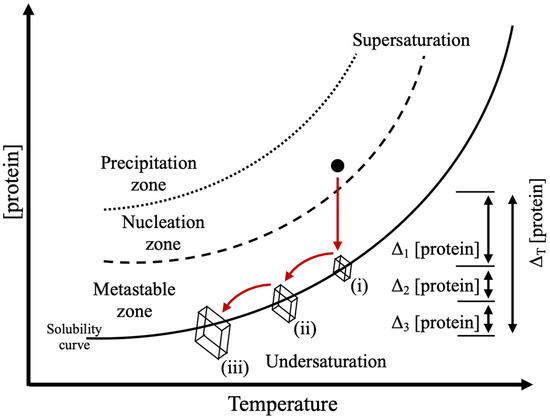
Figure 6.
Concept of producing crystal samples for NMX studies through physiochemical manipulation. A sample’s trajectory through phase space resulting in extended growth is shown (arrows). Protein concentration drops as crystal nuclei undergo an initial growth phase (i), which stops once the solubility limit is reached. Altering the physiochemical parameters of the solution by changing the temperature and/or composition of the reservoir solution lowers solubility and brings the sample into the metastable zone again, restarting crystal growth which continues until the protein concentration reaches the solubility limit again (ii). Large crystals can be obtained by repeating this process (iii), where multiple, successive growth phases (Δ1, Δ2, Δ3) result in a maximal contribution of protein (ΔT).
4.2. Sample Deuteration
Sample deuteration, in which hydrogens (Hs) are exchanged for deuterium (D), is often a prerequisite for NMX experiments. This is due to the fact that, while H has a coherent scattering cross section comparable to that of the other atoms which comprise biological macromolecules (i.e., carbon, oxygen, nitrogen, and sulfur), its large incoherent scattering cross-section produces significant background (Table 2). Another issue arises from the fact that the coherent scattering cross-section of H is negative. For the resolutions at which soft-matter crystals typically diffract (≥2 Å), the negative coherent scattering from H can destructively interfere with positive coherent scattering from the surrounding atoms (e.g., C, N, and O), resulting in cancellation effects which complicate analysis [11,58].

Table 2.
The coherent neutron-scattering length and incoherent neutron-scattering cross-sections for the major elemental constituents of biological macromolecules.
To mitigate the issues listed above, it is common practice to either partially or fully exchange H for D within one’s sample. In comparison to H, D has a coherent scattering cross-section that is both larger in magnitude and positive in sign, while also possessing a significantly reduced incoherent scattering cross-section (Table 1). As a result, for a given sample, H/D exchange results in improved signal-to-noise and reduced background, increasing the resolution limit of the collected data [11,46]. The improvement in data quality following deuteration allows NMX to be performed using either smaller sample sizes (enhancing accessibility) or reduced data-collection times (enhancing throughput).
Practically, there are several ways to incorporate D into biological samples, which able to be implemented at various stages between production and crystallization (and offering various degrees of deuteration). Perdeuteration, where the sample is expressed in D2O-containing media in the presence of a perdeuterated carbon source, is the most complete method; perdeuterated samples can achieve <99% D incorporation, including aliphatic carbon atoms (e.g., CH2 and CH3 groups) [59]. Perdeuterated samples offer the optimal signal-to-noise ratio, allowing faster data collection, higher resolutions, and less-stringent sample requirements (i.e., smaller crystals are needed) [58,60,61,62]. In particular, a major advantage offered by perdeuterated samples is that the aforementioned cancellation effects arising from the negative coherent neutron scattering of hydrogen scattering at moderate resolutions (~2 Å) are completely mitigated (see [58] for a direct comparison between a perdeuterated and partially deuterated sample, and [63] for an example protocol for perdeuterated-protein production). Thus, the improved neutron diffraction data offered by perdeuterated samples make it the preferable option for NMX studies.
However, perdeuteration is relatively expensive, due to the costs associated with using heavy water and a D-labelled carbon source [59]. Furthermore, expression yields are often lower under deuterated conditions [64], and in general the amount of valuable sample available for performing and optimizing NMX experiments is reduced. Thus, due to the challenges involved in perdeuteration, partially deuterated samples are often used. Partial deuteration may be performed at a variety of stages during a project. For example, the sample may be expressed in D2O-containing media in the presence of a hydrogenous carbon source; hydrogenous samples may be purified and/or solubilized in buffers prepared using D2O; samples may be crystallized in solutions prepared using D2O; and finally, crystals may be mounted inside a sealed capillary alongside deuterated precipitant solution (with H/D exchange taking place via vapor diffusion). Though an easier and more convenient method than perdeuteration, partial deuteration exchanges only solvent-accessible, labile hydrogens—i.e., H atoms bonded to oxygens or nitrogens, but not those incorporated into carbons [46]. While not optimal, partial deuteration allows incorporated D atoms to be visualized at resolutions of ~2.5 Å (due to the aforementioned cancellation effects, resolutions of ~1.5 Å are required to visualize H atoms). As biological questions often pertain to protonation states, hydrogen bonds, and coordinated waters at functional sites—often located in well-hydrated regions containing large numbers of polar/charged residues—partial deuteration is usually sufficient.
4.3. NMX, Radiation Damage, and Temperature Variation
While data collection at 100 K has long been standard for X-ray MX, chiefly to protect samples from radiation damage (it is also the standard for both EM and ED), room-temperature analysis is the default for NMX. This is because, in contrast to both electrons and X-rays, the neutrons used in diffraction experiments (with wavelengths generally between 2 and 5 Å) effect virtually no damage due to their low energy and neutral charge. This results in NMX possessing two unique advantages. First, the low radiation damage offered by NMX protects sample integrity and avoids the potential breaking of disulfide bridges, the decarboxylatation of acidic side chains, and the generation of reactive species (such as hydroxyl radicals, hydrated electrons, and hydrogen atoms). [52,65,66]. For example, with respect to metalloproteins, irradiation can alter metal-centre bond lengths, resulting in the gain or loss of ligands, or cause radiolysis [67]. Consequently, radiation damage can significantly complicate structural analyses and/or mechanistic interpretations. Second, the ability to more easily collect data at room—i.e., near physiological—temperature mitigates potential artifacts arising from cooling samples down to 100 K. This is significant, as cryogenic temperatures themselves can alter the specific local conformations required for substrate binding, catalysis, and allosteric regulation [68]. Indeed, this phenomenon was observed for the main protease of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), in which residues at the active site were found to adopt different conformations when 100 K and RT X-ray structures were compared, a finding which has possible implications for the molecular docking studies used in structure-based drug design [69].
