Abstract
In this study, we utilized the human coronavirus 229E (HCoV-229E) for evaluating the in vitro efficacy of some analogues of the ion-channel inhibitors and Amantadine and Rimantadine. The application of these investigated compounds did not result in any detectable cytotoxic effects. Furthermore, we observed that the derivatives of both analogues did not affect the viability of MDBK cells. Amantadine and Rimantadine applied at a concentration of 50 μg/mL inhibited the replication by 47% and 36%, respectively. The derivatives of Amantadine displayed a slightly weaker inhibitory effect in comparison with its own inhibitory effect. The derivative 4R of Rimantadine exhibited the same antiviral effect as the Rimantadine control, inhibiting viral replication by 37%. However, the other two derivatives of Rimantadine demonstrated lower activities. The molecular structures of the newly synthesized compounds were investigated thoroughly using single-crystal X-ray analysis. Molecular docking studies were performed using Autodock Vina. Two of the studied compounds 2A and 4A showed a promising binding affinity (−8.3 and −8.0 kcal/mol) towards SARS-CoV-2 RNA-dependent polymerase RNA site and SARS-CoV-2 Nsp3 (207-379, MES site) respectively.
1. Introduction
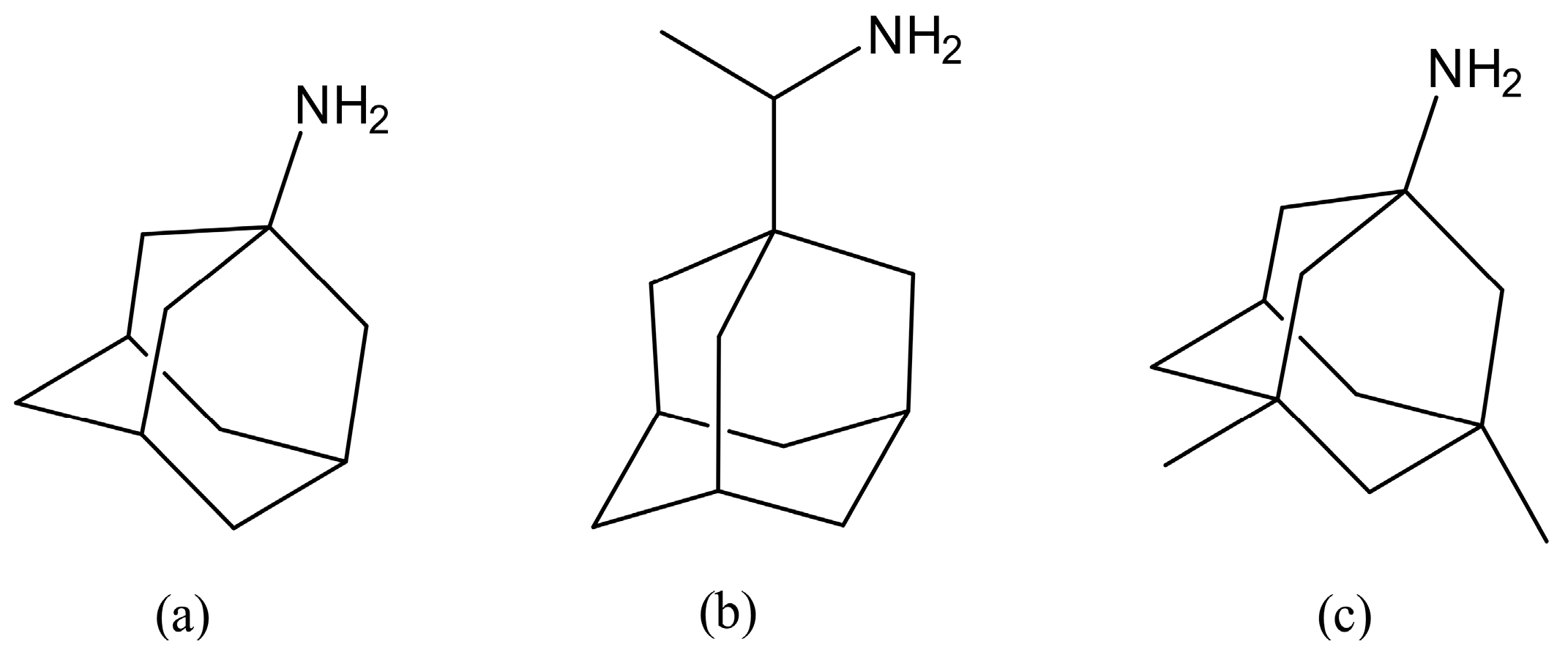
The worldwide effects of the COVID-19 pandemic, initiated by the SARS-CoV-2 virus responsible for severe acute respiratory syndrome, have been substantial. By 27 February 2023, the recorded cases of infection from the disease surpassed 675 million, with the number of attributed deaths exceeding 6.8 million [1]. Originally developed as a treatment for hepatitis C virus (HCV), Remdesivir has exhibited promising outcomes in accelerating the recovery process among adults hospitalized with COVID-19, especially those presenting with symptoms of lower respiratory tract infection. Additionally, Remdesivir is an approved medication strategically formulated to target a specific protein of the SARS-CoV-2 virus for combating COVID-19 [2]. The in vitro effectiveness of ion-channel inhibitors, namely Amantadine, Memantine, and Rimantadine (Figure 1), against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was investigated. Among these inhibitors, Rimantadine demonstrated the highest selectivity index, followed by Amantadine and Memantine. These ion-channel inhibitors interacted with Remdesivir in a comparable antagonistic fashion, and they shared a similar barrier against viral escape [3]. Several adamantanes have proven to be effective against various coronaviruses. For instance, Amantadine, Rimantadine, bananins, and the structurally related Memantine have shown efficacy against human respiratory coronavirus HCoV-OC43, bovine coronavirus, and severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1). Molecular docking studies have suggested that Amantadine might obstruct the viral E protein channel, leading to impaired viral propagation. Furthermore, analogues of Amantadine could potentially hinder the virus’s entry into the host cell by raising the pH of endosomes, thus impeding the activity of host cell proteases like Cathepsin L [4].
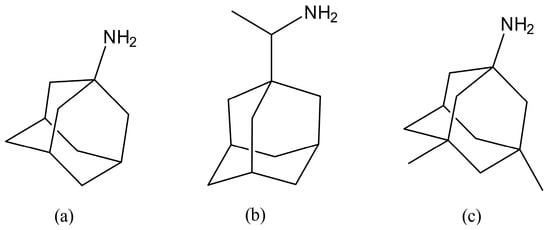
Figure 1.
Structures of Amantadine (a), Rimantadine (b), and the structurally related analogue Memantine (c), which manifest actions against a range of coronaviruses.
Amantadine and Rimantadine might possess the capability to hinder viral entry by interfering with clathrin-mediated endocytosis, the mechanism via which SARS-CoV-2 gains access to host cells. Additionally, they could potentially diminish the activation of polymorphonuclear [PMN] cells by platelets, thereby potentially mitigating cellular damage and inflammation. This is significant because a considerable portion of tissue damage resulting from viral infections stems from the release of tissue-destructive agents by PMN cells [5].
In vitro investigations have revealed that sarcosine boosts the proportion of viable cells against neurotoxicity [6]. Sarcosine is one of the metabolites derived from N,N-dimethylglycine (DMG). Acting as a medication, it holds the potential for addressing depression in middle-aged and elderly individuals, boosting the human immune system, and reducing cholesterol levels within the body. Its application extends to acting as an antioxidant in the food industry as well [7]. In our previous work, we studied the in vivo biological activity of DMG bonded with Amantadine. The results showed that DMG-amantadine reinstated neuromuscular coordination and memory performance in animals treated with MPTP [8]. In our exploration of derivatives of Memantine, we found out that those combined with N-methylglycine (sarcosine) and N,N-dimethylglycine-memantine exhibited the highest activity while maintaining low cytotoxicity. Furthermore, studies on glutamate-induced neurotoxicity in SH-SY5Y cells showed that glycylglycine-memantine treated at 0.032 μM significantly improved the cell viability of SH-SY5Y cells [9]. Interestingly, the analysis conducted by Barberis et al. showed the potential protective role of some molecules, such as monolaurin and N,N-dimethylglycine, which were present at higher levels in the subjects that did not develop the disease. The biosynthesis of some amino acids (glyceric acid, glycine, and N,N-dimethylglycine) was involved in the protection from COVID-19 [10]. In the present work, we utilized the human coronavirus 229E for in vitro evaluation of the efficacy of the newly synthesized analogues of the ion-channel inhibitors and Amantadine and Rimantadine.
2. Materials and Methods
2.1. Chemistry
Amantadine, Rimantadine, amino acids, peptides, 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium tetrafluoroborate (TBTU) coupling reagent, triethylamine (TEA), trifluoroacetic acid (TFA), and all necessary solvents for the synthesis were purchased from Sigma-Aldrich (St. Louis, MO, USA). Aluminum TLC plates and silica gel coated with the fluorescent indicator F254 were obtained from Merck. All NMR spectra were recorded on Bruker AVANCE 500 MHz and Bruker Avance II+ 600 MHz NMR spectrometers in DMSO-d6. MS spectra were recorded on Bruker Esquire 3000 Plus Ion Trap Mass Spectrometer in ESI mode (Supplementary materials).
