Abstract
In the postgenomic age, rapid growth in the number of sequence-known proteins has been accompanied by much slower growth in the number of structure-known proteins (as a result of experimental limitations), and a widening gap between the two is evident. Because protein function is linked to protein structure, successful prediction of protein structure is of significant importance in protein function identification. Foreknowledge of protein structural class can help improve protein structure prediction with significant medical and pharmaceutical implications. Thus, a fast, suitable, reliable, and reasonable computational method for protein structural class prediction has become pivotal in bioinformatics. Here, we review recent efforts in protein structural class prediction from protein sequence, with particular attention paid to new feature descriptors, which extract information from protein sequence, and the use of machine learning algorithms in both feature selection and the construction of new classification models. These new feature descriptors include amino acid composition, sequence order, physicochemical properties, multiprofile Bayes, and secondary structure-based features. Machine learning methods, such as artificial neural networks (ANNs), support vector machine (SVM), K-nearest neighbor (KNN), random forest, deep learning, and examples of their application are discussed in detail. We also present our view on possible future directions, challenges, and opportunities for the applications of machine learning algorithms for prediction of protein structural classes.
1. Introduction
Proteins are macromolecules with a complex structure made up from 20 different types of amino acids, and they play a pivotal role in cellular life. Because protein function is closely related to protein structure, knowledge of protein structure plays an important role in cell biology, pharmacology, molecular biology, and medical science [1]. However, the determination of protein structure remains a grand challenge because of the limitations of experimental methods, including X-ray crystallography and nuclear magnetic resonance, which are expensive and time-consuming [2]. The exponential growth of newly discovered protein sequences by different scientific communities has created a huge knowledge gap between the number of proteins of known sequences and the number of proteins of known structure. Thus, prediction of protein structure from its sequence is one of the most important goals in protein science [3]. Proteins usually consists of multiple domains and protein domains can be classified into distinct classes, named protein structural classes (PSCs), according to their similarities in structure as detailed in Section 2. The ability to predict which classes a given protein domain belongs to from its primary sequence is important, as knowledge of PSC provides useful information towards the determination of protein structure from its primary sequence. For example, knowledge of PSC is useful in finding a proper template for a given query protein in homology modeling. Therefore, a practical, accurate, rapid, and well-developed computational method for identifying the structural classes of proteins from their primary structure is both important and urgently needed.
The general idea in the prediction of PSC is to establish a classification model between the sequence of a protein and its structural class based on data available from proteins of known sequence and known structure. Machine learning methods were a popular choice in this endeavor, and numerous studies on this topic have been published over the past several decades [4,5,6,7,8,9,10,11]. Similarly, machine learning has also been employed successfully to retrieve information from protein sequences for the prediction of protein fold classification [1,2,12,13,14].
Because data plays a foundational role in machine learning (ML), collating data (proteins of known sequences and structures) is the first step in predicting PSCs. Fortunately, the protein data bank (PDB) provides a huge amount of data on the three-dimensional structure of proteins. As demonstrated in the schematic in Figure 1, the next step in the prediction of protein structural classes is the extraction of features from the sequences of these proteins, or the representation of a protein as a vector from a mathematical viewpoint (Step 2 in Figure 1). These feature descriptors should be an accurate representation of the essential information in a protein, and the accuracy of these representations significantly affects the performance of a prediction model. To construct an effective model to represent the protein samples, numerous different features have been exploited, including amino acid composition, dipeptide composition, pseudo-amino-acid composition, functional domain composition, and distance-based features [15,16,17,18,19]. To extract as much as information from the protein sequence as possible, a large number of features are usually constructed. As a consequence, the input space (feature descriptors) is comprised of many dimensions, resulting in limitations to several ML methods. Thus, it is generally necessary to reduce the dimensionality of the feature space by feature selection techniques (Step 3 in Figure 1). After obtaining a feature space of reasonable dimensions, choice of a suitable and practical classifier is the next important step in PSC prediction (Step 4 in Figure 1). Machine learning algorithms are usually the first choice of many researchers, and these algorithms have been extensively applied in classification model construction [8,20,21]. Finally, to appraise the performance of a predictor, a cross validation approach is used (Step 5 in Figure 1).
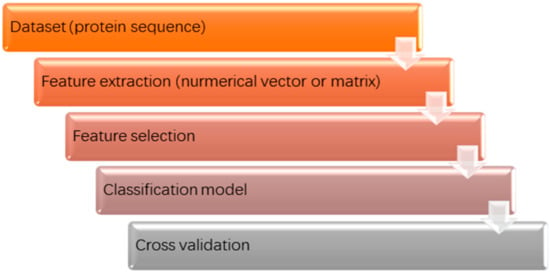
Figure 1.
Schematic showing general procedure for protein structural class prediction by using machine learning (ML) methods.
In this review, we highlight the most recent advances in the prediction of protein structural class, with emphasis on an insightful categorization and classification of new feature descriptors and ML methods. First, we describe the commonly used datasets (Section 2). Next, we examine protein sample representations (Section 3) and review successful approaches to feature selection (Section 4). We then evaluate the different approaches to protein structural class prediction using ML (Section 5). Finally, following a brief introduction to cross validation, future perspectives, challenges, and opportunities are discussed in the conclusion.
2. Datasets
The Protein Data Bank (PDB) is a database containing the three-dimensional structures of proteins previously determined by experimental methods (such as X-ray crystal diffraction, nuclear magnetic resonance, and electron microscopy). Several classification databases have been generated using different structural classification methods, including Structural Classification of Proteins (SCOP), CATH-Gene3D, Families of Structurally Similar Proteins, Homologous Structure Alignment Database, Molecular Modeling Database and 3d Environment. The SCOP database, which comprises protein structural domains that have been classified according to similarities in their structures and amino acid sequences, provides a detailed and comprehensive description of the structural and evolutionary relationships between proteins [12]. SCOP has been used extensively in protein structure prediction studies. In the latest version of SCOP, extended release 2.07 (accessed on 2020-07-16, stable release March 2018), proteins are classified into twelve structural classes: (1) all-alpha proteins (all-α); (2) all-beta proteins (all-β); (3) alpha or beta proteins (α/β); (4) alpha and beta proteins (α + β); (5) multidomain proteins (γ); (6) membrane and cell-surface proteins (δ); (7) small proteins (ζ); (8) coiled coil proteins; (9) low-resolution protein structures; (10) peptides; (11) designed proteins; and (12) artifacts. However, for most practical applications, only the original four categories (all-α, all-β, α/β, α + β), or occasionally seven categories (all-α, all-β, α/β, α + β, γ, δ, ζ), are considered. Several widely used benchmark datasets showing low similarity (Table 1), such as 640 [16], 1189 [22], 25PDB [23], ASTRAL [22], and C204 [24] are used to provide a comprehensive and unbiased comparison with the existing prediction methods [22].

Table 1.
Typical protein datasets with four categories used for benchmarking. The number of proteins in each category and the total number of proteins in a dataset are shown.
