Protein Crystal Instance Segmentation Based on Mask R-CNN
Abstract
1. Introduction
2. Algorithm Design
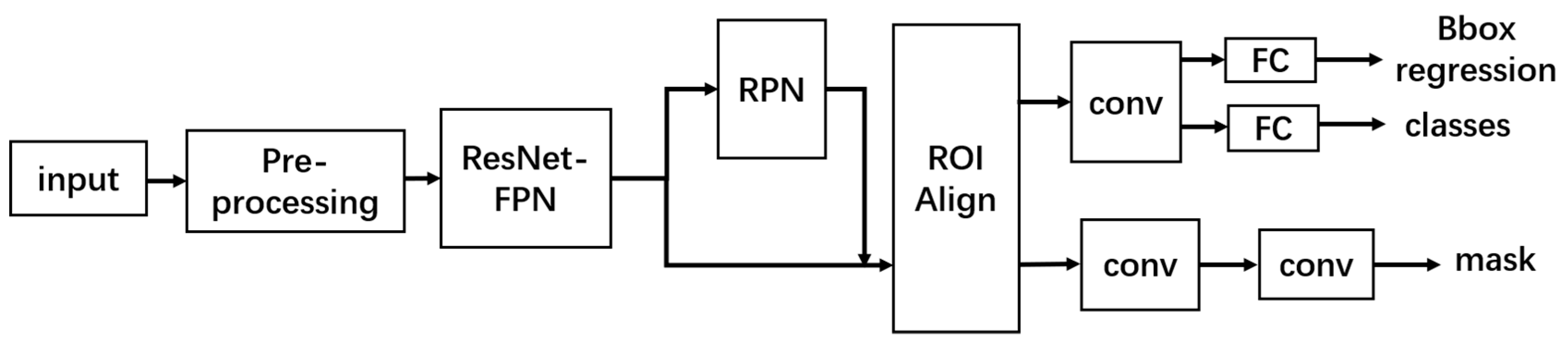
2.1. Network Introduction
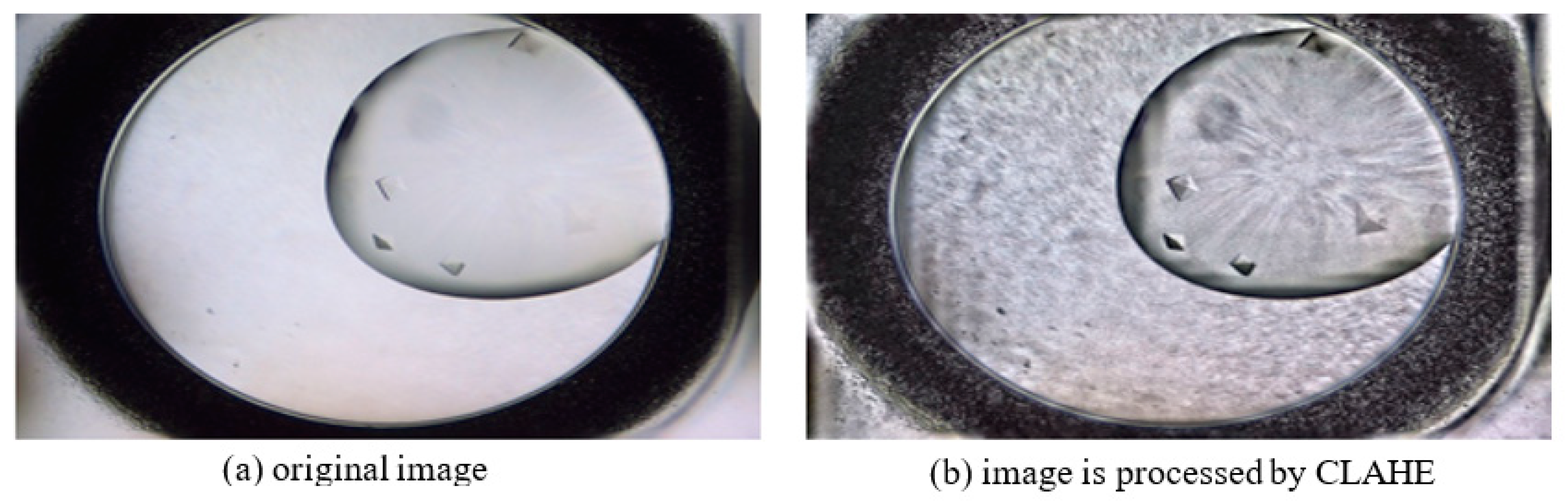
2.2. Pre-Processing Module
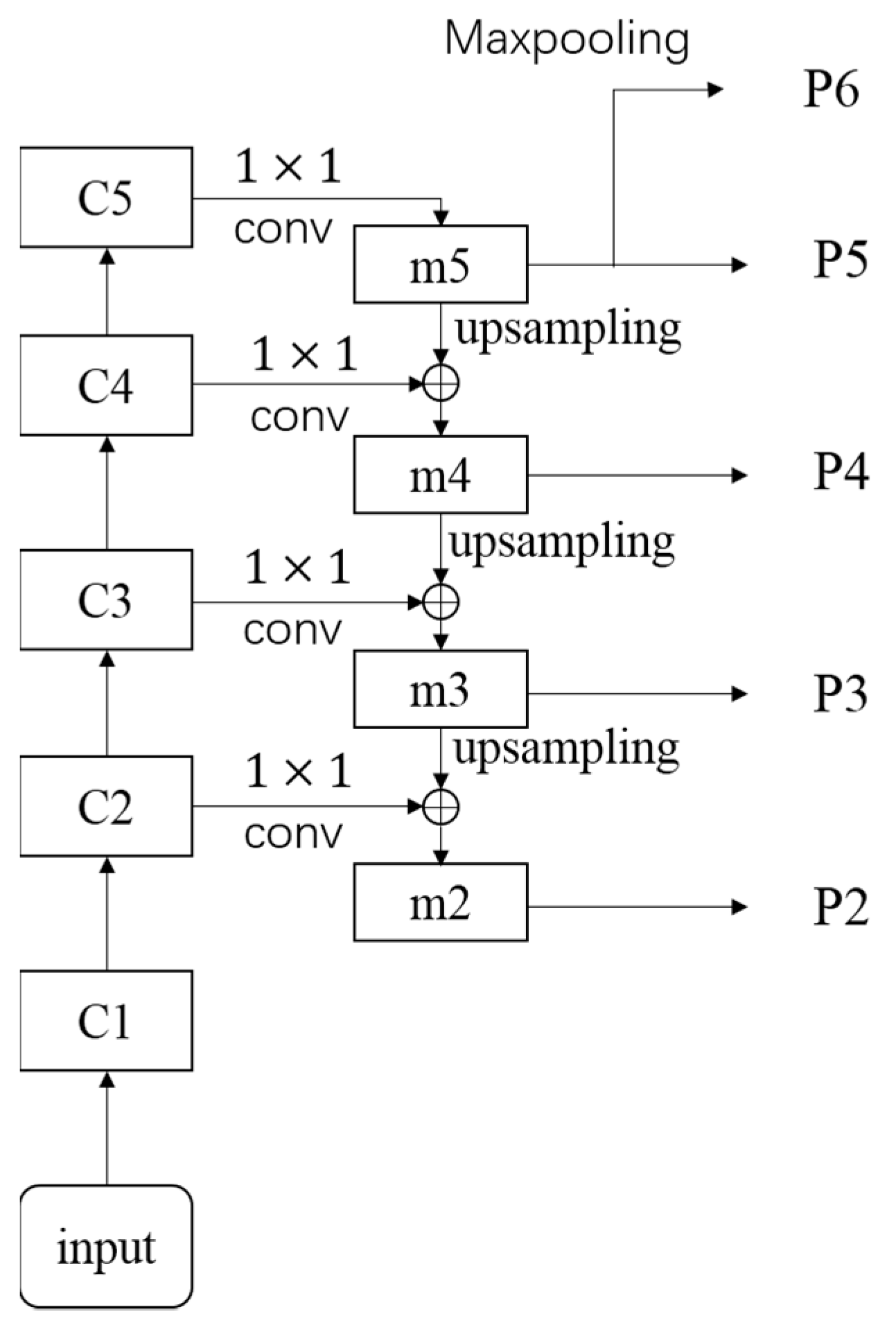
2.3. FPN Module
2.4. ROI Align Module
3. Results and Analysis
3.1. Experiment Platform
3.2. Experiment Dataset
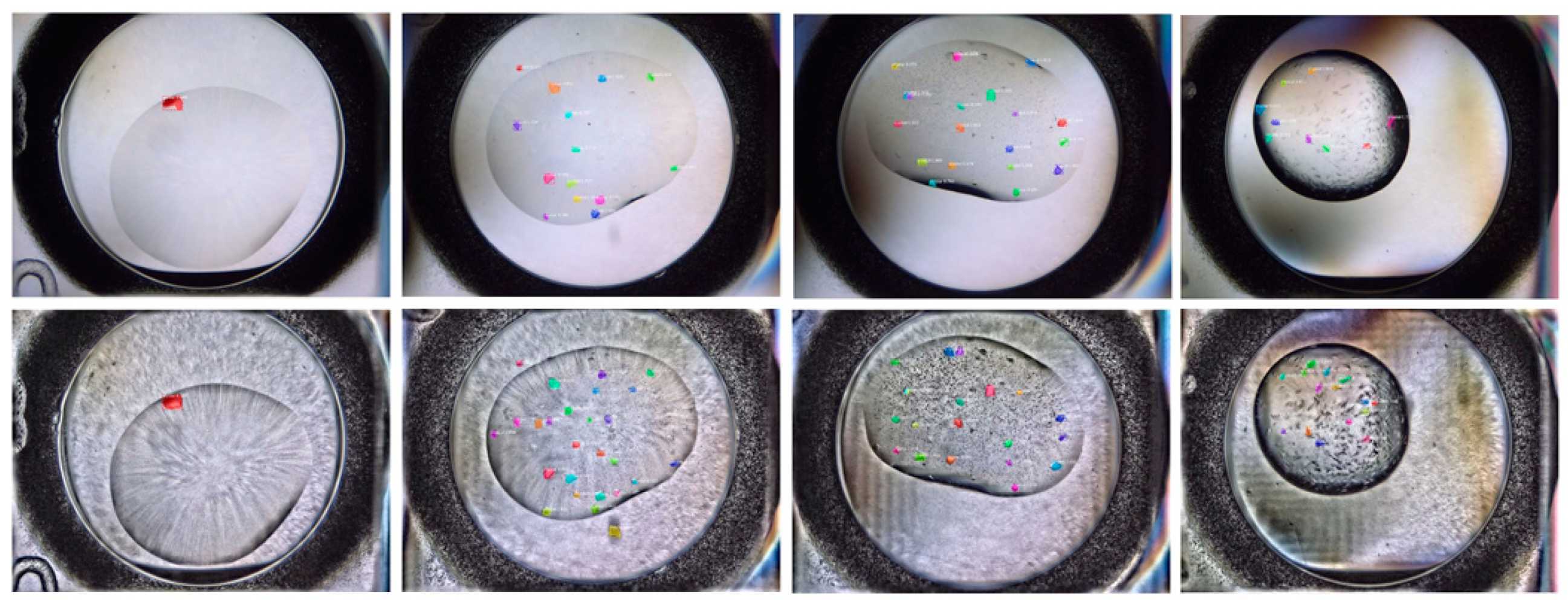
3.3. Experiment Results and Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blundell, T.L. Protein crystallography and drug discovery: Recollections of knowledge exchange between academia and industry. IUCrJ 2017, 4, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Spiliopoulou, M.; Valmas, A.; Triandafillidis, D.-P.; Kosinas, C.; Fitch, A.N.; Karavassili, F.; Margiolaki, I. Applications of X-ray Powder Diffraction in Protein Crystallography and Drug Screening. Crystals 2020, 10, 54. [Google Scholar] [CrossRef]
- Brink, A.; Helliwell, J. Why is interoperability between the two fields of chemical crystallography and protein crystallography so difficult? IUCrJ 2019, 6, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Theveneau, P.; Baker, P.; Barrett, R.; Beteva, A.; Bowler, M.W.; Carpentier, P.; Caserotto, H.; Sanctis, D.; Dobias, F.; Flot, D.; et al. The Upgrade Programme for the Structural Biology beamlines at the European Synchrotron Radiation Facility—High throughput sample evaluation and automation. In Proceedings of the 11th International Conference on Synchrotron Radiation Instrumentation, Lyon, France, 9–13 July 2012. [Google Scholar] [CrossRef]
- Ng, J.T.; Dekker, C.; Reardon, P.; Von Delft, F. Lessons from ten years of crystallization experiments at the SGC. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Tice, J.D.; Roach, L.S.; Ismagilov, R.F. A Droplet-Based, Composite PDMS/Glass Capillary Microfluidic System for Evaluating Protein Crystallization Conditions by Microbatch and Vapor-Diffusion Methods with On-Chip X-ray Diffraction. Angew. Chem. Int. Ed. 2004, 43, 2508–2511. [Google Scholar] [CrossRef]
- Kissick, D.J.; Wanapun, D.; Simpson, G.J. Second-Order Nonlinear Optical Imaging of Chiral Crystals. Annu. Rev. Anal. Chem. 2011, 4, 419–437. [Google Scholar] [CrossRef] [PubMed]