Chemical Synthesis
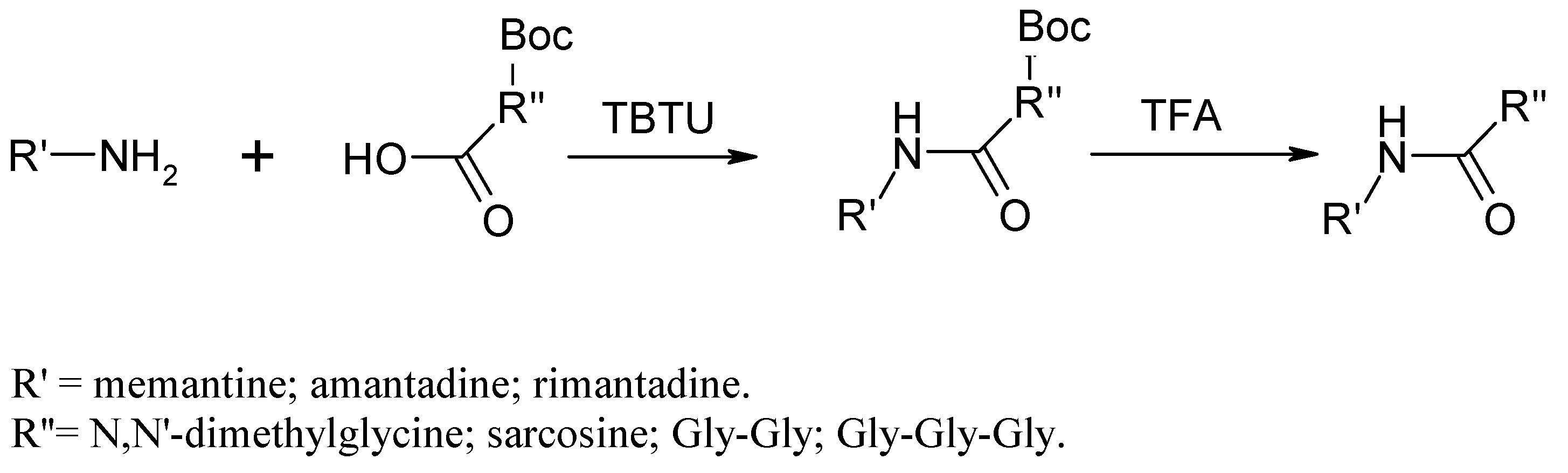
Amide bond formation for all eighth compounds was conducted according to the method described by Knorr et al. [11], shown in Scheme 1.
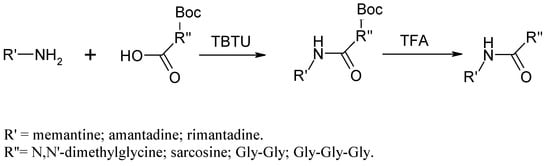
Scheme 1.
Synthesis of Amantadine/Rimantadine derivatives via TBTU as a coupling reagent.
Amantadine, resp. Rimantadine (8 mmol) was dissolved in 5 mL dichloromethane (DCM), and TEA (8 mmol) was added to the solution. In a separate flask, the carboxyl component (8.8 mmol) was dissolved in 10 mL DCM. To the mixture was added TEA (8.8 mmol) and TBTU (12.3 mmol). Both solutions were mixed together after 30 min. The reaction mixture was stirred at room temperature under nitrogen for 24 h. The reaction was monitored by TLC. After 24 h, the reaction mixture was washed with 5% NaHCO3 solution twice and then dried with anhydrous Na2SO4. Sarcosine and the peptides necessitated Boc-protection group removal (DCM/TFA 50:50 for 3 h) before any further purification. The obtained compound was purified by flash chromatography on a silica gel column with an elution system TCM/MeOH 97:3 or Hex/EtOAc 5:4 ratio, depending on the polarity of the compounds.
N,N′-dimethylglycine-amantadine (1A): 1H NMR (600 MHz, DMSO-d6) δ 10.01 (s, 1H), 8.27 (s, 1H), 3.82 (s, 2H), 2.78 (s, 6H), 2.03 (s, 3H), 1.95 (s, 6H), 1.63 (s, 6H). 13C NMR (151 MHz, DMSO-d6) δ 163.56, 58.25, 52.12, 43.37, 41.26, 36.34, 29.20. ESI-MS m/z 237.1964 [M + H]+, calc. 237.1962.
N,N′-dimethylglycine-rimantadine (1R): 1H NMR (600 MHz, DMSO-d6) δ 10.13 (s, 1H), 8.43 (d, J = 9.4 Hz, 1H), 3.97 (d, J = 15.4 Hz, 1H), 3.93 (d, J = 15.4 Hz, 1H), 3.56 (dq, J = 9.3, 6.9 Hz, 1H), 2.80 (s, 6H), 1.94 (s, 3H), 1.66 (d, J = 12.1 Hz, 3H), 1.58 (d, J = 11.0 Hz, 3H), 1.51 (d, J = 12.0 Hz, 3H), 1.46 (d, J = 11.9 Hz, 3H), 0.97 (d, J = 6.9 Hz, 3H). 13C NMR (151 MHz, DMSO-d6) δ 163.91, 57.87, 53.33, 43.37, 38.30, 37.01, 35.98, 28.21, 14.50. ESI-MS m/z 265.2277 [M + H]+, calc. 265.2275.
Sarcosine-amantadine (2A): 1H NMR (600 MHz, DMSO-d6) δ 9.01 (s, 2H), 8.14 (s, 1H), 7.66–7.27 (m, 3H), 3.59 (s, 2H), 2.02 (s, 3H), 1.94 (d, J = 3.0 Hz, 6H), 1.62 (s, 6H). 13C NMR (151 MHz, DMSO) δ 164.31, 51.90, 49.76, 41.35, 36.36, 33.00, 29.21. ESI-MS m/z 223.1806 [M + H]+, calc. 223.1805.
Sarcosine-rimantadine (2R): 1H NMR (600 MHz, DMSO-d6) δ 9.16–8.92 (m, 2H), 8.24 (d, J = 9.4 Hz, 1H), 4.70 (s, 2H), 3.70 (qdd, J = 15.6, 6.7, 5.0 Hz, 2H), 3.55 (dq, J = 9.2, 6.9 Hz, 1H), 2.69 (s, 2H), 2.55–2.50 (m, 3H), 1.98–1.89 (m, 3H), 1.65 (d, J = 12.2 Hz, 3H), 1.58 (d, J = 11.0 Hz, 3H), 1.54–1.43 (m, 6H), 0.96 (d, J = 6.9 Hz, 3H). 13C NMR (151 MHz, DMSO-d6) δ 164.73, 53.24, 49.49, 38.72, 38.29, 37.03, 36.02, 33.06, 28.22, 14.53. ESI-MS m/z 251.2121 [M + H]+, calc. 251.2118.
Glycylglycine-amantadine (3A): 1H NMR (600 MHz, DMSO-d6) δ 8.58 (t, J = 5.7 Hz, 1H), 8.22 (s, 4H), 7.49 (s, 1H), 3.72 (d, J = 5.7 Hz, 2H), 3.57 (q, J = 5.8 Hz, 2H), 2.02–1.98 (m, 3H), 1.92 (d, J = 2.9 Hz, 6H), 1.61 (d, J = 3.3 Hz, 6H). 13C NMR (151 MHz, DMSO-d6) δ 167.47, 166.47, 51.37, 42.77, 41.47, 40.47, 36.45, 29.25. ESI-MS m/z 266.1866 [M + H]+, calc. 266.1863.
Glycylglycine-rimantadine (3R): 1H NMR (600 MHz, DMSO-d6) δ 8.61 (t, J = 5.7 Hz, 1H), 8.18 (s, 4H), 7.44 (s, 1H), 3.73 (d, J = 5.7 Hz, 2H), 3.55 (q, J = 5.8 Hz, 2H), 1.92–1.88 (m, 3H), 1.56 (d, J = 2.9 Hz, 6H), 1.38 (d, J = 3.3 Hz, 6H). 13C NMR (151 MHz, DMSO-d6) δ 169.74, 168.52, 43.89, 42.74, 41.75, 38.05, 30.18. ESI-MS m/z 294.2171 [M + H]+, calc. 294.2177.
Glycylglycylglycine-amantadine (4A): 1H NMR (500.17 MHz, DMSO-d6) δ 8.66 (1H, NH), 7.42 (2H, NH2), 7.32 (1H, NH), 3.82 (2H, CH2), 3.62 (2H, CH2), 3.36 (1H, NH), 2.07 (m, 1H, AdmH), 1.75 (m, 2H, AdmH), 1.56 (q, 4H, AdmH), 1.48–1.36 (CH2), 1.34–1.20 (m, 4H, AdmH), 1.10 (s, 2H, AdmH). 13C NMR (125.77 MHz, DMSO-d6) δ 168.86 (C=O), 167.97 (C=O), 166.84 (C=O), 52.86 (C, AdmC), 50.68 (CH2, AdmC), 47.60 (2CH2, AdmC), 46.50 (CH2), 43.00 (2CH2, AdmC), 42.70 (CH2), 42.00 (CH2), 39.90 (CH2, AdmC), 32.41 (C, AdmC), 32.39 (C, AdmC), 30.12 (CH, AdmC). ESI-MS m/z 323.2075 [M + H]+ calc. 323.2078.
Glycylglycylglycine-rimantadine (4R): 1H NMR (500.17 MHz, DMSO-d6) δ 8.69 (1H, NH), 8.31–8.22 (2H, NH2), 7.42 (1H, NH), 3.88 (2H, CH2), 3.67 (2H, CH2), 3.56 (1H, NH), 2.15 (m, 1H, AdmH), 1.56 (m, 2H, AdmH), 1.33 (q, 4H, AdmH), 1.06–0.91 (CH2). 13C NMR (125.77 MHz, DMSO-d6) δ 164.77 (C=O), 165.99 (C=O), 50.61 (C, AdmC), 45.63 (2CH2, AdmC), 44.55 (CH2), 43.10 (2CH2, AdmC), 42.00 (CH2), 38.90 (CH2, AdmC), 30.12 (CH, AdmC). ESI-MS m/z 351.2390 [M + H]+, calc. 351.2391.
