Identification of the Precursor Cluster in the Crystallization Solution of Proteinase K Protein by Molecular Dynamics Methods
Abstract
:1. Introduction
2. Materials and Methods
3. Results
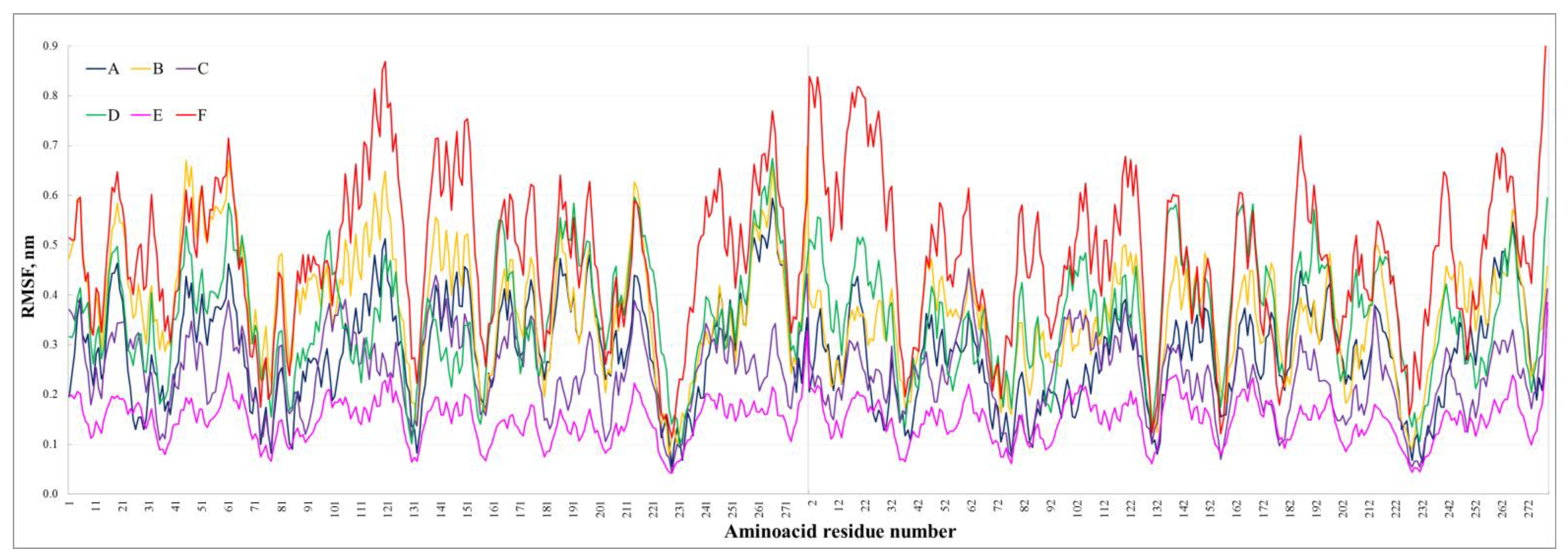
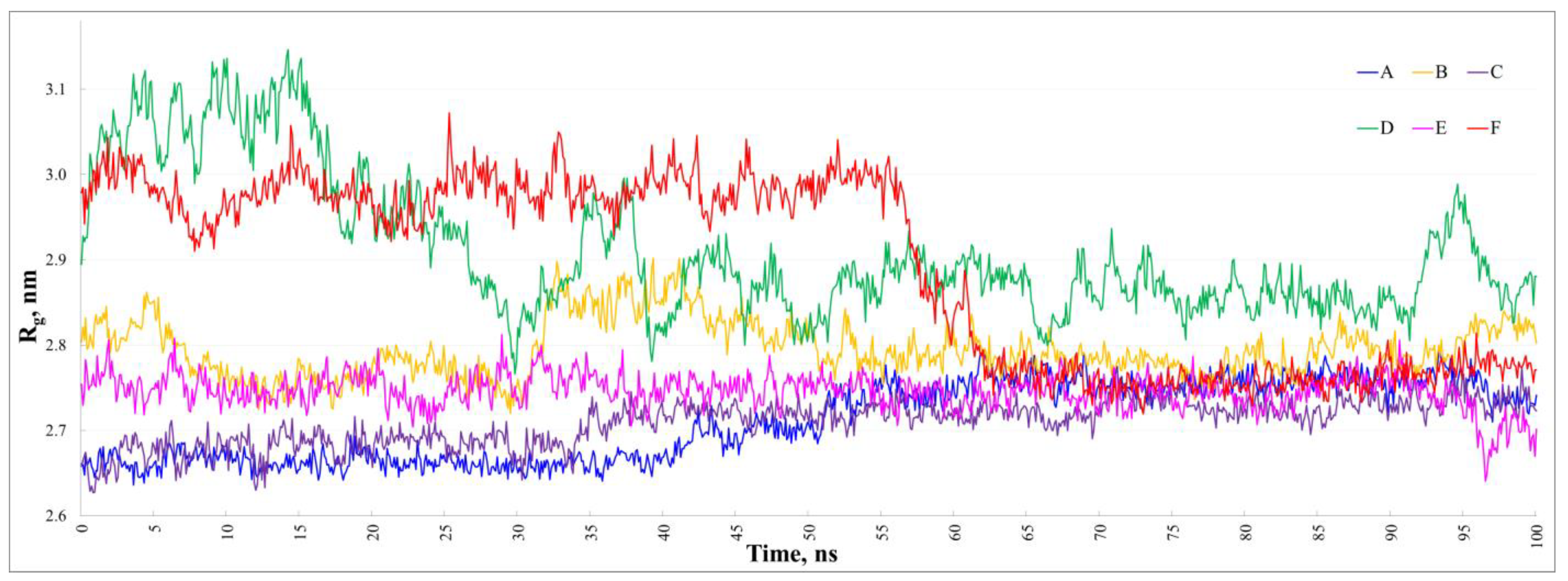
3.1. Characterization of the Dimers’ Stability at 20 °C
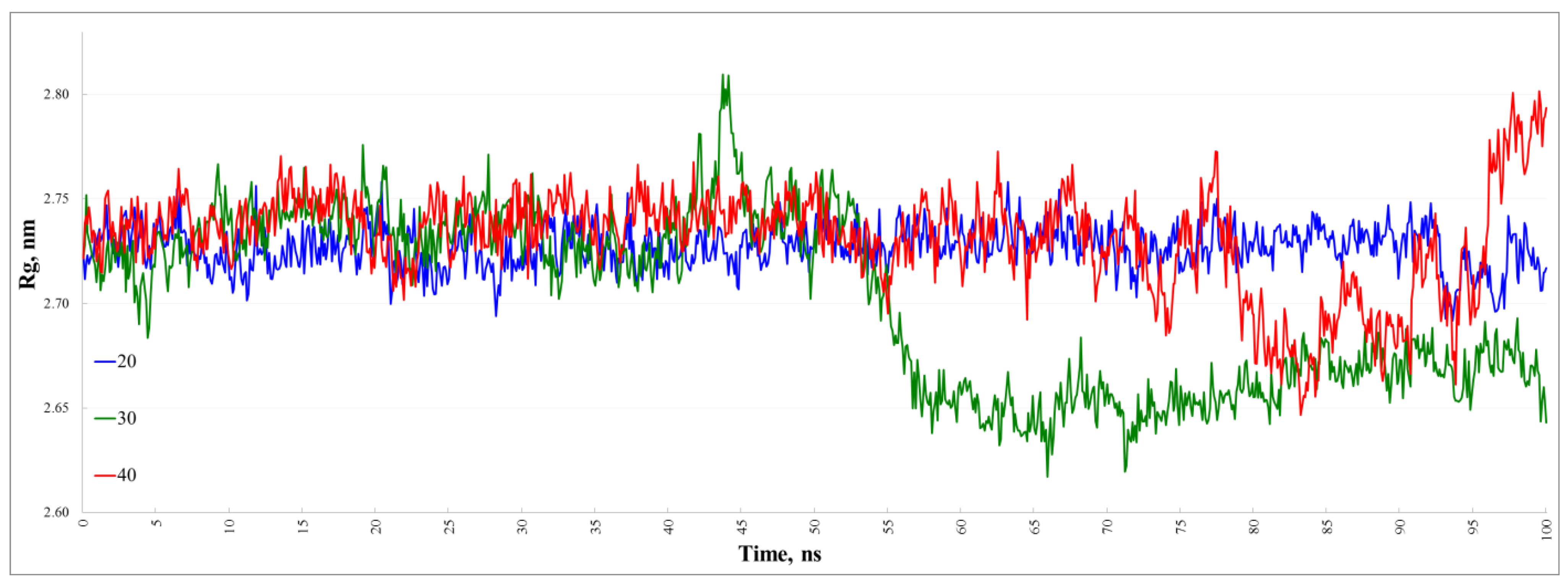
3.2. The Stability of the Dimers at 20, 30, and 40 °C
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gebauer, D.; Kellermeier, M.; Gale, J.D.; Bergström, L.; Cölfen, H. Pre-nucleation clusters as solute precursors in crystallisation. Chem. Soc. Rev. 2014, 43, 2348–2371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vekilov, P.G.; Vorontsova, M.A. Nucleation precursors in protein crystallization. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 271–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalchuk, M.V.; Blagov, A.E.; Dyakova, Y.A.; Gruzinov, A.Y.; Marchenkova, M.A.; Peters, G.S.; Pisarevsky, Y.V.; Timofeev, V.I.; Volkov, V.V. Investigation of the Initial Crystallization Stage in Lysozyme Solutions by Small-Angle X-ray Scattering. Cryst. Growth Des. 2016, 16, 1792–1797. [Google Scholar] [CrossRef]