3. Feature Extraction
In the context of PSC prediction, a specific encoding scheme is used to generate a set of features to represent each protein and these are then used as the inputs for ML algorithms. Generally, feature descriptors should capture the most essential information or properties of a protein, and thus an effective encoding scheme is of vital importance. Over the past three decades, numerous different feature descriptors of proteins have been developed for use in a broad range of predictions, including PSC prediction, protein fold classification, protein subcellular location prediction, and membrane protein type prediction [23,26,29,30,31,32,33,34,35]. Each feature is a representation of a specific piece of information about a protein, generally concerning either composition, order, and/or evolutionary information within a protein sequence. To extract a specific type of information from a protein sequence, several different strategies may be used. In the following sections, feature descriptors are classified and described according to their information types and the concepts of feature design. A list of features applied to represent proteins is shown in Table 2.
3.1. Amino Acid Composition
Each protein is comprised of a set number of amino acids arranged in a particular order, and this arrangement determines the process of folding into a specific spatial structure [29]. In the earliest research, only the amino acid composition (AAC) was utilized as a feature descriptor [8,9,22], yielding a vector containing 20 elements, each of which corresponds to amino acid frequency. Unfortunately, the simple discrete model considering only sequence-composition information did not generate an ideal outcome, as additional important information, including sequence-order and the physicochemical properties of the amino acids, were simply neglected.
In realizing the necessity of incorporating additional information from the protein into the feature space, Zhou proposed the concept of pseudo amino acid composition (PseAAC), which includes, in addition to amino acid composition, important information such as sequence-order, hydrophobicity, hydrophilicity, and side chain mass [36]. The PseAAC-discrete model is defined by 20 + λ discrete numbers:
where the 20 factors represent the occurrence frequencies of the 20 native amino acids, and the factors incorporate additional information beyond AAC. Subsequently, several different variants of PseAAC have been reported, some of which incorporate more complex information [15,26,32,37,38]. For user convenience, Shen and Chou established a web server called “PseAAC” (https://www.csbio.sjtu.edu.cn/bioinf/PseAAC/, accessed on 1 February 2021) from which users can access 63 different kinds of PseAACs [39]. In general, most of the existing feature descriptors can be incorporated into the concept of PseAAC. However, we feel that it is useful to discuss the many different types of features which can be incorporated into the PseAAC, and to review recent developments in the use of PseAAC for PSC prediction.

Table 2.
List of features applied to represent proteins.
Table 2.
List of features applied to represent proteins.
| Feature Types | Description | References |
|---|---|---|
| Amino Acid Composition | Simplest, primary, and fundamental | [8,9,22] |
| Sequence Order | Capture all possible combinations of amino acids in oligomeric proteins, exceptionally large number of features | [40,41,42,43,44,45] |
| Physicochemical Properties | Classify amino acids based on properties; Composition, order, and position-specific information are usually extracted | [1,36,46] |
| Multiprofile Bayes | Incorporate both position-specific information and the posterior probability of each amino acid type | [16,47,48] |
| Secondary Structure Based Features | Classify amino acids according to their tendency to form a specific secondary structural element | [1,23,25,48] |
| PSSM-based Probability | Evolutionary information was included by a position-specific scoring matrix | [16,18,19,49] |
| Fourier Transform Based Feature | Extract low frequency coefficients in frequency domain | [15,27,50,51] |
| Functional Domain Composition | Convert protein sequence into a sequence of functional domain types | [37] |
| Split Amino Acid Composition | Incorporate both position-specific information and amino acid composition | [16] |
3.2. Sequence Order
To capture sequence-order information, the dipeptide composition was incorporated into feature vectors for predicting PSCs by Lin and Li [40]. The dipeptide composition is the absolute occurrence frequency of each pair of adjacent amino acids, totaling 400 possible dipeptide arrangements. The dipeptide composition has previously been applied in the prediction of protein secondary structural components [41,42]. Analogously, polypeptide components, including dipeptide, tripeptide, and tetrapeptide elements, were used by Yu and coworkers to enhance predictive accuracy [43]. Instead of using polypeptide composition directly as features, they assigned each polypeptide into a structural class according to its structural class tendency, and then converted the protein sequence into a structural class tendency sequence, in which each element represents one PSC. The composition components of the structural class tendency in structural class tendency sequences were then used as feature descriptors [43]. Tetrapeptides are known to play an important role in the formation of regular structure, as 60–70% of tetrapeptides encode specific structures [44]. For example, hydrogen bonds in an α-helix connect the i-th residue with the (i + 4)-th residue. Ding and coworkers adapted tetrapeptide signals as feature descriptors which represent proteins [45]. By assuming a binomial distribution, confidence levels of tetrapeptides, larger than a given cutoff, were used as an optimal set of features to represent tetrapeptide information.
3.3. Physicochemical Properties
In addition to features based on amino acid sequence, the physicochemical properties of individual amino acids have also been used in structural class prediction [1,36,46]. Due to differences in their side chains, the 20 natural amino acids are characterized by different physicochemical properties, including isoelectric point, molecular weight, polarity, polarizability, hydrophobicity, normalized van der Waals volume, average flexibility, and surface tension [52,53]. Most of these properties can be accessed in the amino acid index database [54], which is available online at https://www.genome.jp/dbget/aaindex.html, accessed on 1 February 2021.
When PseAAC was first proposed, Chou used a set of sequence order correlation factors to extract information from physicochemical properties [36]. Subsequently, different forms of sequence order correlation factors were also introduced [15,17,21,22]. Using a given protein of sequence and one or a set of properties of each amino acid , the sequence order correlation factors are defined as:
where, N and are the length of the protein and the µ-th rank of the coupling factor, respectively; is the correlation function, which can take various forms; and λ is the maximum correlation length or the maximum rank of the coupling factor. These correlation factors incorporate the sequence-order information to some extent, and they have previously been employed as feature vectors in the prediction of enzyme subfamily class and membrane protein type [55,56].
Prior to the concept of PseAAC, global protein sequence descriptors (GPSD) were proposed to include physicochemical properties such as hydrophobicity and solvent accessibility into the feature space [1]. In GPSD theory, amino acids are classified into two or more types according to their physicochemical properties. For example, based on hydrophobicity, amino acids can be categorized into hydrophobic, neutral, and polar types. By assigning a single letter to all the amino acids of the same type, each amino acid sequence can be converted into a property sequence in which each element represents one amino acid type. The GPSD consists of three descriptors: composition, transition, and distribution. The composition is the occurrence frequency of each amino acid type. The transition characterizes the frequencies with which amino acids changes from one type to a different type along the property sequence, analogous to ‘dipeptide’ information in the property sequence. The distribution describes the distribution pattern of each amino acid type along the sequence, and thus takes into account position-specific information in the property sequence.