2.2. Single Crystal X-ray Diffraction
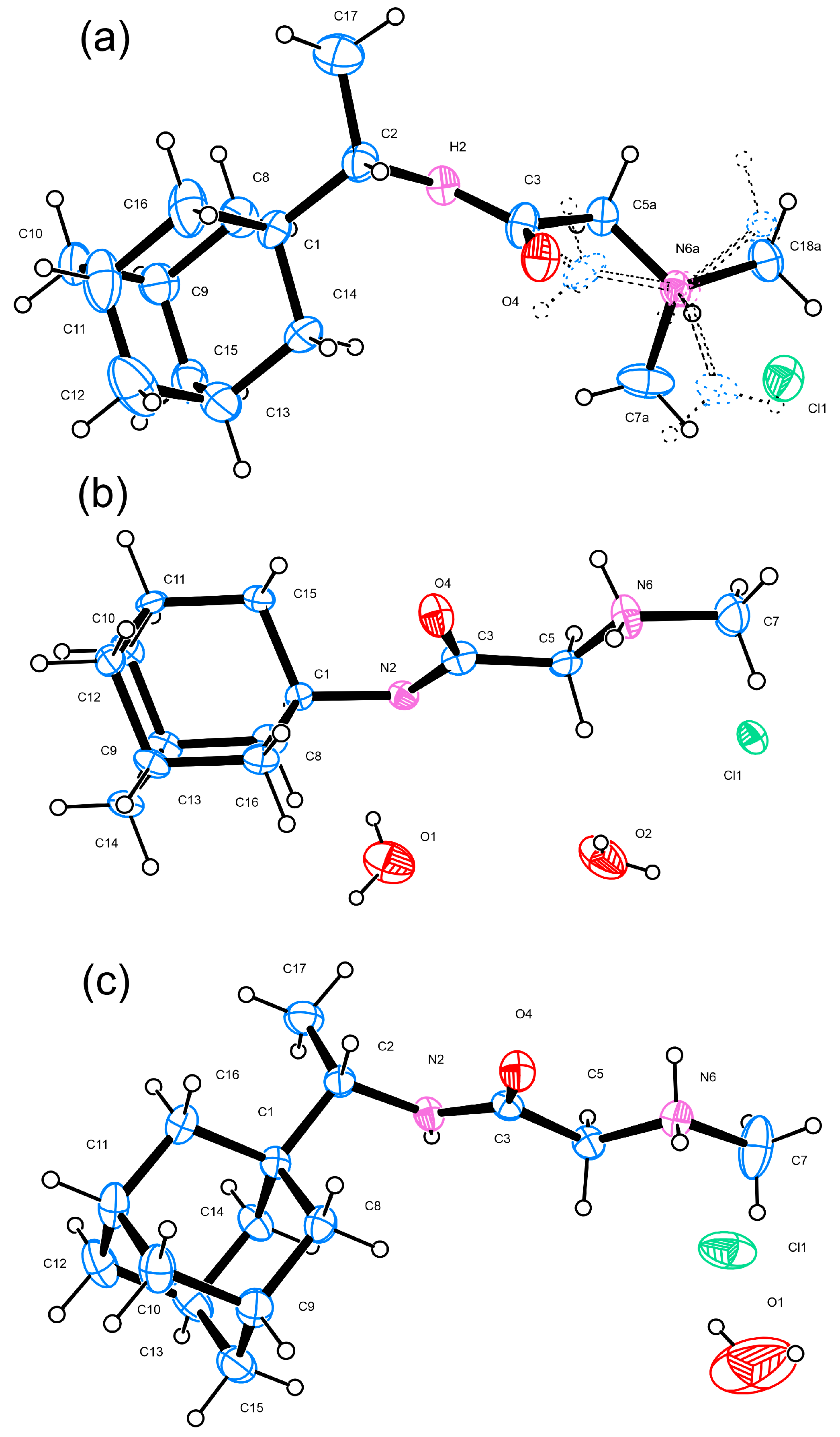
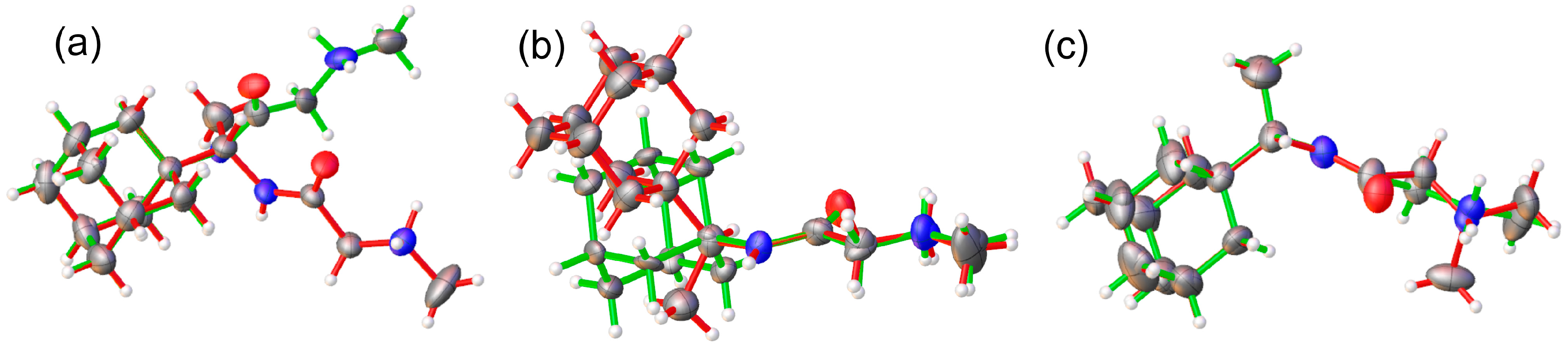
Needle-shaped single crystals of 1R, 2A, and 2R were mounted on a glass capillary. The diffraction data for compounds 1R, 2A, and 2R were collected on a Bruker D8 Venture diffractometer equipped with a PhotonII CMOS detector using micro-focus MoK radiation (λ = 0.71073 Å). Data reduction was performed using APEX4 software [12]. The structures were solved with intrinsic methods using ShelxT [13] and refined by the full-matrix least-squares method on F2 with the ShelxL program [14]. All non-hydrogen atoms were located successfully from the Fourier map and were refined anisotropically. Hydrogen atoms were placed in calculated positions (C–Hmethyl = 0.96 Å, C–Hmethylenic = 0.97 Å) riding on the parent atom (Ueq = 1.2). The most important data collection and crystallographic refinement parameters for 1R, 2A, and 2R are provided in Table 1, and the molecular structures are depicted in Figure 2. Complete crystallographic data for the structure of compounds 1R, 2A, and 2R reported in this paper have been deposited in CIF format with the Cambridge Crystallographic Data Center as 2263306, 2263307, and 2263308. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html, accessed on 16 May 2023 or from the CCDC, 12 Union Road, Cambridge CB2 1EZ, UK; Fax: +44-1223336033; E-mail: deposit@ccdc.cam.ac.uk).

Table 1.
Most important data collection and crystallographic refinement parameters for 1R, 2A, and 2R.
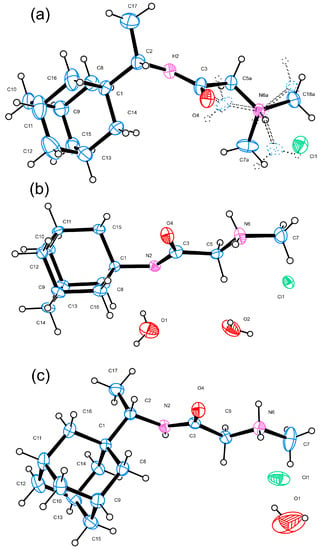
Figure 2.
ORTEP [15] view and atom-numbering scheme of the molecules present in the asymmetric unit (ASU) of (a) 1R, the minor disorder component (~21%) of which is shown with dashed lines, (b) 2A, and (c) 2R; atomic displacement parameters are at 50% probability level.
2.3. Biology
MDBK (Madine and Darby bovine kidney) cell line was cultivated using Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 8% (growth medium), and 4% (maintenance medium) fetal calf serum (FCS with Gentamycin 8 μg/mL and 10 mM HEPES buffer) were used in the experiments. The experiments conducted in this study utilized the human coronavirus 229E (HCoV-229E), which was propagated in MDBK cells and stored at −70 °C until further use.
2.3.1. Cell Viability Assays
The cytotoxicity was determined using a colorimetric MTT assay [16]. Confluent monolayers of MDBK cells in 96-well plates were overlaid with 0.1 mL/well maintenance medium, 0.1 mL/well of serial two-fold dilutions of the compounds (in maintenance medium) or 0.1 mL/well only maintenance medium (in cell controls) and were incubated at 37 °C for 72 h. On the second day, 0.020 mL of MTT (Sigma-Aldrich (5 mg/mL in PBS)) was added to each well and the plates were incubated for 2 h at 37 °C. The optical densities (OD) were determined by a plate reader at λ = 540 nm. The percentage of viable treated cells was calculated by the following formula: [(ODexp.)/(ODcell control)] × 100, where (ODexp.) and (ODcell control) indicate the absorbencies of the test sample and the cell control, respectively. The 50% cytotoxicity concentration (CC50) was calculated via regression analysis of the dose—response curves generated from the data. MTC (maximum tolerated concentration) was determined microscopically on the 72nd hour.
2.3.2. HCoV-229E Replication Inhibition
In the antiviral experiments, the viability of virus-infected cells treated with sub-stances was assessed using the MTT test. For this purpose, we employed a modified version of the MTT assay specifically designed for screening antiviral compounds [17]. Con-fluent monolayers in 96-well plates were overlaid with 0.1 mL/well of a virus suspension. The plates were incubated for 1.5 h at 37 °C and dilutions of the compounds or only the maintenance medium (for virus control) was added after that. Uninoculated cells were used for cell control. On day 5, the plates were treated in the same way described in the method for measuring cell viability. The percentage of protection was calculated using the following formula: [(ODexp.) − (ODvirus control)/(ODcell control) − (ODvirus control)] × 100, where (ODexp.), (ODvirus control), and (ODcell control) indicate the absorbencies of the test sample, the virus control, and the cell control, respectively. A total of 50% effective concentration (EC50) was determined by performing regression analysis on the dose–response curves derived from the collected data. The selectivity index (SI) was calculated using the following formula: CC50/IC50.
2.4. Molecular Docking
Autodock Vina docking engine [18] was used for small molecule docking into selected SARS-CoV-2 and SARS-CoV protein targets (main protease (Mpro)) 6LU7 [19], papain-like protease (PLpro), 6WUU [20], Nsp13 helicase NCB or ADP sites, 6JYT [21], 2XZL [22], ADP-ribose phosphatase 6W6Y [23], RNA polymerase RNA/RTP sites, 7BV2 [24], nonstructural protein 14 (nsp14), 5C8S [25], SARS-CoV-2 nucleocapsid phosphoprotein 6YI3 [26], and nonstructural proteins 16 (Nsp16) S-adenosylmethionine (SAM) site 6WVN [27] as implemented and optimized for docking purposes in the COVID-19 server [28]. The docking box is established by taking the coordinates of the native ligand center and extending its dimensions up to 80 Å × 30 Å × 40 Å to encompass all the residues of the complete cavity. The ligands were obtained from single-crystal structures after removing the solvent and/or counter ions. The most active compound, 4A was prepared extending the side chain of 2A and optimized in the Phenix Elbow ligand builder [29] while MGLTools software [30] was used to create pdbqt files. To further visualize and understand the amino acid–ligand interactions, the most energetic protein–ligand complexes (in the PDB format) were imported in Ligplot+ v2.2 [31] and 2D representation of the weak binding interaction was generated.