- Spraggon, G.; Lesley, S.A.; Kreusch, A.; Priestle, J.P. Computational analysis of crystallization trials. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Snell, E.H.; Luft, J.R.; Potter, S.A.; Lauricella, A.M.; Gulde, S.M.; Malkowski, M.G.; Koszelak-Rosenblum, M.; Said, M.I.; Smith, J.L.; Veatch, C.K.; et al. Establishing a training set through the visual analysis of crystallization trials. Part I: ∼150 000 images. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.E.; Charbonneau, P.; Newman, J.; Snell, E.H.; So, D.R.; Vanhoucke, V.; Watkins, C.J.; Williams, S.; Wilson, J.C. Classification of crystallization outcomes using deep convolutional neural networks. PLoS ONE 2018, 13, e0198883. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.G.; Wrapp, D.; Gilman, M.S.A.; Battles, M.B.; Wang, N.; Sacerdote, S.; Chuang, G.-Y.; Kwong, P.D.; McLellan, J.S. Iterative screen optimization maximizes the efficiency of macromolecular crystallization. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2019, 75, 123–131. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R.B. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.M. Realization of the Contrast Limited Adaptive Histogram Equalization (CLAHE) for Real-Time Image Enhancement. J. VLSI Signal Process. Syst. Signal Image Video Technol. 2004, 38, 35–44. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 937–944. [Google Scholar]


| ResNet101 | CLAHE-ResNet101 | IOU | |
|---|---|---|---|
| 10 images | 0.668 | 0.703 | 0.50 |
| 100 images | 0.302 | 0.430 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, J.; Zhang, Y.; Zhou, H.; Yu, F.; Sun, B.; Wang, Q. Protein Crystal Instance Segmentation Based on Mask R-CNN. Crystals 2021, 11, 157. https://doi.org/10.3390/cryst11020157
Qin J, Zhang Y, Zhou H, Yu F, Sun B, Wang Q. Protein Crystal Instance Segmentation Based on Mask R-CNN. Crystals. 2021; 11(2):157. https://doi.org/10.3390/cryst11020157
Chicago/Turabian StyleQin, Jiangping, Yan Zhang, Huan Zhou, Feng Yu, Bo Sun, and Qisheng Wang. 2021. "Protein Crystal Instance Segmentation Based on Mask R-CNN" Crystals 11, no. 2: 157. https://doi.org/10.3390/cryst11020157
APA StyleQin, J., Zhang, Y., Zhou, H., Yu, F., Sun, B., & Wang, Q. (2021). Protein Crystal Instance Segmentation Based on Mask R-CNN. Crystals, 11(2), 157. https://doi.org/10.3390/cryst11020157