3.4. Multiprofile Bayes
In multiprofile Bayes, the protein sequence is first treated as peptides of fixed length (usually starting from the N- and C- termini). From these fixed-length peptides, the occurrence of a specific type of amino acid at a given position can be estimated. Using this information, a 20 × L posterior probability matrix is then constructed, where L is the length of peptides. Each amino acid of a peptide is thus represented by its corresponding value in the posterior probability matrix, which includes both position-specific information and the posterior probability of each amino acid type. Thus, the peptide is represented by an L-dimensional vector, which is termed a single-profile Bayes. For each structural class, a posterior probability matrix can be constructed from proteins of the same structural class, and thus a multiple profile Bayes can be adapted to describe a single peptide. Multiprofile Bayes is a natural extension of biprofile Bayes, which was first applied by Shao and coworkers to predict methylation sites in proteins [47]. Multiprofile Bayes have also been employed to predict membrane protein type [48]. Recently, Khan and coworkers applied multiprofile Bayes in the prediction of PSC [16]. Since only the N- and C-termini of proteins were used in the construction of these position-specific profiles, sequences in the middle of the protein could simply be ignored.
3.5. Secondary-Structure-Based Features
Amino acids can also be categorized into three groups according their tendency to appear in one of the three major secondary structural elements: helix (H); strand (E); and coil (C) [12]. The protein sequence can thus be converted to a secondary structure sequence, from which GPSD can be used to extract features (similar to the case with physicochemical properties discussed above) [1]. Liu et al. also incorporated the maximum length and the average length of H, E, and C segments in the secondary structure sequence to enhance the predictive power of the classification models [23]. Secondary structure information was also integrated with physicochemical properties to form general PseAAC for PSC prediction [25,53].
3.6. Others
Additional descriptor features have also been constructed to represent proteins in an effort to predict PSC. Examples include position-specific scoring matrix based probability [16,18,19,54], Fourier transform based features [15,55], functional domain composition [37], split amino acid composition [16], approximate entropy [17], and image-based features that are derived from protein tertiary structures [57]. Several of these are discussed in greater detail below.
In PSSM-based probability, the position-specific scoring matrix (PSSM) is an evolutionary profile generated by the Position-Specific Iterative (PSI)-BLAST program using NCBI’s nonredundant (NR) database [18,19]. Hayat and coworkers converted amino acid sequence into a PSSM and then computed bi-gram probabilities descriptors [16]. Such PSSM-based bi-gram probabilities preserve both the order and the evolutionary information of the original sequence. Tao and coworkers extracted tri-gram features from the PSSM to construct a tri-gram occurrence matrix of 8000 elements [19].
In the Fourier transform based feature, a discrete Fourier transform (DFT) was applied to extract periodicities of physicochemical properties from the frequency domain [15,50]. In general, the low frequency components are more informative, because the high frequency components are noisy [50]. Thus, the low frequency coefficients are employed as feature descriptors. Zhang and Ding applied a DFT to an original property series comprised of hydrophobic or hydrophilic values of amino acids [50], while Sahu et al. employed DFT to extract low-frequency Fourier coefficients from a series of correlation factors of different ranks (see equation 2) [15]. While the correlation factors preserve the sequence order information from the protein sequence, the low frequency DFT coefficients preserve the global information from the protein sequence (along with some of the order information). In place of DFT, wavelet transformations were also employed to extract frequency coefficients [27,51].
In functional domain composition, each amino acid in the protein sequence is assigned to its functional domain composition type using the integrated domain and motif database [37]. In split amino acid composition, the protein sequence is divided into different segments and the composition of each segment is treated separately [16]. Thus, split amino acid composition can, to a certain extent, capture the position-specific information in a protein sequence.
4. Feature Selection
The different feature descriptors obtained using the information described in the previous section are frequently comprised of vectors of high dimension. However, there are several issues associated with the use of vectors of high dimension as an input for PSC prediction. First, many ML algorithms have trouble coping with high dimensional data. Second, the feature spaces described by vectors of high dimension show a great amount of redundancy. Third, the use of too many inputs is associated with overfitting or a reduction in prediction accuracy. Therefore, a high-dimensional feature space is commonly reduced into a low-dimensional feature space using feature selection techniques which select only the key features, thereby enhancing the speed and performance of classifiers [58]. Based on the characteristics of the features in the resulting low-dimensional feature space, feature selection approaches can be divided into two different types, type one and type two. In type one methods, a representative subset of the original features are selected. In type two methods, a smaller number of hybrid features are selected, and these can either be a linear combination of the original features or a nonlinear combination. In the following sections, several methods used in recent efforts to predict PSC are briefly introduced. For a comprehensive description of feature selection approaches, the authors are referred to a recent review, in which feature selection methods are classified into three classes based on the selection mechanism [59].
4.1. Minimum Redundancy-Maximum Relevance (mRMR)
The mRMR algorithm first proposed by Peng et al. is a type one method which selects a subset of features that minimizes the redundancy of the original feature space, removing features of low relevance to the target class [60]. This algorithm is especially useful for large-scale feature selection problems. mRMR has been employed by Li and coworkers in combination with a forward feature searching strategy to predict PSC using a dataset of 12,520 inputs from seven structural classes [46]. mRMR has also been used to generate a low-dimensional feature list for accessing the performance of multiple classifiers [29,61].
4.2. Genetic Algorithm
The Genetic Algorithm represents the selected features as a “chromosome”, and this information is then optimized by simulating biological evolution via natural selection and several associated genetic mechanisms [62]. This algorithm employs selection, crossover, and mutation operators to improve the chromosome, and subsequent performance is evaluated by a fitness function. The Genetic Algorithm selects these features in combination with a classification model. It has been coupled with the support vector machine (SVM) to search for an optimized subset of features, in which a fitness function combines the classification accuracy and the number of selected features [26].
4.3. Particle Swarm Optimization (PSO)
PSO is a global optimization algorithm. Introduced by Eberhart and Kennedy in 1995, PSO is a population-based stochastic evolutionary algorithm [63]. PSO is frequently employed in the optimization of parameters in neural network training [64,65]. In PSO, a random population (called a swarm) of candidate solutions (called particles) is first proposed. These particles are then moved around within the parameter space to search for a satisfactory (if not optimal) solution under the guidance of two types of memory: the cognitive memory, which is the optimum solution found by each individual particle; and the so-called “social memory”, which is the optimum solution visited by the whole swarm [66]. In PSC prediction, PSO has been applied in combination with neural networks to construct a set of hybrid descriptors from PSSM-based bi-gram probabilities and multiprofile Bayes [16]. PSO has also been used in the training of flexible neural trees [22].
4.4. Principal Component Analysis (PCA)
PCA is a simple, widely-used technique with many applications, including dimension reduction, lossy data compression, feature extraction, and data visualization [67]. By extracting relevant information from confusing datasets, PCA generates features that are a linear combination of the original features [68]. Moreover, the principal components are always independent of each other, and they always represent a lower dimension. The application of PCA for the prediction of PSCs was demonstrated by Du el al. in 2006 [68,69] and by Wang and coworkers in 2012 [70].