3. Results and Discussion
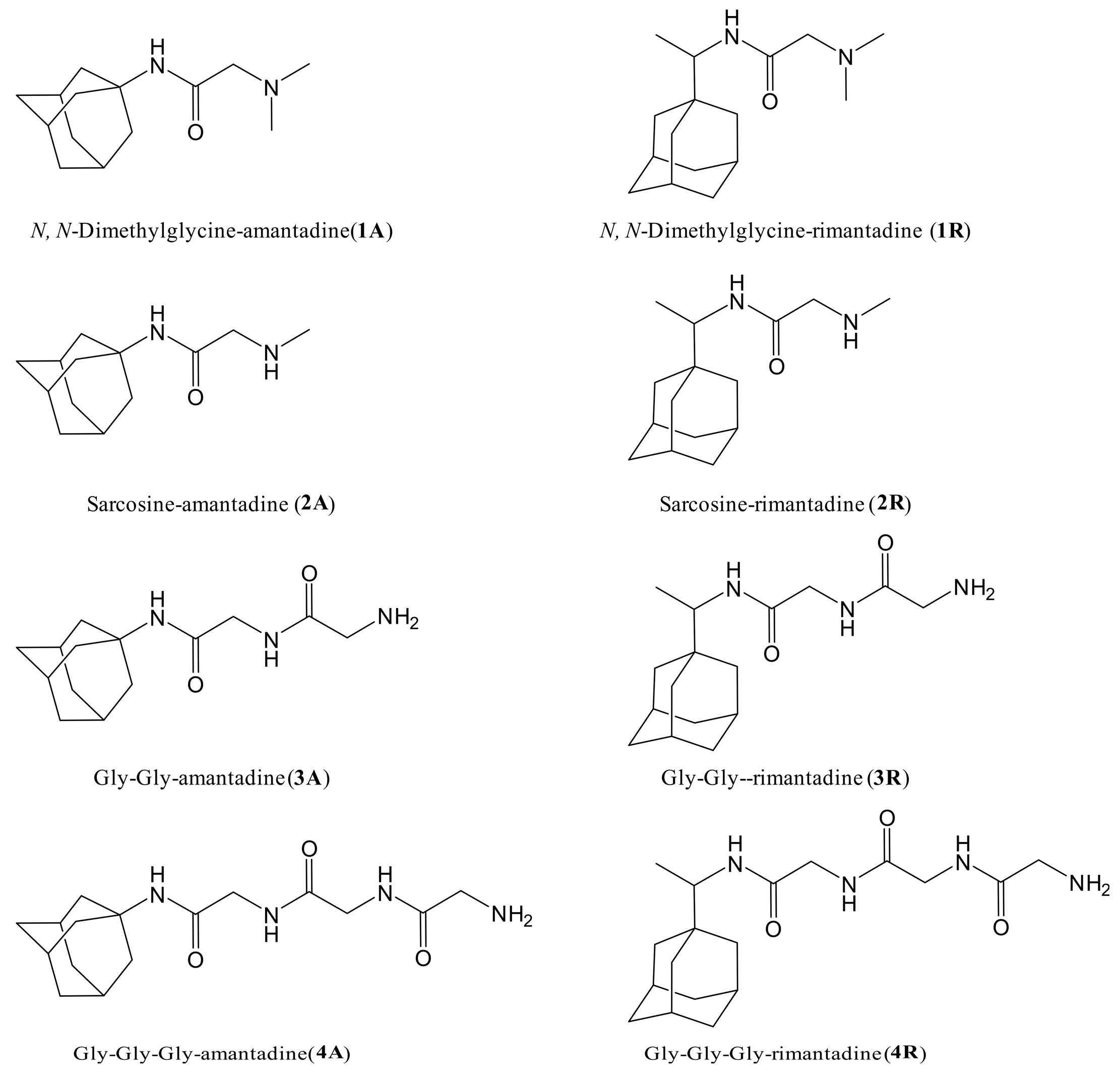
In view of the exhibited neuroprotective activity of Amantadine and Rimantadine the object of this study was to investigate the antiviral activity of newly synthesized derivatives (Figure 3) against HCoV-229E in MDBK cells.
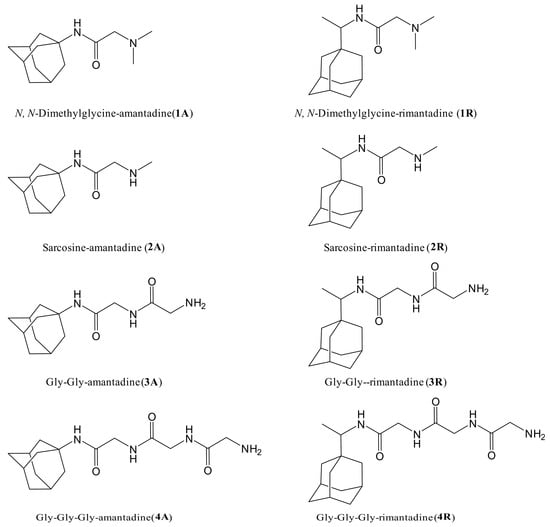
Figure 3.
The structures of newly synthesized derivatives of Amantadine and Rimantadine with N, N-dimethylglycine, sarcosine, Gly-glycine, and Gly-Gly-glycine.
3.1. Single-Crystal Analyses
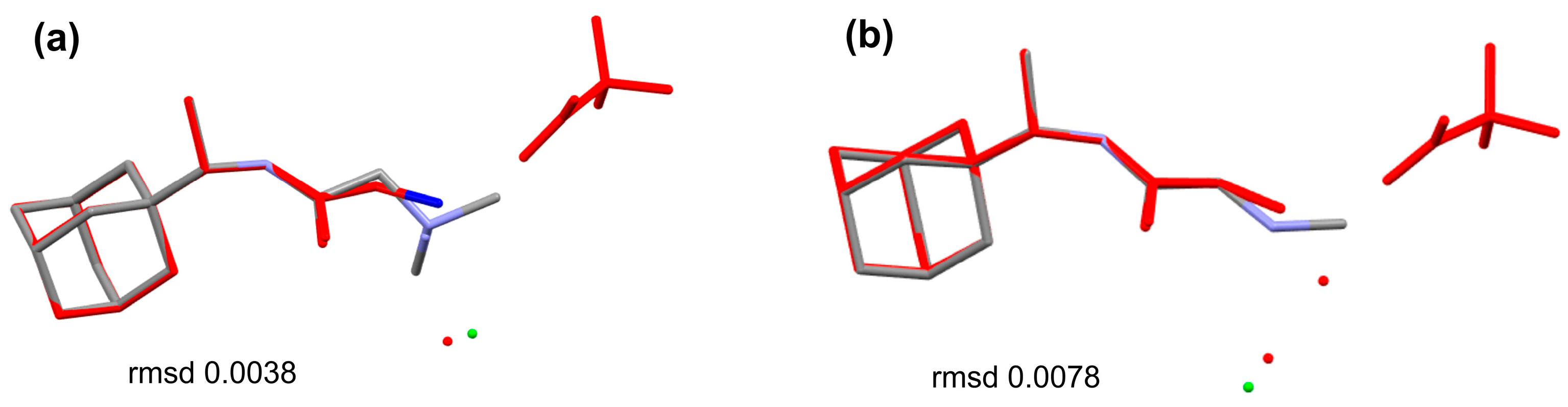
Single-crystal analyses reveal that compound 1R crystallizes in centrosymmetric space group P21/c while compounds 2A and 2R crystallize in a noncentrosymmetric manner in monoclinic and orthorhombic space groups C2 and Iba2, respectively (Table 1). Table 1 provides a summary of the most significant crystal data and refinement indicators. All three compounds feature one molecule in the asymmetric unit along with the expected chlorine counter ion (Figure 2). Both 2A and 2R are solvates, 1.5 and 1/3 hydrates, respectively. In terms of chemical design, compounds 1R, 2A, and 2R are built up by a bulky adamantane moiety (either from Amantadine or Rimantadine) and a tail, e.g.,—N,N-dimethylglycine, sarcosine, glycylglycine, or glycylglycylglycine. The bond lengths and angles of 1R, 2A, and 2R are comparable and are also similar to those observed in analogous compounds [8,18] (Tables S1–S6). The structural features of the adamantane moieties in 1R, 2A, and 2R are highly conserved (bond lengths and angles) and overlay of 1R/2R and 2A/2R (Figure 4). In 1R, the trimethylamine tail end of the molecule is disordered over two positions, the major component being ~79%.
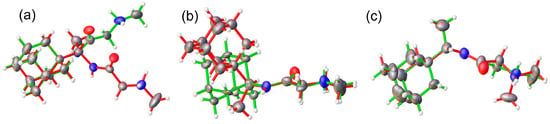
Figure 4.
Overlay of the molecules present in the ASU of (a) 2A (green) vs. 2R (red) based on adamantane (b) 2A (green) vs. 2R (red) based on sarcosine and (c) 1R (red) vs. 2R (green) based on adamantane.
The orientation of the tails (N,N-dimethylglycine and sarcosine) in 1R and 2R, with respect to the bulkier Rimantadine, is conserved, basically simplifying the structure–activity relationship with the substitution effect. On the other hand, the overlay of 2A vs. 2R discloses different relative orientations of the building fragments.