Nevertheless, there are some scenarios in which cryocooling samples is desirable. As demonstrated by X-ray MX, reduced temperatures can increase resolution and data quality by reducing thermal motion [70,71,72,73]. Additionally, rapid freezing affords the possibility of trapping reaction intermediates, which are able to provide valuable mechanistic insights [71,72]. However, performing NMX studies at low temperatures involves some additional challenges compared to X-ray MX. One challenge is the cryocooling of large crystals. Protein crystals consist of ~20–80% aqueous solvent [74,75], and therefore must be cooled at a rate sufficient to avoid destruction due to ice formation [72,76]. Yet, large crystals suffer from a comparatively low surface-area-to-volume ratio, making the cooling process relatively inefficient. As such, one must apply best practice when freezing samples for neutron diffraction experiments, which can include the following: the use of cryoprotectants (preferably perdeuterated to minimise background from incoherent H scattering); selecting sample loops just slightly larger than the crystal itself (to minimise the volume of the surrounding mother liquor); cooling crystals in dewars filled to the brim with liquid nitrogen (to avoid the vapour layer which typically forms above the surface); or immersion in liquid propane (for more efficient heat transfer). A detailed overview of these practicalities can be found in [73]. Another consideration is that the NMX instruments themselves must be capable of maintaining samples at cryogenic temperatures over the multiple days required for data collection. While this was an issue in the past, today most facilities support low- (100 K) or even ultra-low-temperature (<15 K) experiments. For 100 K data collection, LADI-III at ILL, iBIX at J-PARC, BIODIFF at FRM II, and MaNDi at SNS are equipped with nitrogen cryostream-cooled goniostats, while both IMAGINE at HIFR and LADI-III at ILL offer liquid-helium cryorefrigerators for data collection at <15 K [73].
The efficacy of cryogenic NMX has been demonstrated by multiple studies. For example, 100 K neutron studies of haem peroxidases were successful in capturing and identifying intermediates, revealing an Fe=O form and the unexpected protonation of the catalytic histidine for compound I of cytochrome c peroxidase [77], and an Fe–OH form for compound II of ascorbate peroxidase [78]. With respect to ultra-low temperatures, a 10 K neutron structure of the lytic polysaccharide monooxygenase enzyme from Neurospora crassa (NcLPMO9PD) succeeded in capturing activated molecular dioxygen within the active site [79]. Such studies highlight the ability of cryogenic experiments to expand upon the biological questions able to be addressed by NMX.
5. Case Studies Involving the Application of NMX to Biological Macromolecule Targets
5.1. Neutron Structure of Urate Oxidate Resolves a Long-Standing Mechanistic Conundrum and Reveals Unexpected Changes in Protonation
Urate oxidase (Uox) catalyses the oxidation of uric acid to 5-hydroxyisourate (5-HIU), a metastable intermediate which undergoes further degradation towards allantoin via either an enzymatic or non-enzymatic pathway [80,81]. As humans do not possess a functional version of this molecular machinery [82], recombinant Uox from Aspergillus flavus is used as the drug Fasturtec® (Sanofi-Aventis; Montpellier, France) to treat hyperuricemia in patients undergoing chemotherapy [83]. While high-resolution X-ray structures of Uox were able to reveal the active site in detail and to enable a putative catalytic mechanism to be proposed [84,85,86,87,88], it was not until the first neutron structure was obtained that the exact mechanism began to be determined unambiguously [89,90].
Uric acid possesses four protonation sites, resulting in a total of four possible monoanionic and six possible dianionic configurations. Consequently, identifying the correct catalytic mechanism of Uox required determining the protonation state of the bound uric acid substrate (and the resulting hydrogen boding network), information which was lacking in the previously obtained X-ray structures. As Uox demonstrated the ability to form large, well-diffracting crystals in the aforementioned X-ray MX studies, it was a prime candidate for neutron analysis.
Due to the advantages associated with the isotopic replacement of H for D (detailed in Section 4.2), sample crystallizations were performed using hydrogenous, recombinant protein and precipitant solutions prepared in D2O-containing buffer [89]. Initial crystal samples were obtained by using the batch crystallization method. These samples were then used to seed further experiments which employed the advanced temperature-controlled crystallization apparatus described in Section 4.1. For Uox, this particular biological macromolecule showed direct solubility—where an increase in temperature corresponded to an increase in solubility. Thus, a strategy was implemented where seeded crystals were able to be reversibly grown/dissolved by modulating the temperature, eventually producing large, single crystals suitable for neutron diffraction studies (Figure 7).
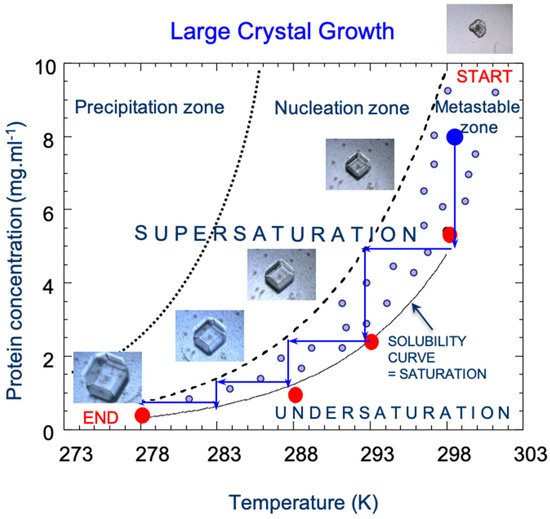
Figure 7.
The experimentally determined phase diagram for Uox demonstrates how manipulating physicochemical conditions (in this case, temperature) can maximize crystal size by extending its trajectory through the metastable (i.e., growth) zone (the blue dot indicates the starting point within the multidimensional phase space; blue arrows indicate the trajectory). The solubility curve was determined by measuring protein concentration at various temperatures (red dots). The crystal was imaged at the following temperatures: 298 K, 293 K, 288 K, 283 K, and 278 K. Figure adapted from Budayova-Spano et al. [91].