5. Classification Models
Given a set of features that capture the relevant information of a protein sequence for the purpose of protein structural classification, a classification model can be built to assign any protein sequence to one of the PSCs. Early efforts in predicting protein structural classes were mainly from the Chou group [7,9,11,37,71]. For each protein class, the geometric center of proteins in the feature space is deemed as the representative position for the protein class. Simple metrics are then used to measure the distance or similarity between the position of a query protein and the representative position for each protein class. The query protein is predicted to belong to the structural class to which it is closest or the structural class with the highest similarity. Several different metrics can be used, including the hamming distance as used in the least hamming distance method [72], the Euclidean distance as in the least Euclidean distance method [73], the Mahalanobis distance [71], or the correlation angle [74]. Alternatively, the position of the query protein can be expressed as a linear combination of the representative positions of all of the protein classes as in the maximum component coefficient method [75], and the predicted structural class is the one for which the component coefficient has the largest value. A representative position of each protein class other than the geometric center can also be used. For example, Zhang and coworkers applied fuzzy clustering to construct the representative positions (also called cluster centroids) [76]. In fuzzy clustering, each protein can belong to more than one structural class, with degrees of membership ranging from one and zero. The summation of membership degrees in all the classes should be one. A given protein is then assigned to the structural class for which its membership degree is maximum. For more details of these methods, the authors are referred to the excellent reviews by Chou and coworkers [7,9,30]. More recently, various ML methods have been applied to learn the statistical laws between feature descriptors of protein sequences in a training dataset and their corresponding structural classes, and to build a probabilistic model for classification purposes, as can been seen in a recent review on protein function prediction [38]. In the following, we focus on the very recent applications of ML methods in the prediction of PSC, which include artificial neural networks [15,22], support vector machine [23,26,50,77], K-nearest neighbor [16,17,46], random forest [4], logistic regression [78,79], and deep learning [80,81,82,83,84]. Table 3 provides a list of machine learning algorithms and their recent variants used as classification models in the prediction of proteins structural classes.

Table 3.
List of machine learning algorithms and their recent variants that are frequently used as classification models in the prediction of proteins structural classes.
5.1. Artificial Neural Networks (ANNs)
ANNs are inspired by the central nervous systems of animals in an attempt to find mathematical representations of information processing in biological systems [86]. ANNs have been successfully applied in medicine, physiology, philosophy, informatics, and many other scientific fields [86,87]. Flexible neural tree (FNT) is a special kind of ANN with flexible tree structures, first proposed by Chen and coworkers [88,89]. Bao and coworkers applied FNTs in the prediction of PSCs using four benchmark datasets: 640 (prediction accuracy, 84.5%); 1189 (prediction accuracy, 82.6%); ASTRAL (prediction accuracy, 83.5%); and C204 (prediction accuracy, 94.6%) [22]. Sahu and Panda employed another kind of neural network classifier, known as radial basis function neural network (RBFNN), in the prediction of PSCs using the standard datasets of C204, 277 domains, and 498 domains [15]. Because of their simple topological structure and their ability to learn in an explicit manner, RBFNN are especially useful for solving function approximation and pattern classification problems [90,91]. By utilizing Fourier transform based features and correlation factors for both hydrophobicity and hydrophilicity, the performance of RBFNN was observed to be better than the performances of the multilayer perceptron and linear discriminant analysis for all three datasets [15].
5.2. Support Vector Machine (SVM)
SVM, first proposed by Vapnik et al. in 1995, is a popular ML method for classification, regression, and abnormal point detection [92,93]. The core idea of SVM is to find a decision boundary where the margin is maximized [94]. In 2001, Cai and coworkers performed pioneering work in applying SVM to the prediction of PSCs, although only the amino acid composition was used and the test data set was rather small [95]. Recent efforts in the use of SVM have focused on larger data sets and/or improved SVM algorithms. The Binary-tree support vector machine (BT-SVM) uses a binary tree structure to organize two classes of SVM and thus form multiple classifiers, avoiding the problem of existing unclassifiable data points. Because of its good performance in solving multiclass classification problems, BT-SVM has become a research hotpot [96]. Zhang et al. formulated a 46-dimensional PseAAC and applied BT-SVM to the C204 dataset, yielding a predictive accuracy rate as high as 92.2% (which was significantly better than the performances of SVMs with a linear kernel function or poly kernel function) [50]. Liu et al. formulated a feature descriptor with 16 secondary structure-based features and applied a genetic algorithm to optimize the coefficients of these features in combination with SVM with a radial-based kernel function. This so-called GASVM algorithm was employed in the prediction of three low-similarity datasets: 25PDB (classification accuracy, 83.3%); 1189 (classification accuracy, 85.4%), and FC369 (classification accuracy, 93.4%) [23]. Li and coworkers formulated a feature vector with 1447 dimensions, and combined an improved genetic algorithm with SVM to construct a novel prediction model for PSC prediction of three datasets: C204 (predictive accuracy, 99.5%); 277 domains (predictive accuracy, 84.5%); and 498 domains (predictive accuracy, 94.2%) [26]. A dual-layer fuzzy support vector machine was also proposed for the classification of protein structure by Ding and coworkers, with an overall accuracy rate on the C204 dataset of 92.6% [77].
5.3. K-Nearest Neighbor (KNN)
As a basic and simple method for classification and regression, the K-nearest neighbor algorithm often uses the majority voting rule in classification problems. The nearest neighbor algorithm (K = 1) was recently employed by Li and coworkers as a classifier to assign proteins to one of the seven structural classes [46]. To enhance the performance of classical K-nearest neighbor algorithms, several different versions of the K-nearest neighbor classifier have been introduced. Successful examples include the optimized evidence-theoretic K-nearest neighbor (OET-KNN) algorithm and the fuzzy K-nearest-neighbor algorithm. The OET-KNN algorithm was shown by Hayat el al. to be a promising classifier, demonstrating high success rates with several datasets: 25PDB (87.0%); 640 (88.4%); and 1189 (86.6%) [16]. The fuzzy K-nearest-neighbor classifier was employed by Zhang and coworkers as a prediction engine, and the predictive accuracy rates of the proposed model for different datasets were: C204 (97.0%); and 1189 (56.9%) [17].
5.4. Random Forest
Random forest is a method of classification and prediction using multiple tree classifiers composed of multiple decision trees. The random forest algorithm has been used for regression, classification, clustering, ecological analysis, and other problems. When the random forest algorithm is used in classification or regression problems, its main idea is to resample using the bootstrap method, thus generating a large number of decision trees [21,97]. By incorporating both sequence and structure information, Wei et al. applied the random forest algorithm in the prediction of PSCs, and the overall accuracies of the proposed model on three benchmark datasets were: 25PDB (93.5%); 640 (92.6%); and 1189 (93.4%) [4]. Random forest was also recently employed as a classifier to predict protein fold types [98].
5.5. Logistic Regression
Kurgan and Chen applied a linear logistic regression classifier to a large set of 1673 twilight zone domains, and the proposed model achieved a prediction accuracy of 62% [78]. Jahandideh et al. used a multinomial logistic regression model in combination with ANN to evaluate the contribution of both AAC and dipeptide features in determining the PSC [79].