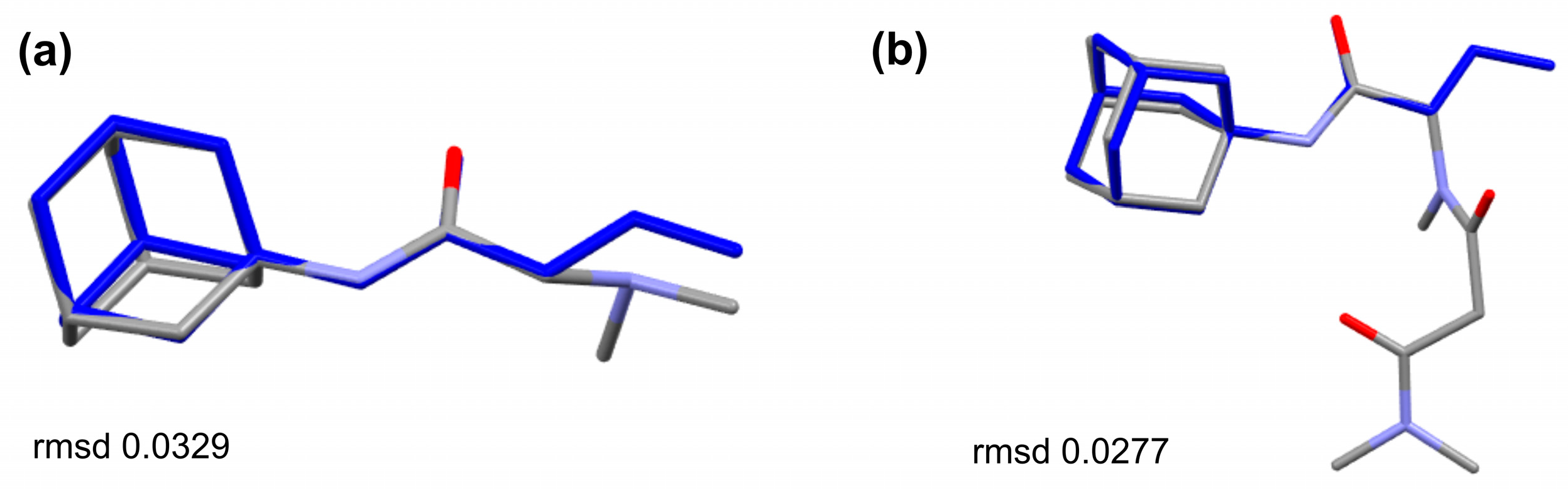
The structural analogues of the studied compounds are present in the CCDC, e.g., SUXXAN [32], FIWJOK [33], NAWKOO [34], ZOZTIR [35], WOVTAF [36], and XUPLIG [37] for 1R or 2R and PAGTUM [38], KETFAM [39], and KEPQAB [8] for 2A. The comparison of SUXXAN against 1R and 2R (Figure 5) shows a conserved molecular geometry for the 1-(dimethylamino)propan-2-one and 1-(methylamino)propan-2-one tails linked to the adamantane. The counter ion in the structures is different; in both cases of TFA for SUXXAN and chlorine for 1R or 2R, the bulkier Cl– basically inhabits the “water molecule” area of the SUXXAN structure. The comparison of the sarcosine–amantadine molecule (2A) looks alike when it is compared to the molecules from KEPQAB and KETFAM (Figure 6). Essentially, the geometry of the linkage between the adamantane and the 1-(methylamino)propan-2-one moiety is highly conserved.
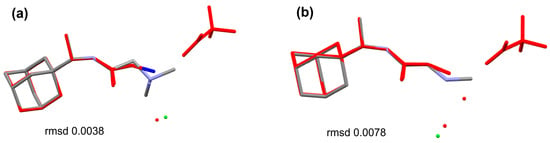
Figure 5.
Overlay of the molecules of (a) 1R and (b) 2R against the molecule of SUXXAN [18] (in red) disclosing stringent molecular geometry; the chlorine counter ion is shown in green.
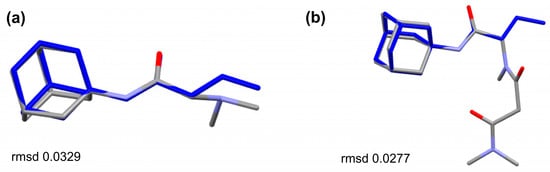
Figure 6.
Overlay of the molecules of (a) KEPQAB [8] and (b) PAGTUM [38] against the molecule of 2A (in blue) disclosing the similarities of the molecular geometry.
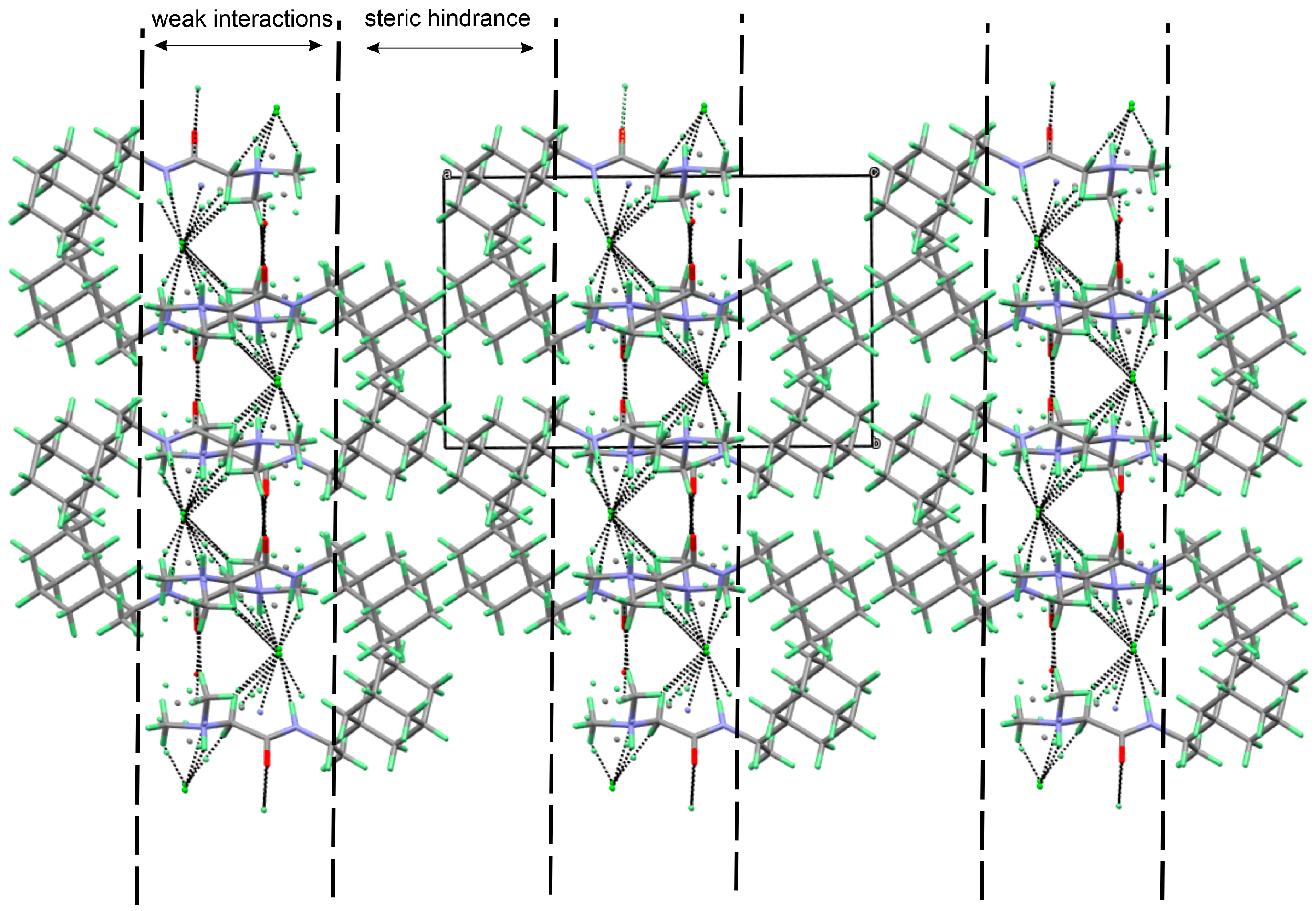
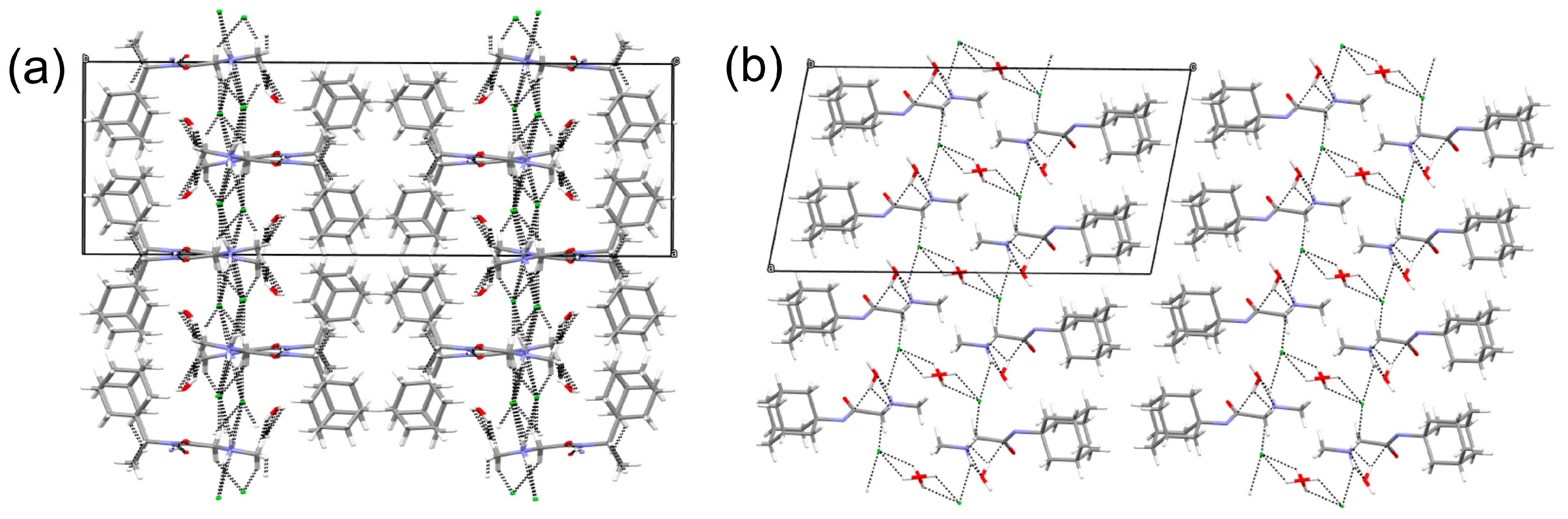
The three-dimensional structure of 1R, 2A, and 2R is stabilized by a multitude of hydrogen bonding, halogen bonding, and C-H…O interactions (Table 2, Table 3 and Table 4). In 1R, only two typical hydrogen bonding interactions, producing chains along the b-axis, are observed. The weak interactions are formed by the tails of the molecules (N,N-dimethylglycine) while the bulkier Rimantadine is not involved in such interactions (Figure 7). The pseudo-layers of alternating tails and Rimantadine are formed.