These crystals were grown in the presence of both the natural substrate uric acid and the inhibitor 8-azaxanthine, which eventually reached 1–4 mm3 in size and were suitable for neutron diffraction analysis [89,92]. Analysis was performed both by neutrons at the LADI-III instrument at the ILL and by X-rays at the macromolecular crystallography beamline (ID14-2) at the European Synchrotron Radiation Facility (ESRF; Grenoble, France). Collecting X-ray diffraction data from the same crystal analysed by neutrons is advantageous for a couple of reasons. The first is that for a partial or fully deuterated sample, strengthened hydrogen bonds and acid dissociation constants (i.e., pKas), may affect biological macromolecular structure [93,94]. Second, as neutron diffraction reveals approximately two-times the number of atoms as X-ray MX (due to the inclusion of hydrogens), the refinement of neutron structures typically suffers from a relatively poor data-to-parameter ratio. Thus, X-ray diffraction data providing information on heavy-element coordinates effectively increase the number of observations able to be refined against model parameters, and can substantially improve the quality of the neutron maps obtained [95]. A joint neutron/X-ray structure (2.30 Å/2.02 Å; PDB ID: 4N9M) and a separate high-resolution X-ray structure (1.06 Å; PDB ID: 4N9S) were acquired for the Uox–substrate complex (inhibited by Cl−), and a joint neutron/X-ray structure (1.90 Å/1.92 Å; PDB ID: 4N3M) and a separate high-resolution X-ray structure (1.10 Å; PDB ID: 4N9V) were acquired for the Uox–inhibitor complex [90]. The key information revealed by the obtained neutron maps was that the chemical species of uric acid in the active site was the N3 monoanion of 8-hydroxyxanthine (Figure 8), rather than a urate anion in the keto tautomeric form as predicted by theoretical calculations [90,96].
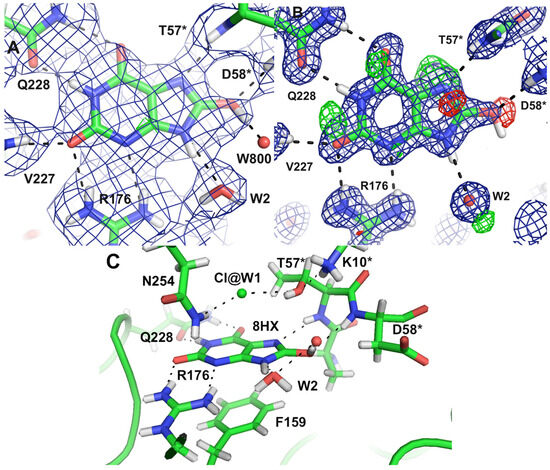
Figure 8.
The substrate 8-hydroxyxanthine within the Uox active site revealed by the (A) joint neutron–X-ray structure and (B) high-resolution X-ray structure. The 2mFO–DFC map is contoured at 1.5 σ (blue mesh) and the mFO–DFC map is contoured at 3.0 σ (positive, green mesh; negative, red mesh). (C) An overview of the catalytic site is shown. The asterisk designation represents residues in contact via crystallographic symmetry. Figure from Oksanen et al. [90].
Vital for identifying this species was a neutron-density OMIT map confirming the presence of a deuteron at O8, which formed a hydrogen bond with a water molecule (W800). This information, when fed into subsequent quantum-mechanical (QM) calculations, allowed a detailed reaction mechanism to be postulated (Figure 9).
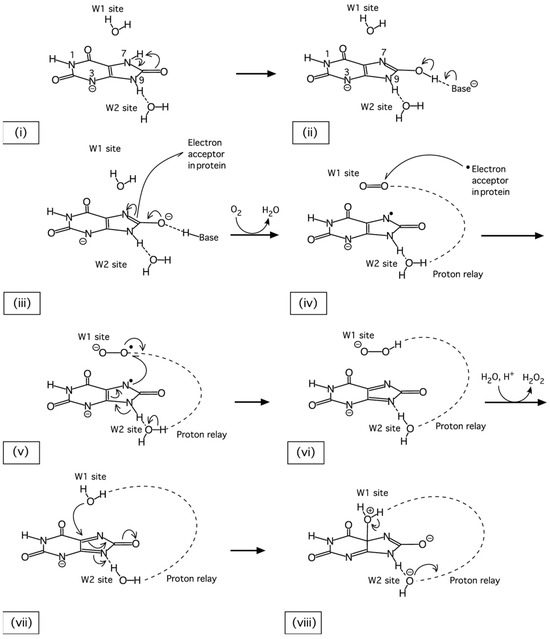
Figure 9.
The postulated first steps of the Uox-catalysed oxidation of uric acid resulting from information obtained by neutron diffraction analysis. Following the binding of a urate N3 monoanion to the Uox active site (i), the substrate rapidly undergoes tautomerization to the 8-hydroxyxanthine species (ii). Next, the deprotonation of 8-hydroxyxanthine occurs, concomitant with an electron transfer to an acceptor in the protein, and creates a triplet state (iii). The presence of molecular oxygen at site W1 accepts first the transferred electron (iv), then an unpaired electron from the urate anion (v), forming a dehydrourate intermediate. After protonation through the proton-relay chain (vi), the hydroperoxide is replaced by the nucleophilic water (vii), which, activated by the proton-relay system, attacks the dehydrourate and generates 5-hydroxyisourate (viii). Figure from Oksanen et al. [90].
Thus, it was not until neutron structures (subsequent work was performed by McGregor et al. [97], who solved a joint neutron/X-ray structure of perdeuterated Uox) were able to provide essential information relating to hydrogen atoms—knowledge which filled in gaps between the high-resolution data on heavy atoms provided by X-ray NMX and quantum chemical calculations—that an unexpected tautomer of uric acid was able to be established as the true substrate of Uox. This study by Oksanen et al. [90] highlights the ability of NMX to answer questions unable to be addressed by other structural determination methods.
5.2. Direct Observation of Protonation-State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography/Unusual Zwitterionic Catalytic Site of SARS-CoV-2 Main Protease Revealed by Neutron Crystallography
A pertinent example of the application of NMX to a biological target is a recent study by Kneller et al. [98], in which a neutron structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; the virus responsible for COVID-19) main protease in complex with an inhibitor was obtained. The main protease of SARS-CoV-2 (Mpro), which is essential for the virus’s replication, has no human counterpart, making it an ideal target for the treatment of COVID-19 [99].
Effectively implementing structural- and computational-based drug design requires atomic-resolution knowledge of both the binding site of the targeted biological macromolecule and the binding mode of the compound-of-interest. With respect to the targeted macromolecule, the functional SARS-CoV-2 protease is a dimer that consists of two identical Mpro protomers (each ~34 kDa in size). The Mpro protomer itself contains three domains: domains I and II are catalytic, while the α-helical domain III is critical for dimerization [98]. A shallow cavity spanning domains I and II comprises the active site, which contains six subsites (S1′ to S5), to which substrate residues and/or inhibitor chemical groups (P1′ to P5) bind, respectively. For the compound-of-interest, the drug Telaprevir belongs to a promising series of α-ketoamide inhibitors which fully occupy the Mpro active site when bound (Figure 10) [100].