5.6. Deep Learning
Although a new ML research field, deep learning [99,100] has already been applied in a variety of protein processing tasks, including protein structure prediction [101,102], protein interaction prediction [103], protein secondary structure prediction [104], and protein subcellular localization prediction [105]. Panda and Majhi employed a deep recurrent neural network in the prediction of PSCs, obtaining high accuracies with small-size datasets (204, 277, and 498) and large-size datasets (PDB25, Protein 640, and FC699): 204 (85.36%); 277 (94.5%); 498 (95.9%); PDB25 (84.2%); Protein 640 (94.31%); and FC699 (93.1%) [105]. Klausen and coworkers have integrated deep learning into a tool (NetSurfP-2.0) to predict protein structural features with high accuracy and low runtime [106]. Nanni et al. rendered proteins into multiview 2D snapshots, to which convolutional neural networks were applied to identify PSCs [80].
In the prediction of PSC, many researchers evaluated the performance of multiple classifiers and chose the best classifier as the final model, while other researchers avoided the selection of classifiers by combining multiple algorithms together in a single prediction model [107,108]. Inspired by the application of majority voting systems in the recognition of handwriting characters [109], Chen and coworkers selected four algorithms from 11 candidates by mRMR and integrated them together through majority voting with weighted majority voting systems. The proposed model achieved a prediction accuracy rate of 68.6%, which was higher than the prediction accuracy rate achieved when only one of the 11 classifiers was used [108]. Durga and coworkers employed different ensemble techniques to integrate four complementary classifiers, and the proposed model provided highly accurate predictions for sequences with homologies ranging from 25% to 90% [107].
6. Cross Validation
To evaluate the performance of predictors, various cross validation approaches have been used. The three methods often used for cross validation in ML are hold-out cross validation (independent dataset), k-fold cross validation (resubstitution test), and leave-one-out cross validation (Jackknife test) [7,15,110]. Of these three methods, leave-one-out cross validation (Jackknife test) is the most objective approach, and is thus used by many researchers for examining the power of various prediction methods [16,26,50,111]. However, leave-one-out cross validation does not always provide better results than the other two methods. When the number of proteins in a given set (N) is not large enough, the leave-one-out method, in which each protein is in turn left out of the set, may result in a severe loss of information. Under such circumstances, the leave-one-out test cannot be utilized [9]. Chou and Zhang have provided a detailed introduction to these three methods [9].
7. Concluding Discussions and Perspectives
In this review, we have introduced the typical processes used in PSC prediction from protein sequences, emphasizing on feature descriptors that capture various kinds of information from protein sequences, and ML algorithms that have been frequently employed in recent publications. The prediction accuracies of existing methods on the benchmark datasets are high, although performance could be improved when predicting seven structural classes in large datasets. The challenge lies in predicting proteins whose structures share low similarities to those used in the dataset to train the prediction model [112]. An additional difficulty arises due to the existence of sequences that share a similar structure but low sequence similarity, these diverse structures share low sequence similarities to those used in the benchmark datasets or the datasets used to train the prediction model. We thus expect further improvements in both the construction of feature descriptors that extract additional information from the protein sequence and the development of new classifiers to train data-driven models. One obvious direction is to employ new features and classifiers that have been used in other classification problems, including the prediction of protein subcellular location [113], rational drug discovery [114,115], and non-coding RNA –protein interactions [116], to improve models for the prediction of PSC. In light of the astonishing success of AlphaFold in protein structure prediction [110], the development of new deep learning algorithms is also a promising approach to further improve predictive models’ performance. Furthermore, the advent of quantum computing could revolutionize the field of PSC prediction in both feature extraction and ML algorithms [117,118].
Many different feature descriptors have been formulated in this field, and a critical assessment of different types of features could be valuable in answering the following questions: Which kind of information is essential for the prediction of a specific subset of a PSC for which the performance of other features is not satisfactory? What are the best possible accuracy rates that can be achieved using particular kinds of features? What kind of information is still missing in the proteins for which all existing models have failed to provide a reliable classification? The answers to these questions may help deepen our understanding of the relationship between protein sequence and structural class.
The knowledge and experiences garnered from PSC prediction studies may shed some light on several different research frontiers in structural and molecular biology, including the prediction of the properties of intrinsically disordered proteins [119,120], transcription factors [121,122,123], splicing factor activities [123], and RNA structures [124,125].
Funding
We thank the financial support from the Natural Science Foundation of Guangdong Province, China (Grant No. 2020A1515010984) and the Start-up Grant for Young Scientists (860-000002110384), Shenzhen University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dubchak, I.; Muchnik, I.; Holbrook, S.R.; Kim, S.H. Prediction of protein folding class using global description of amino acid sequence. Proc. Natl. Acad. Sci. USA 1995, 92, 8700–8704. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Baldi, P. A machine learning information retrieval approach to protein fold recognition. Bioinformatics 2006, 22, 1456–1463. [Google Scholar] [CrossRef]
- Chou, K.-C. Structural Bioinformatics and its Impact to Biomedical Science. Curr. Med. Chem. 2004, 11, 2105–2134. [Google Scholar] [CrossRef]
- Wei, L.; Liao, M.; Gao, X.; Zou, Q. An Improved Protein Structural Classes Prediction Method by Incorporating Both Sequence and Structure Information. IEEE Trans. NanoBiosci. 2015, 14, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, L.; Cios, K.; Chen, K. SCPRED: Accurate prediction of protein structural class for sequences of twilight-zone similarity with predicting sequences. BMC Bioinform. 2008, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.D.; Feng, K.Y.; Lu, W.C.; Chou, K.-C. Using LogitBoost classifier to predict protein structural classes. J. Theor. Biol. 2006, 238, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Progress in protein structural class prediction and its impact to bioinformatics and proteomics. Curr. Protein Pept. Sci. 2005, 6, 423–436. [Google Scholar] [CrossRef]