Table 2.
Halogen bonding and weak interactions observed in 1R.

Table 3.
Weak interactions observed in 2A.

Table 4.
Weak interactions observed in 2R.
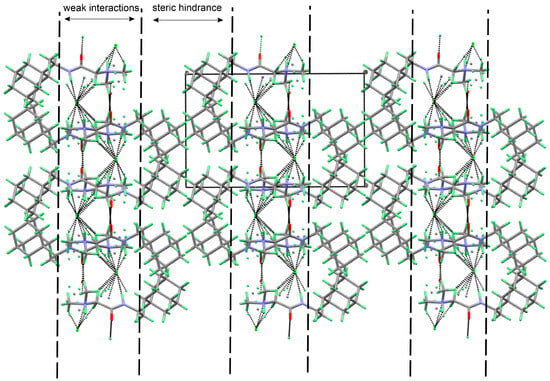
Figure 7.
Visualization of the three-dimensional arrangement of the molecules of 1R, depicting the weak interactions between the tails and the generation of pseudo-layers.
Although compounds 2A and 2R are hydrates, the three-dimensional arrangement of the molecules is similar to 1R (Figure 8 and Table 3 and Table 4).
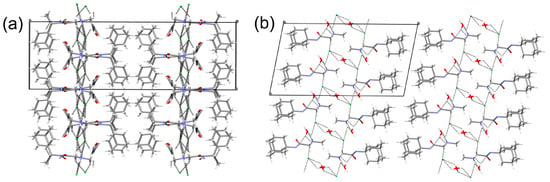
Figure 8.
Visualization of the three-dimensional arrangement of the molecules of (a) 2A and (b) 2R depicting the weak interactions between the tails and the generation of pseudo layers.
3.2. Antiviral Activity
The compounds were administered at concentrations ranging from 800 μg/mL to 100 μg/mL to determine the cytotoxic effect on cells of the MDBK cell line. The established MTC values for the derivative compounds 1A and 1R of the ion-channel inhibitors Amantadine and Rimantadine are 400 μg/mL. The results obtained by us for the toxicity of compounds 1A and 1R are similar to the results obtained by other author groups using another cell line for the toxicity of Amantadine and Rimantadine [40]. The determined CC50 is above 400 μg/mL. Amantadine derivative compound 4A was found to show lower cytotoxicity compared to Amantadine. MTC of 4A is 800 μg/mL and CC50 > 800 μg/mL. Derivatives 3A and 3R exhibit cytotoxicity identical to that of substances 1A and 1R. The remaining derivatives of the two antiviral preparations have higher cytotoxicity. The MTC values determined were 200 μg/mL.
To determine the inhibitory effect on viral replication, the compounds were applied to HCoV-229E-infected cells in MTC and three two-fold dilutions. The results of the experiments showed that the tested compounds have a weak antiviral potential that is dependent on the applied dose. In continuation of the conducted experiments on the influence of the substances on viral replication, we found that the derivatives 1A and 1R applied in a concentration of 400 μg/mL inhibited the viral replication by 42% and 39%, respectively. The Amantadine derivative compound 4A has a significant antiviral effect compared to the other tested substances. HCoV-229E displays that the replication was inhibited by 60% when substance 4A was administered in MTC. The remaining derivatives of Amantadine and Rimantadine administered in MTC show a weaker inhibitory effect. The results are presented in Table 5.

Table 5.
Antiviral activity of Amantadine and Rimantadine derivatives against HCoV-229E in MDBK cells.
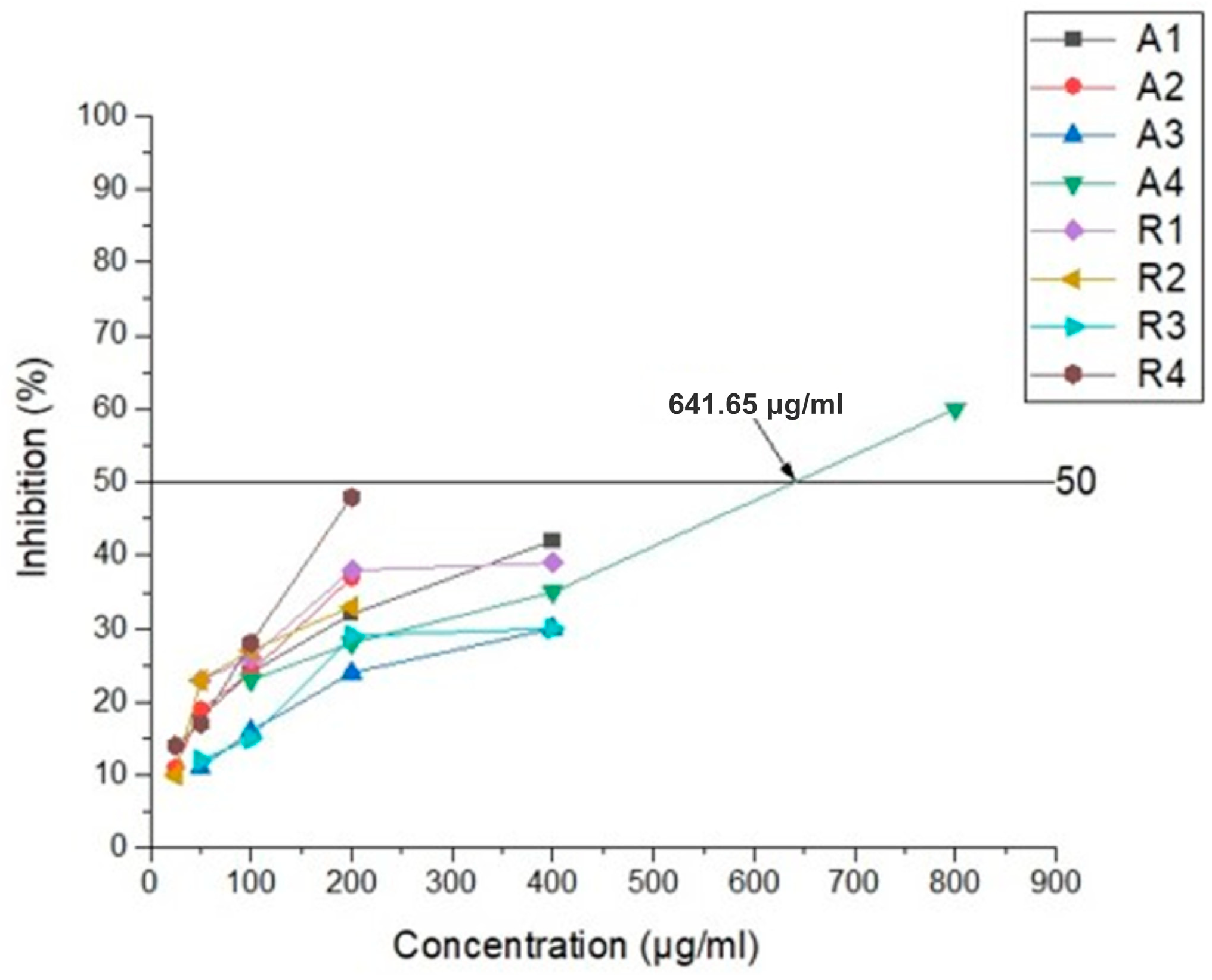
The results of the conducted antiviral experiments are presented in Figure 9. Only the Amantadine derivative compound, 4A, achieved and graphically determined the IC50 value of 641.65 μg/mL. The remaining derivatives of the substances applied in MTC exhibited a weaker inhibitory effect, making it impossible to determine IC50. The calculated selectivity index (SI) for substance 4A is 1.25. Substance 4A demonstrated the most potent impact on virus replication among all the substances studied.
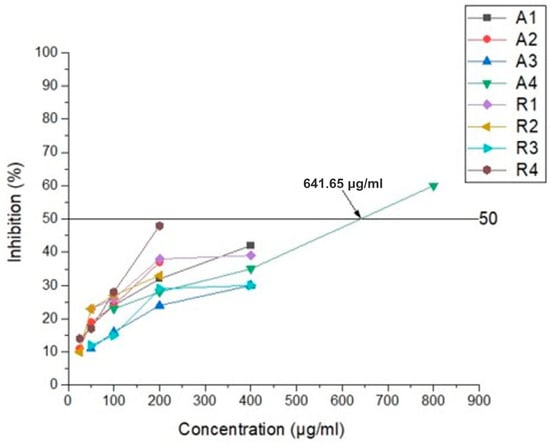
Figure 9.
Antiviral activity of Amantadine derivatives and Rimantadine compounds against HCoV-229E.
The obtained docking scores for the complexes between the selected SARS-CoV-2 and SARS-CoV proteins and compounds 1R, 2A, 2R, and 4A are shown in Table 6. Most of the docking scores for the binding affinity are in the range of 4.8 to 6.0 kcal/mol and thus correspond to lower moderate binding affinity. The docking score for the complexes 2A/RdRp (RNA site) and 4A/Nsp3 (207-379, MES site) showed a docking value of 8.30 and 8.00 kcal/mol, respectively. Although the binding affinities for those complexes are quite pronounced, one should keep in mind the experimental data for antiviral activity where 4A is the most prominent compound.

Table 6.
Obtained docking scores as binding affinity (kcal/mol) and random forest (RF) score value (pKd) [41] for the protein–ligand complexes.