- Marchenkova, M.A.; Volkov, V.V.; Blagov, A.E.; Dyakova, Y.A.; Ilina, K.B.; Tereschenko, E.Y.; Timofeev, V.I.; Pisarevsky, Y.V.; Kovalchuk, M.V. In situ study of the state of lysozyme molecules at the very early stage of the crystallization process by small-angle X-ray scattering. Crystallogr. Rep. 2016, 61, 5–10. [Google Scholar] [CrossRef]
- Boikova, A.S.; D’yakova, Y.A.; Il’ina, K.B.; Konarev, P.V.; Kryukova, A.E.; Marchenkova, M.A.; Blagov, A.E.; Pisarevskii, Y.V.; Koval’chuk, M.V. Small-angle X-ray scattering study of the influence of solvent replacement (from H2O to D2O) on the initial crystallization stage of tetragonal lysozyme. Crystallogr. Rep. 2017, 62, 876–881. [Google Scholar] [CrossRef]
- Boikova, A.S.; Dyakova, Y.A.; Ilina, K.B.; Konarev, P.V.; Kryukova, A.E.; Kuklin, A.I.; Marchenkova, M.A.; Nabatov, B.V.; Blagov, A.E.; Pisarevsky, Y.V.; et al. Octamer formation in lysozyme solutions at the initial crystallization stage detected by small-angle neutron scattering. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 591. [Google Scholar] [CrossRef]
- Dyakova, Y.A.; Boikova, A.S.; Ilina, K.B.; Konarev, P.V.; Marchenkova, M.A.; Pisarevsky, Y.V.; Timofeev, V.I.; Kovalchuk, M.V. Study of the Influence of a Precipitant Cation on the Formation of Oligomers in Crystallization Solutions of Lysozyme Protein. Crystallogr. Rep. 2019, 64, 11–15. [Google Scholar] [CrossRef]