- Bonetta, R.; Valentino, G. Machine learning techniques for protein function prediction. Proteins Struct. Funct. Bioinform. 2020, 88, 397–413. [Google Scholar] [CrossRef]
- Chou, K.-C.; Zhang, C.-T. Prediction of Protein Structural Classes. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 275–349. [Google Scholar] [CrossRef]
- Wang, Z.-X.; Yuan, Z. How good is prediction of protein structural class by the component-coupled method? Proteins Struct. Funct. Bioinform. 2000, 38, 165–175. [Google Scholar] [CrossRef]
- Liu, W.M.; Chou, K.-C. Prediction of protein structural classes by modified mahalanobis discriminant algorithm. J. Protein Chem. 1998, 17, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zou, Y.; Qin, J.; Liu, X.; Jiang, Y.; Ke, C.; Zou, Q. Hierarchical Classification of Protein Folds Using a Novel Ensemble Classifier. PLoS ONE 2013, 8, e56499. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, C.; Burge, L.; Mahmood, M.; Southerland, W.; Gloster, C. Protein Fold Classification with Genetic Algorithms and Feature Selection. J. Bioinform. Comput. Biol. 2009, 7, 773–788. [Google Scholar] [CrossRef]
- Chen, K.; Kurgan, L. PFRES: Protein fold classification by using evolutionary information and predicted secondary structure. Bioinformatics 2007, 23, 2843–2850. [Google Scholar] [CrossRef]
- Sahu, S.S.; Panda, G. A novel feature representation method based on Chou’s pseudo amino acid composition for protein structural class prediction. Comput. Biol. Chem. 2010, 34, 320–327. [Google Scholar] [CrossRef]
- Hayat, M.; Tahir, M.A.; Khan, S.A. Prediction of protein structure classes using hybrid space of multi-profile Bayes and bigram probability feature spaces. J. Theor. Biol. 2014, 346, 8–15. [Google Scholar] [CrossRef]
- Zhang, T.L.; Ding, Y.S.; Chou, K.-C. Prediction protein structural classes with pseudo-amino acid composition: Approximate entropy and hydrophobicity pattern. J. Theor. Biol. 2008, 250, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zheng, X.; Wang, J.; Chen, M.; Zhou, C. Prediction of protein structural class based on linear predictive coding of psi-blast profiles. Open Life Sci. 2015, 10, 529–536. [Google Scholar] [CrossRef]
- Tao, P.; Liu, T.; Li, X.; Chen, L. Prediction of protein structural class using trigram probabilities of position-specific scoring matrix and recursive feature elimination. Amino Acids 2015, 47, 461–468. [Google Scholar] [CrossRef]
- Kotsiantis, S.B. Supervised machine learning: A review of classification techniques. Informatica 2007, 31, 249–268. [Google Scholar]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef]
- Bao, W.; Chen, Y.; Wang, D. Prediction of protein structure classes with flexible neural tree. Bio-Med. Mater. Eng. 2014, 24, 3797–3806. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ma, M.; Zhao, T. A GASVM algorithm for predicting protein structure classes. J. Comput. Commun. 2016, 4, 46–53. [Google Scholar] [CrossRef][Green Version]
- Xiao, X.; Lin, W.Z.; Chou, K.-C. Using grey dynamic modeling and pseudo amino acid composition to predict protein structural classes. J. Comput. Chem. 2008, 29, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Brahnam, S.; Lumini, A. Prediction of protein structure classes by incorporating different protein descriptors into general Chou’s pseudo amino acid composition. J. Theor. Biol. 2014, 360, 109–116. [Google Scholar] [CrossRef]
- Li, Z.C.; Zhou, X.B.; Lin, Y.R.; Zou, X.Y. Prediction of protein structure class by coupling improved genetic algorithm and support vector machine. Amino Acids 2008, 35, 581–590. [Google Scholar] [CrossRef]
- Li, Z.-C.; Zhou, X.-B.; Dai, Z.; Zou, X.-Y. Prediction of protein structural classes by Chou’s pseudo amino acid composition: Approached using continuous wavelet transform and principal component analysis. Amino Acids 2008, 37, 415–425. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, S.; Zhang, L.; Qin, J.; Wang, J.; Tang, K. Prediction of protein structural class with Rough Sets. BMC Bioinform. 2006, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Pearl, F.M.; Sillitoe, I.; Orengo, C.A. Protein Structure Classification; American Cancer Society: Atlanta, GA, USA, 2015. [Google Scholar]
- Chou, K.-C. WITHDRAWN: An insightful recollection for predicting protein subcellular locations in multi-label systems. Genomics 2019. [Google Scholar] [CrossRef]
- Chou, K.-C. Some remarks on protein attribute prediction and pseudo amino acid composition. J. Theor. Biol. 2011, 273, 236–247. [Google Scholar] [CrossRef]
- Chou, K.-C. Retracted article: An insightful 20-year recollection since the birth of pseudo amino acid components. Amino Acids 2020, 52, 847. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Chou, K.-C.; Elrod, D.W. Protein subcellular location prediction. Protein Eng. Des. Sel. 1999, 12, 107–118. [Google Scholar] [CrossRef]
- Chou, K.-C.; Elrod, D.W. Prediction of membrane protein types and subcellular locations. Proteins 1999, 34, 137–153. [Google Scholar] [CrossRef]
- Chou, K.-C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct. Funct. Bioinform. 2001, 43, 246–255. [Google Scholar] [CrossRef]
- Chou, K.-C.; Cai, Y.-D. Predicting protein structural class by functional domain composition. Biochem. Biophys. Res. Commun. 2004, 321, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, J.S. A Review of Protein Function Prediction under Machine Learning Perspective. Recent Pat. Biotechnol. 2013, 7, 122–141. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.-B.; Chou, K.-C. PseAAC: A flexible web server for generating various kinds of protein pseudo amino acid composition. Anal. Biochem. 2008, 373, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, Q.-Z. Using pseudo amino acid composition to predict protein structural class: Approached by incorporating 400 dipeptide components. J. Comput. Chem. 2007, 28, 1463–1466. [Google Scholar] [CrossRef]
- Chou, K.-C. Using paircoupled amino acid composition to predict protein secondary structure content. Protein J. 1999, 18, 473–480. [Google Scholar] [CrossRef]
- Liu, W.-M.; Chou, K.-C. Prediction of protein secondary structure content. Protein Eng. Des. Sel. 1999, 12, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Sun, Z.B.; Sang, J.P.; Huang, S.Y.; Zou, X.W. Structural class tendency of polypeptide: A new conception in predicting protein structural class. Phys. Part A Stat. Mech. Appl. 2007, 386, 581–589. [Google Scholar] [CrossRef]
- Rackovsky, S. On the nature of the protein folding code. Proc. Natl. Acad. Sci. USA 1993, 90, 644–648. [Google Scholar] [CrossRef]
- Ding, H.; Lin, H.; Chen, W.; Li, Z.-Q.; Guo, F.-B.; Huang, J.; Rao, N. Prediction of protein structural classes based on feature selection technique. Interdiscip. Sci. Comput. Life Sci. 2014, 6, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lin, K.; Feng, K.; Cai, Y. Prediction of protein structural classes using hybrid properties. Mol. Divers. 2008, 12, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Xu, N.; Tsai, S.-N.; Wang, Y.; Ngai, S.-M. Computational Identification of Protein Methylation Sites through Bi-Profile Bayes Feature Extraction. PLoS ONE 2009, 4, e4920. [Google Scholar] [CrossRef] [PubMed]