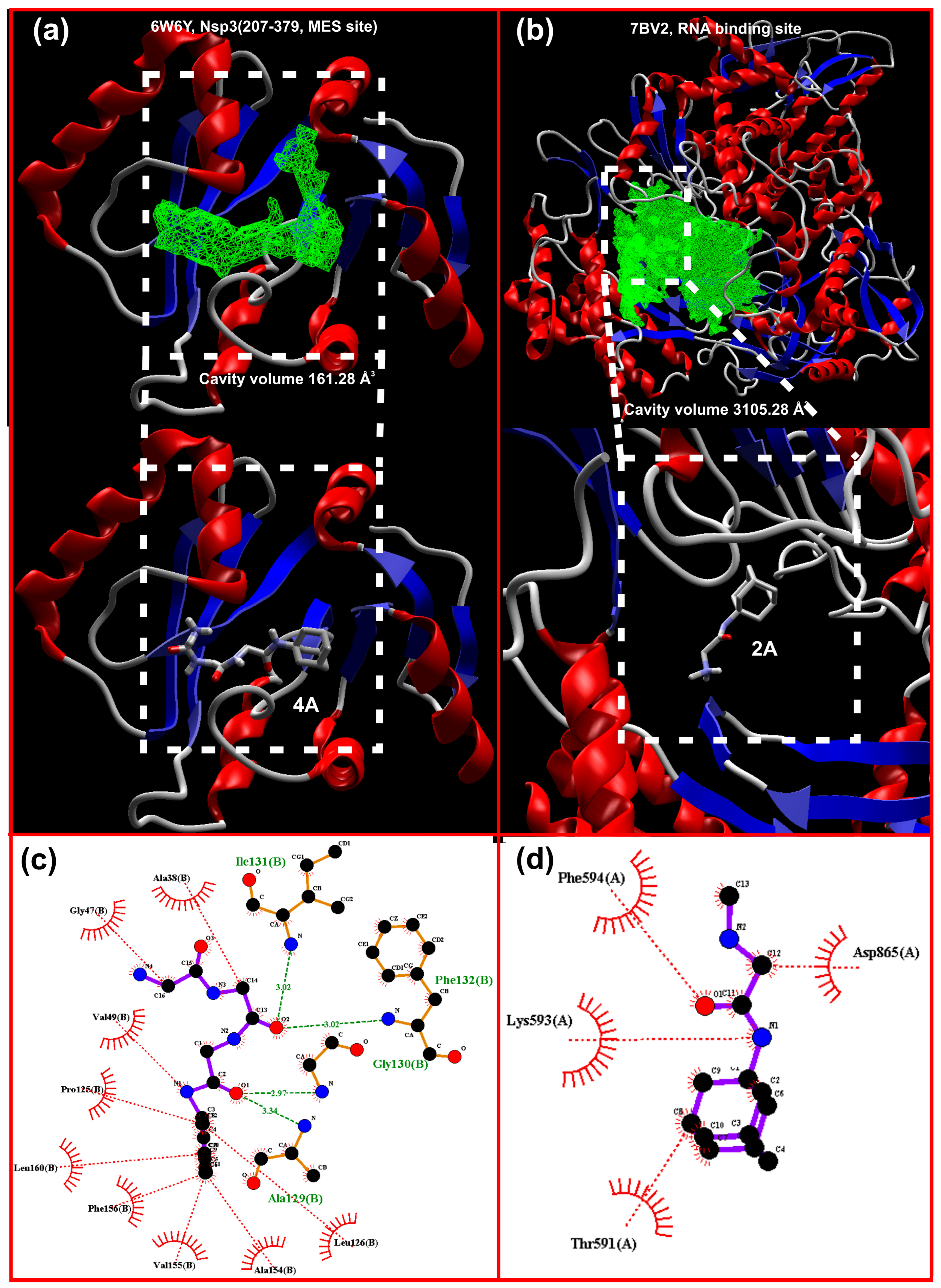
A visualization of the docking results of 2A and 4A in the active site of 7BV2 and 6W6Y is shown in Figure 10a,b. From these results, one can deduce that the stabilization for the 2A–7BV2 complex is through hydrophobic interactions (Figure 10d, a manual inspection of the results disclosed just a plausible interaction N…Lys593). For 4A–6W6Y, in addition to the hydrophobic interactions mainly due to the bulky adamantane moiety, four typical hydrogen bonds are detected (Figure 10c). One has to note that in both cases, the “binding site/cavity” volume is greater than the one achieved by the ligand. This is especially interesting for 4A where the “V shape” of the active site suggests that a molecule with “V-shape” (Figure 11b) may produce a binding with higher affinity and probably better selectivity. In addition, the core of the site is hydrophobic while the extensions are subject to hydrogen bonding and stabilization.
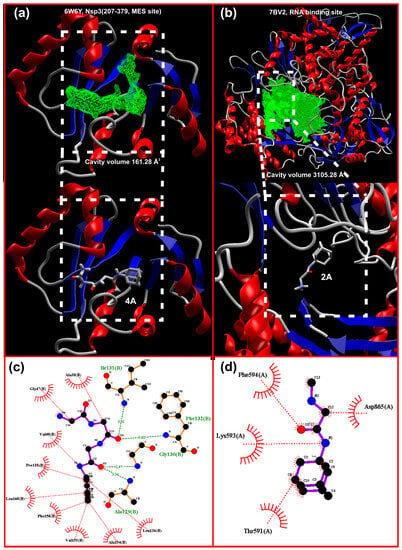
Figure 10.
Display of the docking of (a) 4A in Nsp3 (207-379, MES site) 6W6Y, the cavity of which is shown as green mesh, (b) 2A in RdRp (RNA site) 7BV2, the cavity of which is shown as green mesh, (c) amino acid interactions between 4A and Nsp3, and (d) hydrophobic interactions between 2A and 7BV2; the hydrogen bonds are shown as green dashed lines while hydrophobic interactions are depicted as red dotted lines.
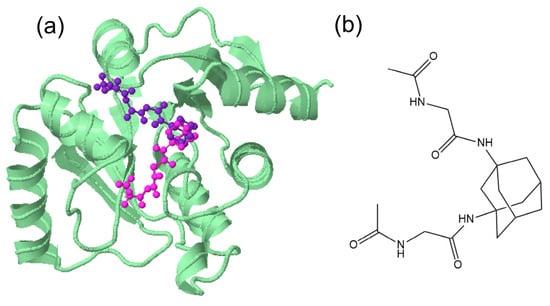
Figure 11.
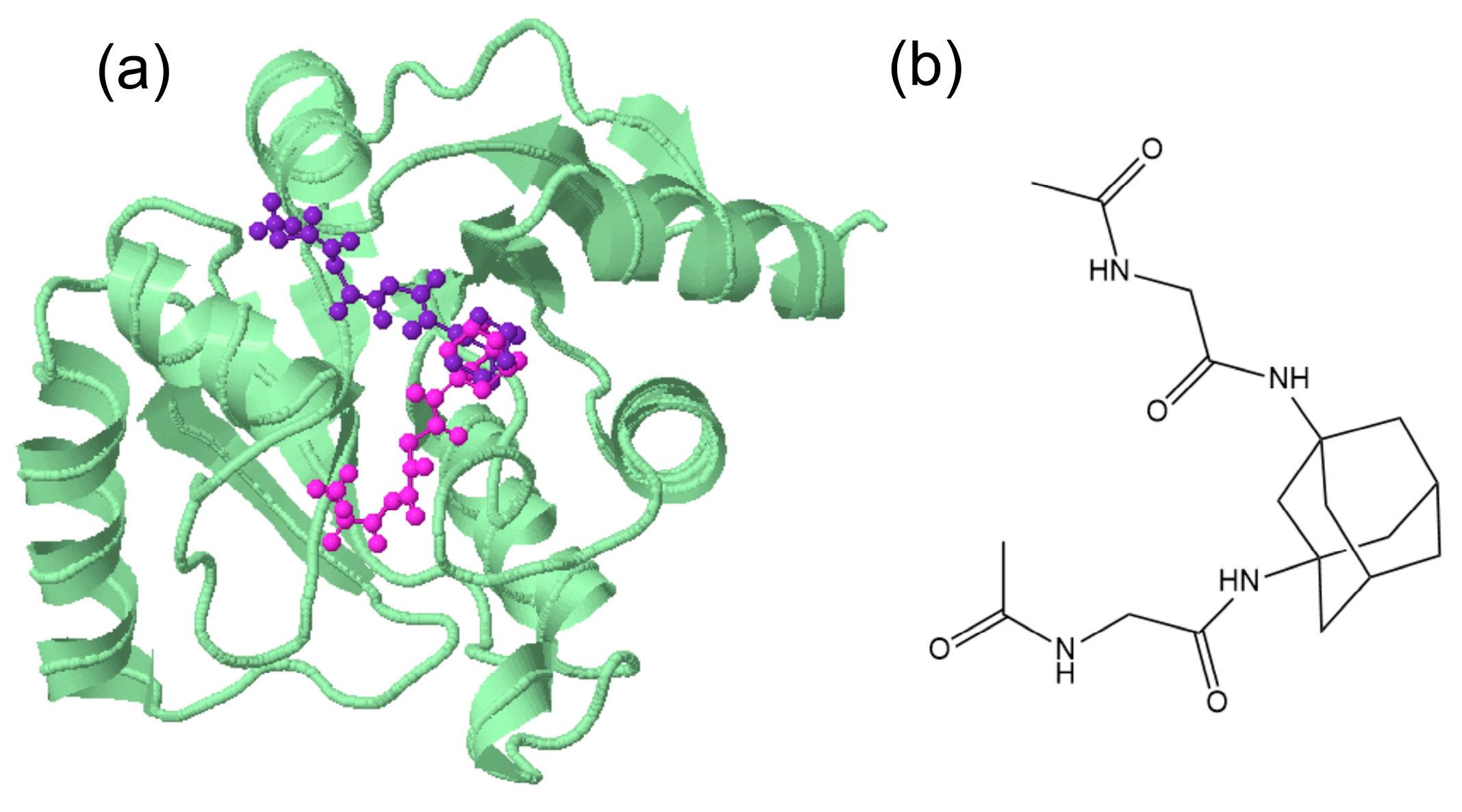
Visualization of (a) the two of the poses (in magenta and violet) of 4A in Nsp3 (in green) (207-379, MES site) 6W6Y suggesting a “V-shape” design of the ligand (b) e.g., N,N′-(adamantane-1,3-diyl)bis(2-acetamidoacetamide).