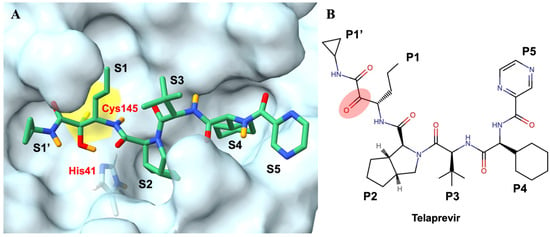
Figure 10.
(A) Active site of the SARS-CoV-2 3CL Mpro with covalently bound telaprevir (PBD ID 7LB7). The six subsites of the binding cavity are labelled (S1′–S5), with the catalytic residues Cys145 and His41 in yellow surface and light-blue sticks respectively, and bound telaprevir in green sticks (D atoms are in orange, with H atoms omitted for clarity). (B) The chemical structure of telaprevir with chemical-group positions is labelled (P1′–P5) and the α-ketoamide warhead is highlighted in red. Figure adapted from Kneller et al. [98].
In an effort to gain insights into the binding of Telaprevir to Mpro, molecular dynamics simulations performed by Pavlova et al. [101] indicated that the stability of the bound inhibitor and of the binding site itself is heavily dependent upon the protonation states of the surrounding histidine residues and the hydrogen bonding network that results. Furthermore, there appeared to be a distinction between the protonation states favoured by the apo- versus the α-ketoamide-bound state. These results highlight the importance of correctly assigning protonation states for both apo- and inhibitor-bound Mpro in drug design efforts—information able to be provided by NMX.
This is the question addressed in some studies by Kneller et al., who aimed to solve neutron structures of Mpro in both apo form [11] and in complex with telaprevir [98]. To grow the large-crystal samples of Mpro, first the initial crystallization conditions were identified by broadly screening the conditions [102], then a sitting-drop vapour-diffusion setup was used to produce crystalline material for seeding larger-scale experiments. After an incubation period of several weeks, either at constant or gradually reducing temperature, large crystals (0.3 and 0.5 mm3 for the apo- and telaprevir-bound Mpro structures, respectively) were harvested and subjected to H/D exchange by mounting the crystals in quartz capillaries adjacent to the deuterated precipitant solution. Neutron diffraction data were collected using the MaNDi instrument at SNS, and X-ray diffraction data were collected using a Rigaku HighFlux HomeLab instrument. Subsequent joint neutron/X-ray analysis resulted in a 2.50 Å/2.30 Å apo Mpro structure (PBD ID: 7JUN) and a 2.40 Å/2.00 Å telaprevir-Mpro structure (PDB ID: 7LB7).
Comparison of the apo- and telaprevir-bound Mpro neutron structures revealed that the protonation states of several key histidine residues at the substrate binding cavity are altered in response to telaprevir binding. Upon binding, the reactive carbonyl of the telaprevir α-ketoamide warhead undergoes a nucleophilic attack by the thiolate of the Mpro Cys145−His41 catalytic dyad, forming a reversible hemithioketal, which possesses a protonated hydroxyl (Figure 11). In contrast to the apo structure, His41 is deprotonated in the telaprevir-Mpro complex, indicating that His41 directly protonates the hemithioketal oxygen. Analysis also revealed that while the hydrogen bond between Nδ1 of His41 and a water molecule found in the apo structure is maintained, the hydrogen bond between this conserved water and the main-chain ND of His41 is not present in the telaprevir-bound structure. Furthermore, the hydrogen bond between His164 and Thr175 is broken in the inhibitor-bound structure, due to a deprotonation at His164 Nε2. Thus, in the telaprevir-Mpro complex, the reduced number of positively charged histidine residues serves to substantially alter the electrostatics at the active site.
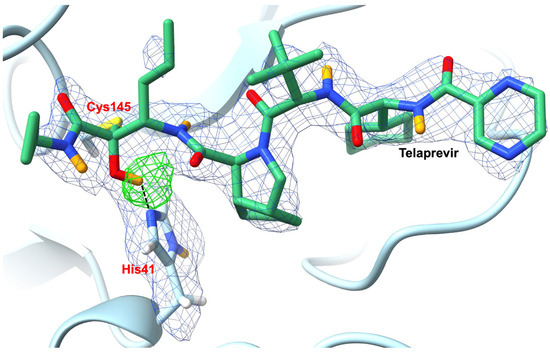
Figure 11.
Upon binding, telaprevir forms a hemithioketal with SARS-CoV-2 3CL Mpro which contains a protonated hydroxyl as revealed by NMX (PBD ID 7LB7). The 2FO−FC (1.0 σ; blue mesh) and OMIT (3.0 σ; green mesh) nuclear density maps clearly reveal the protonation states of the telaprevir-Mpro complex. Telaprevir is shown in green sticks with H atoms omitted for clarity. D atoms are shown in orange. Figure adapted from Kneller et al. [98].
Further changes in response to telaprevir binding occur at subsite S1 of the Mpro active site, which is typically selective for chemical groups at P1 which are able to engage His163 in a hydrogen bond. As telaprevir possesses a hydrophobic norvaline substituent incapable of forming hydrogen bonds at this location, the inhibitor-bound structure contains a recruited water molecule which forms a hydrogen bond with a doubly protonated, positively charged His163 (Figure 12A). This is in contrast with apo Mpro, in which His163 only possesses a proton at Nδ1. The double protonation of His163 may be common in inhibitor binding—with the protonation of Nε2 allowing hydrogen bond formation with inhibitor groups occupying P1—as a separate study by Zhang et al. [100] obtained an inhibitor-Mpro structure (PDB ID: 6Y2F) where a carbonyl oxygen of the bound compound overlays with the hydrogen bond accepting the water molecule found in the telaprevir-Mpro complex. Furthermore, this protonation of His163 upon inhibitor binding (resulting in the addition of a positive charge) attracts the Glu166 side chain and leads to a strengthening of its hydrogen bond with His172 and the N-terminus of the second Mpro protomer (Figure 12B).
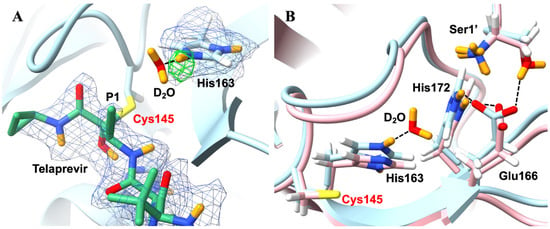
Figure 12.