- Hayat, M.; Khan, A. Memphybrid: Hybrid features-based prediction system for classifying membrane protein types. Anal. Biochem. 2012, 424, 35–44. [Google Scholar] [CrossRef]
- Xia, X.-Y.; Ge, M.; Wang, Z.-X.; Pan, X.-M. Accurate Prediction of Protein Structural Class. PLoS ONE 2012, 7, e37653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.-L.; Ding, Y.-S. Using pseudo amino acid composition and binary-tree support vector machines to predict protein structural classes. Amino Acids 2007, 33, 623–629. [Google Scholar] [CrossRef]
- Yu, B.; Lou, L.; Li, S.; Zhang, Y.; Qiu, W.; Wu, X.; Wang, M.; Tian, B. Prediction of protein structural class for low-similarity sequences using Chou’s pseudo amino acid composition and wavelet denoising. J. Mol. Graph. Model. 2017, 76, 260–273. [Google Scholar] [CrossRef]
- Dubchak, I.; Muchnik, I.; Mayor, C.; Dralyuk, I.; Kim, S.H. Recognition of a protein fold in the context of the Structural Classification of Proteins (SCOP) classification. Proteins Struct. Funct. Bioinform. 1999, 35, 401–407. [Google Scholar] [CrossRef]
- Anand, A.; Pugalenthi, G.; Suganthan, P.N. Predicting protein structural class by svm with class-wise optimized features and decision probabilities. J. Theor. Biol. 2008, 253, 375–380. [Google Scholar] [CrossRef]
- Kawashima, S.; Kanehisa, M. AAindex: Amino acid index database. Nucleic Acids Res. 2000, 28, 374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.B.; Chen, C.; Li, Z.C.; Zou, X.Y. Using Chou’s amphiphilic pseudo-amino acid composition and support vector machine for prediction of enzyme subfamily classes. J. Theor. Biol. 2007, 248, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2004, 21, 10–19. [Google Scholar] [CrossRef]
- Sadique, N.; Ahmed, A.A.N.; Islam, T.; Pervage, N.; Shatabda, S. Image-based effective feature generation for protein structural class and ligand binding prediction. PeerJ Comput. Sci. 2020, 6, e253. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. Feature Selection for High-Dimensional Data; Springer International Publishing: Cham, Switzerland, 2015. [Google Scholar]
- Cao, D.S.; Xu, Q.S.; Liang, Y.Z. propy: A tool to generate various modes of chou’s Pse-AAc. Bioinformatics 2013, 29, 960–962. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Ni, Q.; Chen, L. A feature and algorithm selection method for improving the prediction of protein structural class. Comb. Chem. High Throughput Screen. 2017, 20, 1. [Google Scholar] [CrossRef]
- Jalali-Heravi, M.; Kyani, A. Application of genetic algorithm-kernel partial least square as a novel nonlinear feature selection method: Activity of carbonic anhydrase II inhibitors. Eur. J. Med. Chem. 2007, 42, 649–659. [Google Scholar] [CrossRef]
- Kennedy, J. Particle swarm optimization. In Proceedings of the ICNN′95—International Conference on Neural Networks, Perth, Australia, 27 November–1 December 2011; Volume 4, pp. 1942–1948. [Google Scholar]
- Kaminski, M. Neural Network Training Using Particle Swarm Optimization—A Case Study. In Proceedings of the 2019 24th International Conference on Methods and Models in Automation and Robotics (MMAR), Miedzyzdroje, Poland, 26–29 August 2019; pp. 115–120. [Google Scholar]
- Meissner, M.; Schmuker, M.; Schneider, G. Optimized Particle Swarm Optimization (OPSO) and its application to artificial neural network training. BMC Bioinform. 2006, 7, 125. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Ji, G. A comprehensive survey on particle swarm optimization algorithm and its applications. Math. Probl. Eng. 2015, 2015, 931256. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal component analysis. J. Mark. Res. 2002, 87, 513. [Google Scholar]
- Jolliffe, I.T. Graphical Representation of Data Using Principal Components. In Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002; pp. 78–110. [Google Scholar]
- Du, Q.-S.; Jiang, Z.-Q.; He, W.-Z.; Li, D.-P.; Chou, K.-C. Amino Acid Principal Component Analysis (AAPCA) and Its Applications in Protein Structural Class Prediction. J. Biomol. Struct. Dyn. 2006, 23, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hu, X.; Cao, X. Identifying Protein Structural Classes Using MVP Algorithm. Int. J. Wirel. Microw. Technol. 2012, 2, 8–12. [Google Scholar] [CrossRef][Green Version]
- Chou, K.-C. A novel approach to predicting protein structural classes in a (20-1)-D amino acid composition space. Proteins Struct. Funct. Bioinform. 1995, 21, 319–344. [Google Scholar] [CrossRef]
- Chou, K.-C.; Maggiora, G.M. Domaˍin structural class prediction. Protein Eng. 1998, 11, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, H.; Nishikawa, K.; Ooi, T. The Folding type of a protein is relevant to the amino acid composition. J. Biochem. 1986, 99, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Prediction of protein folding types from amino acid composition by correlation angles. Amino Acids 1994, 6, 231–246. [Google Scholar] [CrossRef]
- Chou, K.-C.; Zhang, C.-T. A correlation-coefficient method to predicting protein-structural classes from amino acid compositions. JBIC J. Biol. Inorg. Chem. 1992, 207, 429–433. [Google Scholar] [CrossRef]
- Zhang, C.-T.; Chou, K.-C.; Maggiora, G.M. Predicting protein structural classes from amino acid composition: Application of fuzzy clustering. Protein Eng. 1995, 8, 425–435. [Google Scholar]
- Ding, Y.S.; Zhang, T.L.; Chou, K.C. Prediction of protein structure classes with pseudo amino acid composition and fuzzy support vector machine network. Protein Pept. Lett. 2007, 14, 811–815. [Google Scholar] [CrossRef]
- Kurgan, L.; Chen, K. Prediction of protein structural class for the twilight zone sequences. Biochem. Biophys. Res. Commun. 2007, 357, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Jahandideh, S.; Abdolmaleki, P.; Jahandideh, M.; Hayatshahi, S.H.S.; Hayatshahi, H.S. Novel hybrid method for the evaluation of parameters contributing in determination of protein structural classes. J. Theor. Biol. 2007, 244, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Lumini, A.; Pasquali, F.; Brahnam, S. iProStruct2D: Identifying protein structural classes by deep learning via 2D representations. Expert Syst. Appl. 2020, 142, 113019. [Google Scholar] [CrossRef]
- Jaiswal, M.; Saleem, S.; Kweon, Y.; Draizen, E.J.; Veretnik, S.; Mura, C.; Bourne, P.E. Deep learning of protein structural classes: Any evidence for an ‘Ur-fold’? In Proceedings of the 2020 Systems and Information Engineering Design Symposium (SIEDS), Charlottesville, VA, USA, 24 April 2020; IEEE: Charlottesville, VA, USA, 2020; pp. 1–6. [Google Scholar]
- Gao, M.; Zhou, H.; Skolnick, J. DESTINI: A deep-learning approach to contact-driven protein structure prediction. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Newaz, K.; Ghalehnovi, M.; Rahnama, A.; Antsaklis, P.J.; Milenkovic, T. Network-based protein structural classification. R. Soc. Open Sci. 2020, 7, 191461. [Google Scholar] [CrossRef]