4. Conclusions
A screening for antiviral activity against human coronavirus 229E (HCoV-229E) of Rimantadine, Amantadine, and newly synthesized and hitherto unexplored Rimantadine and Amantadine derivatives was performed. The experimental results indicated that the tested compounds exhibited a limited antiviral potential against HCoV-229E, which was dose-dependent. It was found that the substances affect the stage of viral replication and have no effect on the glycoproteins of the supercapsid of the free virion and the viral receptors on the surface of the host cell. None of the amantadine/rimantadine derivatives demonstrated a significant antiviral effect. However, among all the substances examined, substance 4A exhibited the most pronounced impact on virus replication and the SI was calculated. At this stage, it was not established whether the substance affects the viral RNA polymerase or affects the synthesis of the viral RNA by another mechanism. The docking results suggest that for maximizing the ligand—Nsp3 (207-379, MES site) interaction, the active molecule may be designed with a “V-shape” molecular form, basically around a bulkier hydrophobic core (adamantane or other) and with side chains bearing hydrogen bonding sites.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst13091374/s1, Figures S1–S8: Mass Spectrum for 1A, 2A, 3A, 4A, 1R, 2R, 3R and 4R; Tables S1–S3: Bond Lengths for 1R, 2A and 2R; Tables S4–S6: Bond Angles for 1R, 2A and 2R.
Author Contributions
Formal analysis, B.S., R.R., H.S.-D., R.C., A.S. and K.S.; Investigation, B.S., R.R., H.S.-D., S.S., R.C. and K.S.; Writing—original draft, R.C.; Writing—review & editing, B.S.; Supervision, B.S., I.S. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
We express our sincere gratitude for the financial support provided by South-West University “Neofit Rilski”.
Data Availability Statement
The data of the current study are available from the corresponding authors upon reasonable request.
Acknowledgments
The authors acknowledge the technical support from the project PERIMED BG05M2OP001-1.002-0005/29.03.2018 (2018–2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Johns Hopkins University. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available online: https://coronavirus.jhu.edu/map.htmL (accessed on 27 February 2023).
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gammeltoft, K.A.; Galli, A.; Offersgaard, A.; Fahnøe, U.; Ramirez, S.; Gottwein, J.M. Efficacy of ion-channel inhibitors amantadine, memantine and rimantadine for the treatment of SARS-CoV-2 In Vitro. Viruses 2021, 13, 2082. [Google Scholar] [CrossRef] [PubMed]
- Butterworth, R.F. Potential for the Repurposing of Adamantane Antivirals for COVID-19. Drugs RD 2021, 21, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.R.; Butterworth, R.F. Repurposing of Adamantanes with Transmitter Receptor Antagonist Properties for the Prevention/Treatment of COVID-19. J. Pharmaceu Pharmacol. 2020, 8, 4. [Google Scholar] [CrossRef]
- Tanas, A.; Tozlu, Ö.Ö.; Gezmiş, T.; Hacimüftüoğlu, A.; Ceylan, O.; Türkez, H. In Vitro and In Vivo Neuroprotective Effects of Sarcosine. BioMed Res. Int. 2022, 2022, 5467498. [Google Scholar] [CrossRef]
- Zheng, Y.; Jin, Y.; Zhang, N.; Wang, D.; Yang, Y.; Zhang, M.; Qu, W. Recovery of N, N-dimethylglycine (DMG) from dimethylglycine hydrochloride by bipolar membrane electrodialysis. Chem. Eng. Process.-Process Intensif. 2022, 176, 108943. [Google Scholar] [CrossRef]
- Chayrov, R.; Kalfin, R.; Lazarova, M.; Tancheva, L.; Sbirkova-Dimitrova, H.; Shivachev, B.; Stankova, I. Development of N, N-Dimethylglycine-Amantadine for Adjunctive Dopaminergic Application: Synthesis, Structure and Biological Activity. Crystals 2022, 12, 1227. [Google Scholar] [CrossRef]
- Chayrov, R.; Volkova, T.; Perlovich, G.; Zeng, L.; Li, Z.; Štícha, M.; Liu, R.; Stankova, I. Synthesis, Neuroprotective Effect and Physicochemical Studies of Novel Peptide and Nootropic Analogues of Alzheimer Disease Drug. Pharmaceuticals 2022, 15, 1108. [Google Scholar] [CrossRef]
- Barberis, E.; Amede, E.; Tavecchia, M.; Marengo, E.; Cittone, M.G.; Rizzi, E.; Sainaghi, P.P. Understanding protection from SARS-CoV-2 using metabolomics. Sci. Rep. 2021, 11, 13796. [Google Scholar] [CrossRef]
- Knorr, R.; Trzeciak, A.; Bannwarth, W.; Gillessen, D. New coupling reagents in peptide chemistry. Tetrahedron Lett. 1989, 30, 1927–1930. [Google Scholar] [CrossRef]
- APEX4 Data Collection Software, Version 2021.4-0; Bruker AXS Inc.: Madison, WI, USA, 2021.
- Sheldrick, G.M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G. SHELXL-97. Program for Crystal-Structure Refinement; ScienceOpen, Inc.: Burlington, MA, USA, 1997. [Google Scholar]
- Farrugia, L. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Sudo, K.; Konno, K.; Yokota, T.; Shigeta, S. A sensitive assay system screening antiviral compounds against herpes simplex virus type 1 and type 2. J. Virol. Methods 1994, 49, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z.; et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef]
- Michalska, K.; Kim, Y.; Jedrzejczak, R.; Maltseva, N.I.; Stols, L.; Endres, M.; Joachimiak, A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: From the apo form to ligand complexes. IUCrJ 2020, 7, 814–824. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef]
- Dinesh, D.C.; Chalupska, D.; Silhan, J.; Koutna, E.; Nencka, R.; Veverka, V.; Boura, E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020, 16, e1009100. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Lemus, M.; Minasov, G.; Shuvalova, L.; Inniss, N.L.; Kiryukhina, O.; Brunzelle, J.; Satchell, K.J. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020, 13, eabe1202. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Yang, G.; Xue, R.; Liu, M.; Wang, F.; Hu, J.; Guo, X.; Chang, S. COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020, 36, 5109–5111. [Google Scholar] [CrossRef]
- Moriarty, N.W.; Grosse-Kunstleve, R.W.; Adams, P.D. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. Sect. D 2009, 65, 1074–1080. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Chayrov, R.; Parisis, N.A.; Chatziathanasiadou, M.V.; Vrontaki, E.; Moschovou, K.; Melagraki, G.; Sbirkova-Dimitrova, H.; Shivachev, B.; Schmidtke, M.; Mitrev, Y.; et al. Synthetic Analogues of Aminoadamantane as Influenza Viral Inhibitors—In Vitro, In Silico and QSAR Studies. Molecules 2020, 25, 3989. [Google Scholar] [CrossRef]
- Kapon, M.; Reisner, G.M. Space-group changes—Revised structures of seven compounds. Acta Crystallogr. Sect. C 1990, 46, 349–350. [Google Scholar] [CrossRef]
- Kuznetsov, N.Y.; Tikhov, R.M.; Godovikov, I.A.; Medvedev, M.G.; Lyssenko, K.A.; Burtseva, E.I.; Kirillova, E.S.; Bubnov, Y.N. Stereoselective synthesis of novel adamantane derivatives with high potency against rimantadine-resistant influenza A virus strains. Org. Biomol. Chem. 2017, 15, 3152–3157. [Google Scholar] [CrossRef] [PubMed]
- Hamodrakas, S.J.; Perrakis, A.; Antoniadou-Vyzas, E. A de novo Designed Possible 5-Lipoxygenase Inhibitor: [alpha]-Acetoxy-N-[1-(1-tricyclo [3.3.1.13,7]dec-1-yl)ethyl]benzeneacetamide. Acta Crystallogr. Sect. C 1996, 52, 677–679. [Google Scholar] [CrossRef]
- Ponath, S.; Menger, M.; Grothues, L.; Weber, M.; Lentz, D.; Strohmann, C.; Christmann, M. Mechanistic Studies on the Organocatalytic α-Chlorination of Aldehydes: The Role and Nature of Off-Cycle Intermediates. Angew. Chem. 2018, 130, 11857–11861. [Google Scholar] [CrossRef]
- Kukushkin, M.E.; Skvortsov, D.A.; Kalinina, M.A.; Tafeenko, V.A.; Burmistrov, V.V.; Butov, G.M.; Zyk, N.V.; Majouga, A.G.; Beloglazkina, E.K. Synthesis and cytotoxicity of oxindoles dispiro derivatives with thiohydantoin and adamantane fragments. Phosphorus Sulfur. Silicon Relat. Elem. 2020, 195, 544–555. [Google Scholar] [CrossRef]
- Dado, G.P.; Desper, J.M.; Holmgren, S.K.; Rito, C.J.; Gellman, S.H. Effects of covalent modifications on the solid-state folding and packing of N-malonylglycine derivatives. J. Am. Chem. Soc. 1992, 114, 4834–4843. [Google Scholar] [CrossRef]
- Ma, X.; Deng, S.; Liang, J.; Chen, J.; Su, J.; Huang, H.; Song, Q. Unsymmetric monothiooxalamides from S8, bromodifluoro reagents and anilines: Synthesis and applications. Tetrahedron Chem. 2022, 3, 100026. [Google Scholar] [CrossRef]
- Arias-Arias, J.L.; Vega-Aguilar, F.; Picado-Soto, D.; Corrales-Aguilar, E.; Loría, G.D. In Vitro Inhibition of Zika Virus Replication with Amantadine and Rimantadine Hydrochlorides. Microbiol. Res. 2021, 12, 727–738. [Google Scholar] [CrossRef]
- Ballester, P.J. Machine learning scoring functions based on random forest and support vector regression. In Proceedings of the Pattern Recognition in Bioinformatics: 7th IAPR International Conference, PRIB 2012, Tokyo, Japan, 8–10 November 2012; Proceedings 7. pp. 14–25. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).