Protonation states and arrangement of the SARS-CoV-2 3CL Mpro are altered in response to telaprevir binding (PBD ID 7LB7). (A) The 2FO−FC (1.0 σ; blue mesh) and OMIT (3.0 σ; green mesh) nuclear-density maps clearly reveal the double protonation of His163, which recruits a water molecule due to the hydrophobic nature of the P1 group of telaprevir (a norvaline substituent). Telaprevir is shown in green sticks, with H atoms omitted for clarity. D atoms are shown in orange. (B) Comparison of the telaprevir-Mpro complex in light blue with the apo Mpro (PBD ID 7JUN) in light pink reveals structural differences at the active site. D atoms are shown in orange. Figure adapted from Kneller et al. [98].
These NMX studies by Kneller et al. provided crucial information revealing how the binding of α-ketoamide inhibitors affect the protonation states of key histidines within the Mpro active site. Such knowledge of how ionizable residues within the binding site(s) of potential therapeutic targets respond to promising drug candidates is critical for further developments driven by structural- and computational-based drug design.
6. NMX for Biological Samples—Conclusions and Future Perspectives
In spite of its currently niche status, NMX has the potential to become a fundamental pillar of structural biology, alongside X-ray MX, EM, and ED, by providing essential information on protonation states, hydrogen bonding networks, and coordinated waters. Indeed, despite rapid advancement of the electron-based methods, due to the nature of how neutrons interact with elements, NMX, if feasible, will remain the most powerful experimental method for unambiguously determining hydrogen locations within one’s sample.
Critical for the future role of NMX in the life sciences is the upcoming next-generation neutron facility being built in Lund, Sweden—the European Spallation Source (ESS)—which is expected to greatly enhance the accessibility of the method. This facility will make use of a powerful linear proton accelerator, an 11-ton tungsten spallation target, and a liquid-hydrogen/water-based modulator and beryllium-lined reflector system to produce the world’s most brilliant neutron beam (up to ~100 times as bright as current neutron sources) [103]. To analyse samples, the ESS will employ an NMX instrument combining the high-flux neutron beam with a quasi-Laue, TOF diffractometer in order to significantly reduce the stringent sample requirements which typically plague neutron diffraction studies. Once online, the ESS will allow data collection to be performed on biological macromolecular crystals with volumes as small as~0.01 mm3 and with unit cells up to ~200 Å along each axis [104]. This represents a substantial increase in the capabilities of current facilities, and will greatly expand the number of feasible targets (including membrane proteins, which are not only challenging to crystallize, but also typically possess relatively large unit cells).
In a future where the major limitation of NMX studies—i.e., the size of the crystal samples required—has to a large extent been addressed, the method will serve to complement, rather than compete with, other means of structural determination. As highlighted in this paper, neutron diffraction data provides the light-element information missing in even the highest-resolution X-ray structures, whilst neutron structures greatly benefit from being jointly refined against X-ray structures which provide precise heavy-element coordinates (greatly improving data-to-parameter ratios). Regarding the electron-based methods, ED and EM will become valuable tools for detecting hydrogens in samples unable to produce large crystals (or be crystallized at all). However, due to the sensitivity of neutrons towards light elements, NMX, if feasible, will provide the best chance for mapping hydrogens for a given project (especially at the resolutions at which biological macromolecules are typically able to be resolved).
In conclusion, given the essential role played by hydrogens in many fundamental biological processes, and with new upcoming facilities that will significantly alleviate the previous limitations, it is sensible to expect NMX to become increasingly important in the field of structural biology over the next decade.
Author Contributions
Writing—original draft preparation, S.J.H.-J. and M.B.-S.; writing—review and editing, S.J.H.-J. and M.B.-S. All authors have read and agreed to the published version of the manuscript.
Funding
M.B.-S. was supported by the French National Agency through Contract ANR-22-CE30-0024-02. S.J.H.-J. was funded by the same grant.
Data Availability Statement
Figure 3, Figure 5, Figure 8 and Figure 9 are reproduced under the Creative Commons CC BY [https://creativecommons.org/licenses/by/4.0/ (accessed on 15 November 2023)] which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Acknowledgments
IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Brooks-Bartlett, J.C.; Garman, E.F. The Nobel Science: One Hundred Years of Crystallography. Interdiscip. Sci. Rev. 2015, 40, 244–264. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kermani, A.A. A guide to membrane protein X-ray crystallography. FEBS J. 2021, 288, 5788–5804. [Google Scholar] [CrossRef]
- Holcomb, J.; Spellmon, N.; Zhang, Y.; Doughan, M.; Li, C.; Yang, Z. Protein crystallization: Eluding the bottleneck of X-ray crystallography. AIMS Biophys. 2017, 4, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Nannenga, B.L.; Gonen, T. Protein structure determination by MicroED. Curr. Opin. Struct. Biol. 2014, 27, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Liebschner, D.; Croll, T.I.; Williams, C.J.; McCoy, A.J.; Poon, B.K.; Afonine, P.V.; Oeffner, R.D.; Richardson, J.S.; Read, R.J.; et al. AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination. Nat. Methods 2024, 21, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Cryst. 2019, D75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Kneller, D.W.; Phillips, G.; Weiss, K.L.; Pant, S.; Zhang, Q.; O’Neill, H.M.; Coates, L.; Kovalevsky, A. Unusual zwitterionic catalytic site of SARS-CoV-2 main protease revealed by neutron crystallography. J. Biol. Chem. 2020, 295, 17365–17373. [Google Scholar] [CrossRef]
- O’Dell, W.B.; Bodenheimer, A.M.; Meilleur, F. Neutron protein crystallography: A complementary tool for locating hydrogens in proteins. Arch. Biochem. Biophys. 2016, 602, 48–60. [Google Scholar] [CrossRef]
- Wider, G. Structure Determination of Biological Macromolecules in Solution Using Nuclear Magnetic Resonance Spectroscopy. BioTechniques 2000, 29, 1278–1294. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR with Biological Macromolecules in Solution; World Scientific: Singapore, 2021. [Google Scholar]
- El Omari, K.; Duman, R.; Mykhaylyk, V.; Orr, C.M.; Latimer-Smith, M.; Winter, G.; Grama, V.; Qu, F.; Bountra, K.; Kwong, H.S.; et al. Experimental phasing opportunities for macromolecular crystallography at very long wavelengths. Commun. Chem. 2023, 6, 219. [Google Scholar] [CrossRef] [PubMed]
- Read, R. Protein Crystallography Course. 2005. Available online: https://www-structmed.cimr.cam.ac.uk/course.html (accessed on 11 October 2023).
