Abstract
The light-driven sodium pump KR2, found in 2013 in the marine bacteria Krokinobacter eikastus, serves as a model protein for the studies of the sodium-pumping microbial rhodopsins (NaRs). KR2 possesses a unique NDQ (N112, D116, and Q123) set of the amino acid residues in the functionally relevant positions, named the NDQ motif. The N112 was shown to determine the Na+/H+ selectivity and pumping efficiency of the protein. Thus, N112A mutation converts KR2 into an outward proton pump. However, no structural data on the functional conversions of the light-driven sodium pumps are available at the moment. Here we present the crystal structure of the N112A mutant of KR2 in the ground state at the resolution of 2.4 Å. The structure revealed a minor deflection in the central part of the helix C and a double conformation of the L74 residue in the mutant. The organization of the retinal Schiff base and neighboring water molecules is preserved in the ground state of KR2-N112A. The presented data provide structural insights into the effects of the alterations of the characteristic NDQ motif of NaRs. Our findings also demonstrate that for the rational design of the KR2 variants with modified ion selectivity for optogenetic applications, the structures of the intermediate states of both the protein and its functional variants are required.
Keywords:
microbial rhodopsin; retinal; ion transport; mutation; NDQ motif; sodium pump; X-ray crystallography 1. Introduction
Microbial rhodopsins (MRs) are light-sensitive membrane proteins, which use a retinal molecule as a cofactor. All MRs share a similar seven transmembrane (TM) α-helical fold [1]. The helices (named from A to G) are interconnected via extracellular and cytoplasmic loops, in many cases containing β-strands [2,3,4] or small helical regions [5,6,7,8]. The retinal is covalently bound to the highly-conserved lysine residue in the TM helix G via a protonated Schiff base (RSB).
The first MR, bacteriorhodopsin from the archaeon Halobacterium salinarum (HsBR), was discovered in 1971 [9]. HsBR is a light-driven proton pump, translocating proton outside of the cell and thus creating a transmembrane electrochemical gradient for the ATP synthesis [10]. Since 1971, many MRs with diverse functions were found and characterized [11,12]. The most significant discoveries were archaeal light-driven chloride pumps (halorhodopsins, HRs) [13] and sensory rhodopsins (SRs) [14], bacterial proton pumps (proteorhodopsins, PRs) [15], cation and anion channelrhodopsins (ChRs) [16,17,18,19,20], bacterial sodium and chloride pumps (NaRs and ClRs) [21,22], native inward proton pumps (xenorhodopsins, XeRs and schizorhodopsins, SzRs) [23,24,25,26], heliorhodopsins (HeRs) [27], rhodopsin genes in giant viruses [6,8,28,29], enzymorhodopsins [30,31], etc. It was recently shown the MRs are the major solar energy capturing proteins in the ocean [32]. The discovery of new MRs and the growth of the family led to the development of their biotechnological applications, where the most significant one is the optogenetics – able to control living tissues, such as neural or muscle, with light [33,34].
Despite the increasing number of MRs with novel properties, there is only one known group of the active non-proton cation transporters. They can pump Li+ and Na+ ions outside of the cell but are converted into the outward proton pumps in the presence of larger cations [21] or at low pH [35,36]. It was shown that under physiological conditions in the ocean (pH 8.0, ~500 mM NaCl), the proteins act almost exclusively as light-driven sodium pumps [36]; therefore, they were denoted as sodium-pumping rhodopsins (NaRs). First, described in 2013, a light-driven NaR from the marine bacteria Krokinobacter eikastus (KR2) [21], has yet become the most characterized representative of this group of MRs. KR2 remains the only NaR, for which the high-resolution structures are available [2,35,37].
Upon light illumination, KR2 undergoes a photocycle, comprised of the distinct K, L, M, and O intermediate states [21]. The L and M states of KR2 photocycle coexist, with a minor fraction of the M-state and the significant portion of the L. The sodium is not bound inside the protein in the ground state. The sodium uptake occurs with the decay of the M-state as it was also reported for another NaR from Gillisia limnaea (GLR) [38]. Thus, the late red-shifted O-state is the only one, where the ion is bound in the core of the KR2 protomer [21,38].