- Bankapur, S.; Patil, N. An Enhanced Protein Fold Recognition for Low Similarity Datasets Using Convolutional and Skip-Gram Features With Deep Neural Network. IEEE Trans. NanoBioscience 2021, 20, 42–49. [Google Scholar] [CrossRef]
- Panda, B.; Majhi, B. A novel improved prediction of protein structural class using deep recurrent neural network. Evol. Intell. 2018, 4096, 1–8. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural networks for pattern recognition. Agric. Eng. Int. CIGR J. Sci. Res. Dev. Manuscr. PM 1995, 12, 1235–1242. [Google Scholar]
- Judith, E.D.; Deleo, J.M. Artificial neural networks. Cancer 2001, 91, 1615–1635. [Google Scholar]
- Chen, Y.; Yang, B.; Dong, J.; Abraham, A. Time-series forecasting using flexible neural tree model. Inf. Sci. 2005, 174, 219–235. [Google Scholar] [CrossRef]
- Yang, B.; Chen, Y.; Jiang, M. Reverse engineering of gene regulatory networks using flexible neural tree models. Neurocomputing 2013, 99, 458–466. [Google Scholar] [CrossRef]
- Park, J.; Sandberg, I.W. Approximation and Radial-Basis-Function Networks. Neural Comput. 1993, 5, 305–316. [Google Scholar] [CrossRef]
- Samantaray, S.; Dash, P.; Panda, G. Fault classification and location using HS-transform and radial basis function neural network. Electr. Power Syst. Res. 2006, 76, 897–905. [Google Scholar] [CrossRef]
- Cortes, C. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Cai, Y.-D.; Liu, X.-J.; Xu, X.-B.; Zhou, G.-P. Support Vector Machines for predicting protein structural class. BMC Bioinform. 2001, 2, 3. [Google Scholar] [CrossRef]
- Fei, B.; Liu, J. Binary tree of SVM: A new fast multiclass training and classification algorithm. IEEE Trans. Neural Networks 2006, 17, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.M.; Nasser, M.; Pal, B.; Ahmad, S. Support Vector Machine and Random Forest Modeling for Intrusion Detection System (IDS). J. Intell. Learn. Syst. Appl. 2014, 6, 45–52. [Google Scholar] [CrossRef]
- Li, J.; Wu, J.; Chen, K. PFP-RFSM: Protein fold prediction by using random forests and sequence motifs. J. Biomed. Sci. Eng. 2013, 6, 1161–1170. [Google Scholar] [CrossRef]
- Deng, L.; Yu, D. Deep learning: Methods and applications. Found. Trends Signal Process. 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Sidi, T.; Keasar, C. Redundancy-weighting the PDB for detailed secondary structure prediction using deep-learning models. Bioinformatics 2020, 36, 3733–3738. [Google Scholar] [CrossRef]
- Sun, T.; Zhou, B.; Lai, L.; Pei, J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinform. 2017, 18, 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, H.; Yi, Z. Protein secondary structure prediction by using deep learning method. Knowl. Based Syst. 2017, 118, 115–123. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.B.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.S.; Jespersen, M.C.; Nielsen, H.; Jensen, K.K.; Jurtz, V.I.; Sønderby, C.K.; Sommer, M.O.A.; Winther, O.; Nielsen, M.; Petersen, B.; et al. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Proteins Struct. Funct. Bioinform. 2019, 87, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Kedarisetti, K.D.; Kurgan, L.; Dick, S. Classifier ensembles for protein structural class prediction with varying homology. Biochem. Biophys. Res. Commun. 2006, 348, 981–988. [Google Scholar] [CrossRef]
- Chen, L.; Lu, L.; Feng, K.; Li, W.; Song, J.; Zheng, L.; Yuan, Y.; Zeng, Z.; Feng, K.; Lu, W.; et al. Multiple classifier integration for the prediction of protein structural classes. J. Comput. Chem. 2009, 30, 2248–2254. [Google Scholar] [CrossRef]
- Rahman, A.F.R.; Alam, H.; Fairhurst, M.C. Multiple classifier combination for character recognition: Revisiting the majority voting system and its variations. In International Workshop on Document Analysis Systems; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Larrañaga, P.; Calvo, B.; Santana, R.; Bielza, C.; Galdiano, J.; Inza, I.; Lozano, J.A.; Armañanzas, R.; Santafé, G.; Pérez, A.; et al. Machine learning in bioinformatics. Brief. Bioinform. 2006, 7, 86–112. [Google Scholar] [CrossRef]
- Kurgan, L.A.; Homaeian, L. Prediction of structural classes for protein sequences and domains—Impact of prediction algorithms, sequence representation and homology, and test procedures on accuracy. Pattern Recognit. 2006, 39, 2323–2343. [Google Scholar] [CrossRef]
- Zhu, X.-J.; Feng, C.-Q.; Lai, H.-Y.; Chen, W.; Hao, L. Predicting protein structural classes for low-similarity sequences by evaluating different features. Knowl. Based Syst. 2019, 163, 787–793. [Google Scholar] [CrossRef]
- Zhang, T.-H.; Zhang, S.-W. Advances in the Prediction of Protein Subcellular Locations with Machine Learning. Curr. Bioinform. 2019, 14, 406–421. [Google Scholar] [CrossRef]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Zhang, S.W.; Fan, X.N. Computational methods for predicting ncrna-protein interactions. Med. Chem. 2017, 13, 515–525. [Google Scholar] [CrossRef]
- Outeiral, C.; Strahm, M.; Shi, J.; Morris, G.M.; Benjamin, S.C.; Deane, C.M. The prospects of quantum computing in computational molecular biology. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11. [Google Scholar] [CrossRef]
- Mulligan, V.K.; Melo, H.; Merritt, H.I.; Slocum, S.; Weitzner, B.D.; Watkins, A.M.; Renfrew, P.D.; Pelissier, C.; Arora, P.S.; Bonneau, R. Designing Peptides on a Quantum Computer. bioRxiv 2019. [Google Scholar] [CrossRef]
- Li, J.; Feng, Y.; Wang, X.; Li, J.; Liu, W.; Rong, L.; Bao, J. An overview of predictors for intrinsically disordered proteins over 2010–2014. Int. J. Mol. Sci. 2015, 16, 23446–23462. [Google Scholar] [CrossRef] [PubMed]
- Vullo, A.; Bortolami, O.; Pollastri, G.; Tosatto, S.C.E. Spritz: A server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006, 34, W164–W168. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-L.; Su, W.; Wang, J.-S.; Yang, Y.-H.; Yang, H.; Lin, H. Predicting Preference of Transcription Factors for Methylated DNA Using Sequence Information. Mol. Ther. Nucleic Acids 2020, 22, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.; Eils, R.; König, R. RIP: The regulatory interaction predictor—A machine learning-based approach for predicting target genes of transcription factors. Bioinformatics 2011, 27, 2239–2247. [Google Scholar] [CrossRef]
- Mao, M.; Hu, Y.; Yang, Y.; Qian, Y.; Wei, H.; Fan, W.; Yang, Y.; Li, X.; Wang, Z. Modeling and Predicting the Activities of Trans-Acting Splicing Factors with Machine Learning. Cell Syst. 2018, 7, 510–520.e4. [Google Scholar] [CrossRef]
- Walia, R.R.; Caragea, C.; Lewis, B.A.; Towfic, F.; Terribilini, M.; El-Manzalawy, Y.; Dobbs, D.; Honavar, V. Protein-RNA interface residue prediction using machine learning: An assessment of the state of the art. BMC Bioinform. 2012, 13, 89. [Google Scholar] [CrossRef]
- Walia, R.R.; Xue, L.C.; Wilkins, K.; El-Manzalawy, Y.; Dobbs, D.; Honavar, V. RNABindRPlus: A Predictor that Combines Machine Learning and Sequence Homology-Based Methods to Improve the Reliability of Predicted RNA-Binding Residues in Proteins. PLoS ONE 2014, 9, e97725. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).