- Nam, K.H. Serial X-ray Crystallography. Crystals 2022, 12, 99. [Google Scholar] [CrossRef]
- Skopintsev, P.; Ehrenberg, D.; Weinert, T.; James, D.; Kar, R.K.; Johnson, P.J.M.; Ozerov, D.; Furrer, A.; Martiel, I.; Dworkowski, F.; et al. Femtosecond-to-millisecond structural changes in a light-driven sodium pump. Nature 2020, 583, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Orville, A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Curr. Opin. Struct. Biol. 2020, 65, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Svensson, O.; Malbet-Monaco, S.; Popov, A.; Nurizzo, D.; Bowler, M.W. Fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1757–1767. [Google Scholar] [CrossRef]
- Bezerra, G.A.; Ohara-Nemoto, Y.; Cornaciu, I.; Fedosyuk, S.; Hoffmann, G.; Round, A.; Márquez, J.A.; Nemoto, T.K.; Djinović-Carugo, K. Bacterial protease uses distinct thermodynamic signatures for substrate recognition. Sci. Rep. 2017, 7, 2848. [Google Scholar] [CrossRef] [PubMed]
- Zander, U.; Hoffmann, G.; Cornaciu, I.; Marquette, J.P.; Papp, G.; Landret, C.; Seroul, G.; Sinoir, J.; Röwer, M.; Felisaz, F.; et al. Automated harvesting and processing of protein crystals through laser photoablation. Acta Crystallogr. D Struct. Biol. 2016, 72, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Márquez, J.A.; Cipriani, F. CrystalDirect™: A novel approach for automated crystal harvesting based on photoablation of thin films. Methods Mol. Biol. 2014, 1091, 197–203. [Google Scholar]
- Junius, N.; Jaho, S.; Sallaz-Damaz, Y.; Borel, F.; Salmon, J.B.; Budayova-Spano, M. A microfluidic device for both on-chip dialysis protein crystallization and in situ X-ray diffraction. Lab Chip 2020, 20, 296–310. [Google Scholar] [CrossRef]
- Junius, N.; Oksanen, E.; Terrien, M.; Berzin, C.; Ferrer, J.L.; Budayova-Spano, M. A crystallization apparatus for temperature-controlled flow-cell dialysis with real-time visualization. J. Appl. Crystallogr. 2016, 49, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Rosier, D.D.J.; Klug, A. Reconstruction of Three Dimensional Structures from Electron Micrographs. Nature 1968, 217, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y. Single-particle cryo-EM—How did it get here and where will it go. Science 2018, 361, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.C. Seeing Atoms by Single-Particle Cryo-EM. Trends Biochem. Sci. 2021, 46, 253–254. [Google Scholar] [CrossRef]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.M.G.E.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef]
- Nannenga, B.L.; Gonen, T. The cryo-EM method microcrystal electron diffraction (MicroED). Nat. Methods 2019, 16, 369–379. [Google Scholar] [CrossRef]
- Martynowycz, M.W.; Clabbers, M.T.B.; Unge, J.; Hattne, J.; Gonen, T. Benchmarking the ideal sample thickness in cryo-EM. Proc. Natl. Acad. Sci. USA 2021, 118, e2108884118. [Google Scholar] [CrossRef]
- Bentley, G.A.; Mason, S.A. Neutron diffraction studies of proteins. Philos. Trans. R. Soc. B Biol. 1980, 209, 505–510. [Google Scholar]
- LADI-III: Quasi-Laue Diffractometer LADI-III. Available online: https://www.ill.eu/users/instruments/instruments-list/ladi/description/instrument-layout (accessed on 13 October 2023).
- Helliwell, J.R. Chapter One—Fundamentals of neutron crystallography in structural biology. Meth. Enzymol. 2020, 634, 1–19. [Google Scholar]
- Tanaka, I.; Niimura, N. Design of a neutron diffractometer on biological marcomolecules. In 15th Meeting of the International Collaboration on Advanced Neutron Sources; Itoh, S., Susuki, J.J., Eds.; Japan Atomic Energy Research Institute: Tsukuba, Japan, 2000; pp. 488–491. [Google Scholar]
- Neutron Spectroscopy. Available online: https://www.ill.eu/neutrons-for-society/neutron-techniques/neutron-spectroscopy (accessed on 14 November 2023).
- Pelican—Time-of-Flight Spectrometer. Available online: https://www.ansto.gov.au/our-facilities/australian-centre-for-neutron-scattering/neutron-scattering-instruments/pelican-time (accessed on 14 November 2023).
- TOF-ND General Purpose Time-of-Flight Neutron Diffractometer. Available online: https://www.bnc.hu/?q=tof-nd (accessed on 14 November 2023).
- Sosnowska, I.M. The birth of time-of-flight (TOF) neutron powder diffraction at pulsed neutron source (invited). Cryst. Res. Technol. 2015, 50, 705–715. [Google Scholar] [CrossRef]
- Kono, F.; Kurihara, K.; Tamada, T. Current status of neutron crystallography in structural biology. Biophys. Physicobiol. 2022, 19, e190009. [Google Scholar] [CrossRef] [PubMed]
- de Broglie, L. Recherches sur la théorie des quanta. Ann. Phys. 1924, 10, 22–128. [Google Scholar] [CrossRef]
- List of Conversion Factors for Neutron Scattering. Available online: https://www.ncnr.nist.gov/instruments/dcs/dcs_usersguide/Conversion_Factors.pdf (accessed on 20 September 2023).