- Boikova, A.; Marchenkova, M. Pre-crystallization phase formation of thermolysin hexamers in solution close to crystallization conditions. J. Biomol. Struct. Dyn. 2019, 37, 3058–3064. [Google Scholar] [CrossRef]
- Marchenkova, M.A.; Konarev, P.V.; Rakitina, T.V.; Timofeev, V.I.; Boikova, A.S.; Dyakova, Y.A.; Ilina, K.B.; Korzhenevskiy, D.A.; Yu Nikolaeva, A.; Pisarevsky, Y.V.; et al. Dodecamers derived from the crystal structure were found in the pre-crystallization solution of the transaminase from the thermophilic bacterium Thermobaculum terrenum by small-angle X-ray scattering. J. Biomol. Struct. Dyn. 2020, 38, 2939–2944. [Google Scholar] [CrossRef]
- Boikova, A.S.; D’yakova, Y.A.; Il’ina, K.B.; Konarev, P.V.; Kryukova, A.E.; Marchenkova, M.A.; Pisarevskii, Y.V.; Koval’chuk, M.V. Investigation of the Pre-crystallization Stage of Proteinase K in Solution (Influence of Temperature and Precipitant Type) by Small-Angle X-Ray Scattering. Crystallogr. Rep. 2018, 63, 865–870. [Google Scholar] [CrossRef]
- Kordonskaya, Y.V.; Timofeev, V.I.; Dyakova, Y.A.; Marchenkova, M.A.; Pisarevsky, Y.V.; Podshivalov, D.D.; Kovalchuk, M.V. Study of the Behavior of Lysozyme Oligomers in Solutions by the Molecular Dynamics Method. Crystallogr. Rep. 2018, 63, 947–950. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinforma. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, H.W.; Swope, W.C.; Pitera, J.W.; Madura, J.D.; Dick, T.J.; Hura, G.L.; Head-Gordon, T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J. Chem. Phys. 2004, 120, 9665–9678. [Google Scholar] [CrossRef] [PubMed]
- Sousa Da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982, 76, 2662–2666. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-Frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8592. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Barberato, C.; Henri, M.; Koch, J.; Svergun, D.; Barberato, C.; Koch, M.H.J. CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
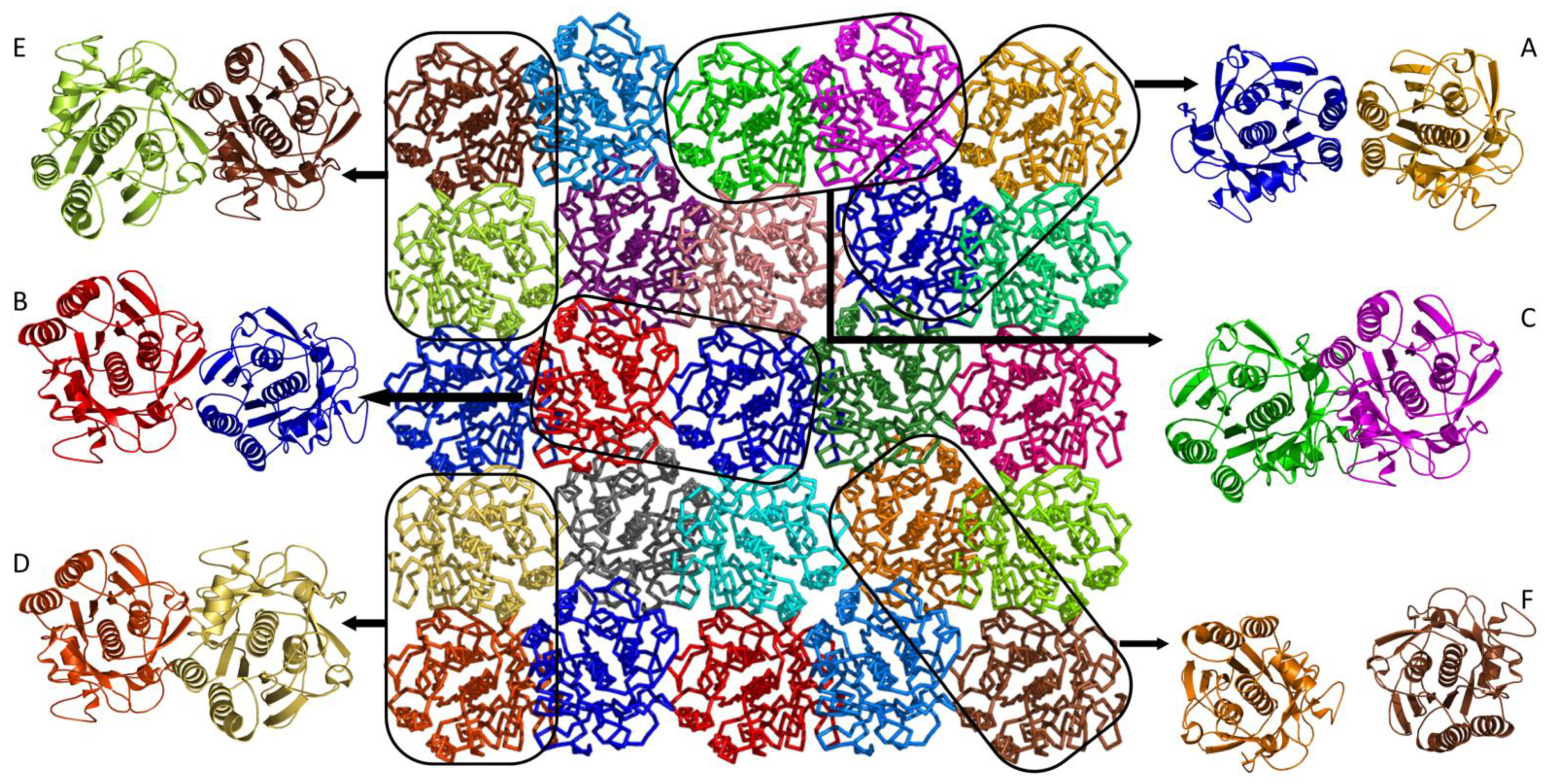



| Dimer | A | B | C | D | E | F |
|---|---|---|---|---|---|---|
| Average RMSF, nm | 0.28 | 0.37 | 0.24 | 0.36 | 0.15 | 0.48 |
| T, °C | Monomer + Dimer A | Monomer + Dimer C | Monomer + Dimer E | |||
|---|---|---|---|---|---|---|
| χ2 | v, % | χ2 | v, % | χ2 | v, % | |
| 30 | 1.30 | 6.6 | 1.36 | 6.1 | 1.18 | 6.5 |
| 20 | 0.75 | 7.4 | 0.76 | 6.9 | 0.71 | 7.3 |
| 10 | 0.78 | 8.7 | 0.80 | 8.2 | 0.74 | 8.5 |
| T, °C | SAXS Data | RMSF, nm | RMSD, nm | ||
|---|---|---|---|---|---|
| χ2 | Monomer, % | Dimer E, % | Dimer E | Dimer E | |