- Ohhara, T.; Kusaka, K.; Hosoya, T.; Kurihara, K.; Tomoyori, K.; Niimura, N.; Tanaka, I.; Suzuki, J.; Nakatani, T.; Otomo, T.; et al. Development of data processing software for a new TOF single crystal neutron diffractometer at J-PARC. Nucl. Instrum. Methods Phys. Res. A Accel. Spectrom. Detect. Assoc. Equip. 2009, 600, 195–197. [Google Scholar] [CrossRef]
- Arnold, O.; Bilheux, J.C.; Borreguero, J.M.; Buts, A.; Campbell, S.I.; Chapon, L.; Doucet, M.; Draper, N.; Ferraz Leal, R.; Gigg, M.A.; et al. Mantid—Data analysis and visualization package for neutron scattering and μ SR experiments. Nucl. Instrum. Methods Phys. Res. A Accel. Spectrom. Detect. Assoc. Equip. 2014, 764, 156–166. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 2017, 73, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Blakeley, M.P.; Hasnain, S.S.; Antonyuk, S.V. Sub-atomic resolution X-ray crystallography and neutron crystallography: Promise, challenges and potential. IUCrJ 2015, 2, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.F. Electron scattering from atomic hydrogen. III. Absolute differential cross sections for elastic scattering of electrons of energies from 20 to 680 eV. J. Phys. B Atom. Mol. Phys. 1975, 8, 2191–2199. [Google Scholar] [CrossRef]
- Carter, C.; March, N.H.; Vincent, D. X-ray and Electron Scattering by Molecular Hydrogen. Proc. Phys. Soc. 1958, 71, 2–16. [Google Scholar] [CrossRef]
- Maki-Yonekura, S.; Kawakami, K.; Takaba, K. Measurement of charges and chemical bonding in a cryo-EM structure. Commun. Chem. 2023, 6, 98. [Google Scholar] [CrossRef] [PubMed]
- Clabbers, M.T.B.; Martynowycz, M.W.; Hattne, J.; Gonen, T. Hydrogens and hydrogen-bond networks in macromolecular MicroED data. J. Struct. Biol. X 2022, 6, 100078. [Google Scholar] [CrossRef] [PubMed]
- Martynowycz, M.W.; Clabbers, M.T.B.; Hattne, J. Ab initio phasing macromolecular structures using electron-counted MicroED data. Nat. Methods 2022, 19, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Hattne, J.; Shi, D.; Glynn, C.; Zee, C.T.; Gallagher-Jones, M.; Martynowycz, M.W.; Rodriguez, J.A.; Gonen, T. Analysis of Global and Site-Specific Radiation Damage in Cryo-EM. Structure 2018, 26, 759–766.e4. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.A.; Schneider, T.R.; Sieker, L.C.; Dauter, Z.; Lamzin, V.S.; Wilson, K.S. Refinement of triclinic hen egg-white lysozyme at atomic resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 522–546. [Google Scholar] [CrossRef]
- Yamashita, K.; Palmer, C.M.; Burnley, T.; Murshudov, G.N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. Acta Crystallogr. D Struct. Biol. 2021, 77, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Zaccai, N.R.; Coquelle, N. Opportunities and challenges in neutron crystallography. EPJ Web Conf. 2020, 236, 02001. [Google Scholar] [CrossRef]
- Chayen, N.E. Turning protein crystallisation from an art into a science. Curr. Opin. Struct. Biol. 2004, 14, 577–583. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A. Protein Crystallization. Methods Mol. Biol. 2017, 1607, 17–50. [Google Scholar]
- Fisher, S.J.; Blakeley, M.P.; Howard, E.I.; Petit-Haertlein, I.; Haertlein, M.; Mitschler, A.; Cousido-Siah, A.; Salvay, A.G.; Popov, A.; Muller-Dieckmann, C.; et al. Perdeuteration: Improved visualization of solvent structure in neutron macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 3266–3272. [Google Scholar] [CrossRef]
- Koruza, K.; Lafuma, B.; Végvari, A.; Knecht, W.; Fisher, S.Z. Deuteration of human carbonic anhydrase for neutron crystallography: Cell culture media, protein thermostability, and crystallization behavior. Arch. Biochem. Biophys. 2018, 645, 26–33. [Google Scholar] [CrossRef]
- Munshi, P.; Chung, S.-L.; Blakeley, M.P.; Weiss, K.L.; Myles, D.A.A.; Meilleur, F. Rapid visualization of hydrogen positions in protein neutron crystallographic structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, M.G.; Mason, S.A.; Blakeley, M.P.; Mitchell, E.P.; Haertlein, M.; Forsyth, V.T. Near-atomic resolution neutron crystallography on perdeuterated Pyrococcus furiosus rubredoxin: Implication of hydronium ions and protonation state equilibria in redox changes. Angew. Chem. Int. Ed. Engl. 2013, 52, 1022–1025. [Google Scholar] [CrossRef] [PubMed]
- Howard, E.I.; Blakeley, M.P.; Haertlein, M.; Petit-Haertlein, I.; Mitschler, A.; Fisher, S.J.; Cousido-Siah, A.; Salvay, A.G.; Popov, A.; Muller-Dieckmann, C.; et al. Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface. J. Mol. Recognit. 2011, 24, 724–732. [Google Scholar] [CrossRef]
- Aggarwal, S.; von Wachenfeldt, C.; Fisher, S.Z.; Oksanen, E. A protocol for production of perdeuterated OmpF porin for neutron crystallography. Protein Expr. Purif. 2021, 188, 105954. [Google Scholar] [CrossRef] [PubMed]
- Meilleur, F.; Weiss, K.L.; Myles, D.A. Deuterium labeling for neutron structure-function-dynamics analysis. Methods Mol. Biol. 2009, 544, 281–292. [Google Scholar] [PubMed]
- Garman, E.F. Radiation damage in macromolecular crystallography: What is it and why should we care? Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Leapman, R.D.; Sun, S. Cryo-electron energy loss spectroscopy: Observations on vitrified hydrated specimens and radiation damage. Ultramicroscopy 1995, 59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Schroder, G.C.; Meilleur, F. Metalloprotein catalysis: Structural and mechanistic insights into oxidoreductases from neutron protein crystallography. Acta Cryst. 2021, D77, 1251–1269. [Google Scholar] [CrossRef]
- Fraser, J.S.; van den Bedem, H.; Samelson, A.J.; Lang, P.T.; Holton, J.M.; Echols, N.; Alber, T. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. USA 2011, 108, 16247–16252. [Google Scholar] [CrossRef]
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, D.W. Cryocrystallography. Structure 1994, 2, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Schröder, G.C.; O’Dell, W.B.; Myles, D.A.A.; Kovalevsky, A.; Meilleur, F. Imagine: Neutrons reveal enzyme chemistry. Acta Cryst. 2018, D74, 778–786. [Google Scholar] [CrossRef]
- Myles, D.A.A.; Dauvergne, F.; Blakeley, M.P.; Meilleur, F. Neutron protein crystallography at ultra-low (<15 K) temperatures. J. Appl. Cryst. 2012, 45, 686–692. [Google Scholar]
- Kwon, H.; Langan, P.S.; Coates, L.; Raven, E.L.; Moody, P.C.E. The rise of neutron cryo-crystallography. Acta Cryst. 2018, D74, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. X-ray crystallographic studies of proteins. Annu. Rev. Phys. Chem. 1976, 27, 493–523. [Google Scholar] [CrossRef]
- Teng, T.-Y.; Moffat, K. Cooling Rates During Flash Cooling. J. Appl. Cryst. 1998, 31, 252–257. [Google Scholar] [CrossRef]
- Casadei, C.M.; Gumiero, A.; Metcalfe, C.L.; Murphy, E.J.; Basran, J.; Consilio, M.G.; Teixeira, S.C.M.; Schrader, T.E.; Fielding, A.J.; Ostermann, A.; et al. Neutron cryo-crystallography captures the protonation state of ferryl heme in a peroxidase. Science 2014, 345, 193–197. [Google Scholar] [CrossRef]
- Kwon, H.; Basran, J.; Casadei, C.M.; Fielding, A.J.; Schrader, T.E.; Ostermann, A.; Devos, J.M.; Aller, P.; Blakeley, M.P.; Moody, P.C.E.; et al. Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme. Nature Commun. 2016, 7, 13445. [Google Scholar] [CrossRef]
- O’Dell, W.B.; Agarwal, P.K.; Meilleur, F. Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase. Angew. Chem. Int. Ed. 2017, 56, 767–770. [Google Scholar] [CrossRef]
- Kahn, K.; Serfozo, P.; Tipton, P.A. Identification of the True Product of the Urate Oxidase Reaction. J. Am. Chem. Soc. 1997, 119, 5435–5442. [Google Scholar] [CrossRef]
- Sarma, A.D.; Serfozo, P.; Kahn, K.; Tipton, P.A. Identification and purification of hydroxyisourate hydrolase, a novel ureide-metabolizing enzyme. J. Biol. Chem. 1999, 274, 33863–33865. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.W.; Lee, C.C.; Muzny, D.M.; Caskey, C.T. Urate oxidase: Primary structure and evolutionary implications. Proc. Natl. Acad. Sci. USA 1989, 86, 9412–9416. [Google Scholar] [CrossRef] [PubMed]
- Giffard, M.; Ferté, N.; Ragot, F.; El-Hajji, M.; Castro, B.; Bonneté, F. Urate oxidase purification by salting-in crystallization: Towards an alternative to chromatography. PLoS ONE 2011, 6, e19013. [Google Scholar] [CrossRef]
- Colloc’h, N.; El-Hajji, M.; Bachet, B.; L’Hermite, G.; Schiltz, M.; Castro, B.; Mornon, J.P. Crystal structure of the protein drug urate oxidase-inhibitor complex at 2.05 A resolution. Nat. Struct. Biol. 1997, 4, 947–952. [Google Scholar] [CrossRef]
- Gabison, L.; Chiadmi, M.; Colloc’h, N.; Castro, B.; El-Hajji, M.; Prangé, T. Recapture of [S]-allantoin, the product of the two-step degradation of uric acid, by urate oxidase. FEBS Lett. 2006, 580, 2087–2091. [Google Scholar] [CrossRef]
- Gabison, L.; Prangé, T.; Colloc’h, N.; El-Hajji, M.; Castro, B.; Chiadmi, M. Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: Mechanistic implications. BMC Struct. Biol. 2008, 8, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Retailleau, P.; Colloc’h, N.; Vivarès, D.; Bonneté, F.; Castro, B.; El-Hajji, M.; Prangé, T. Urate oxidase from Aspergillus flavus: New crystal-packing contacts in relation to the content of the active site. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 218–229. [Google Scholar] [CrossRef]
- Retailleau, P.; Colloc’h, N.; Vivarès, D.; Bonneté, F.; Castro, B.; El-Hajji, M.; Mornon, J.P.; Monard, G.; Prangé, T. Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: A reassignment of the active-site binding mode. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 453–462. [Google Scholar] [CrossRef]
- Oksanen, E.; Blakeley, M.P.; Bonneté, F.; Dauvergne, M.T.; Dauvergne, F.; Budayova-Spano, M. Large crystal growth by thermal control allows combined X-ray and neutron crystallographic studies to elucidate the protonation states in Aspergillus flavus urate oxidase. J. R. Soc. Interface 2009, 6, S599–S610. [Google Scholar] [CrossRef]
- Oksanen, E.; Blakeley, M.P.; El-Hajji, M.; Ryde, U.; Budayova-Spano, M. The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation. PLoS ONE 2014, 9, e86651. [Google Scholar] [CrossRef]
- Budayova-Spano, M.; Dauvergne, F.; Audiffren, M.; Bactivelane, T.; Cusack, S. A methodology and an instrument for the temperature-controlled optimization of crystal growth. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Budayova-Spano, M.; Bonneté, F.; Ferté, N.; El-Hajji, M.; Meilleur, F.; Blakeley, M.P.; Castro, B. A preliminary neutron diffraction study of rasburicase, a recombinant urate oxidase enzyme, complexed with 8-azaxanthin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 306–309. [Google Scholar] [CrossRef]
- Hattori, A.; Crespi, H.L.; Katz, J.J. Effect of side-chain deuteration on protein stability. Biochemistry 1965, 4, 1213–1225. [Google Scholar] [CrossRef]
- Liu, X.; Hanson, B.L.; Langan, P.; Viola, R.E. The effect of deuteration on protein structure: A high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 1000–1008. [Google Scholar] [CrossRef]
- Afonine, P.V.; Mustyakimov, M.; Grosse-Kunstleve, R.W.; Moriarty, N.W.; Langan, P.; Adams, P.D. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Altarsha, M. Modélisation du mécanisme catalytique de l’urate oxydase, in Chimie Informatique et Theorique. Ph.D. Thesis, Université Henri Poincaré—Nancy 1, Nancy, France, 2005. [Google Scholar]
- McGregor, L.; Földes, T.; Bui, S.; Moulin, M.; Coquelle, N.; Blakeley, M.P.; Rosta, E.; Steiner, R.A. Joint neutron/X-ray crystal structure of a mechanistically relevant complex of perdeuterated urate oxidase and simulations provide insight into the hydration step of catalysis. IUCrJ 2021, 8, 46–59. [Google Scholar] [CrossRef]
- Kneller, D.W.; Phillips, G.; Weiss, K.L.; Zhang, Q.; Coates, L.; Kovalevsly, A. Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography. J. Med. Chem. 2021, 64, 4991–5000. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, B.; Jiang, X.-M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, A.; Lynch, D.L.; Daidone, I.; Zanetti-Polzi, L.; Smith, M.D.; Chipot, C.; Kneller, D.W.; Kovalevsky, A.; Coates, L.; Golosov, A.A.; et al. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2020, 12, 1513–1527. [Google Scholar] [CrossRef] [PubMed]
- Luft, J.R.; Collins, R.J.; Fehrman, N.A.; Lauricella, A.M.; Veatch, C.K.; DeTitta, G.T. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 2003, 142, 170–179. [Google Scholar] [CrossRef]
- European Spallation Source. Available online: https://europeanspallationsource.se (accessed on 20 October 2023).
- Markó, M.; Nagy, G.; Aprigliano, G.; Oksanen, E. Neutron macromolecular crystallography at the European spallation source. Meth. Enzymol. 2020, 634, 125–151. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).