| 40 | — | — | — | 0.24 | 0.22 |
| 30 | 1.18 | 93.5 | 6.5 | 0.33 | 0.40 |
| 20 | 0.71 | 92.7 | 7.3 | 0.15 | 0.16 |
| 10 | 0.74 | 91.5 | 8.5 | — | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kordonskaya, Y.V.; Timofeev, V.I.; Marchenkova, M.A.; Konarev, P.V. Identification of the Precursor Cluster in the Crystallization Solution of Proteinase K Protein by Molecular Dynamics Methods. Crystals 2022, 12, 484. https://doi.org/10.3390/cryst12040484
Kordonskaya YV, Timofeev VI, Marchenkova MA, Konarev PV. Identification of the Precursor Cluster in the Crystallization Solution of Proteinase K Protein by Molecular Dynamics Methods. Crystals. 2022; 12(4):484. https://doi.org/10.3390/cryst12040484
Chicago/Turabian StyleKordonskaya, Yuliya V., Vladimir I. Timofeev, Margarita A. Marchenkova, and Petr V. Konarev. 2022. "Identification of the Precursor Cluster in the Crystallization Solution of Proteinase K Protein by Molecular Dynamics Methods" Crystals 12, no. 4: 484. https://doi.org/10.3390/cryst12040484
APA StyleKordonskaya, Y. V., Timofeev, V. I., Marchenkova, M. A., & Konarev, P. V. (2022). Identification of the Precursor Cluster in the Crystallization Solution of Proteinase K Protein by Molecular Dynamics Methods. Crystals, 12(4), 484. https://doi.org/10.3390/cryst12040484