KR2, like all other NaRs, possesses a characteristic NDQ motif (N112, D116, and Q123 residues at the functionally relevant positions, corresponding to D85, T89, and D96 belonging to the helix C in HsBR) [4,39]. Some light was shed on the particular role of each of these amino acid residues. For instance, it was shown that D116 is a proton acceptor from the RSB and forms a direct hydrogen bond with it in the ground, K, L, and O-states [2,35,37,40]. The bond is absent only in the M-state when the proton is translocated from the RSB to the D116 side chain [40]. Recently it was shown that D116 is also a part of the transient sodium binding site near the RSB, formed in the O-state of KR2 photocycle [41]. It was demonstrated that the substitution of the D116 with other residues lead to the loss of the sodium-pumping function of KR2 [21]. The Q123 is a part of a gate at the cytoplasmic side of the protein, separating a large ion-uptake cavity from the RSB region in the ground state [2,35,37]. However, Q123A/V mutants retain sodium-pumping activity, although it is much lower than that of the wild type (WT) protein [37,42]. Thus, it was suggested that the role of Q123 is in the optimization of the sodium pathway inside the protein, and in the creation of the proper inter-helical interactions in the protein [43].
The role of the N112 was investigated by an extensive mutational analysis [44]. It was shown that N112D/G/S/T mutants are able to pump sodium, while N112A/C/P/V/E/Q/L/I/M/F/W mutations convert KR2 into an outward proton pump. N112H/K/Y/R substitutions lead to the loss of any pumping activity of the protein [44]. At the same time, Fourier-transformed infrared (FTIR) spectroscopy data on the N112A mutant of KR2 showed no alterations of the RSB region in the ground and the K-state of the mutant [45]. The N112 is a part of the transient sodium binding site in the O intermediate state [41]. Thus, the N112 of the NDQ motif is believed to determine the ion selectivity of KR2. However, no structural information on the N112X variants of KR2 is available at the moment, which limits our understanding of the N112 role in functional conversions of NaRs.
The goal of this study is to add important structural information on the functional Na+-to-H+ pump conversion of KR2. For that we present a crystal structure of the N112A mutant of KR2 in the ground state at 2.4 Å resolution. We show that N112A mutation changes neither the oligomeric form nor the internal organization of the protein. However, in KR2-N112A, the helix C is slightly deflected, and the side chain of the L74 residue adopts two alternative conformations. The lack of asparagine at the 112th position of KR2 leads to the enlargement of the polar cavity near the RSB. The structure provides insights into the rearrangements in the NaRs upon alteration of the characteristic NDQ motif. It might also contribute to the rational design of the KR2 variants with modified ion selectivity.
2. Materials and Methods
2.1. Cloning, Protein Expression, and Purification
Cloning, protein expression, and purification were performed as described previously [2,35]. KR2 gene, synthesized commercially (Eurofins Scientific, Luxembourg), was subcloned into the pET15bmod (Apr) expression plasmid using the restriction sites for XbaI and XhoI enzymes (Thermo Fisher Scientific, Waltham, MA, USA). A point N112A mutation was introduced into the KR2-gene-containing plasmid by PCR mutagenesis. E. coli cells of strain C41 (DE3) (Lucigen, Middleton, WI, USA) were transformed with the KR2-N112A expression plasmid. Transformed cells were grown at 37 °C in shaking baffled flasks in an autoinducing medium (ZYP-5052 [46]) containing 100 mg/L ampicillin and were induced at optical density OD600 of 0.7–0.9 with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 10 μM all-trans-retinal. 3 h after induction, the cells were collected by centrifugation at 4000× g for 30 min. Harvested cells were disrupted in M-110P Lab Homogenizer (Microfluidics, Westwood, MA, USA) at 25,000 psi in a buffer containing 20 mM Tris-HCl pH 8.0, 5% glycerol, 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MI, USA) and 50 mg/L DNase I (Sigma-Aldrich, St. Louis, MI, USA). The membrane fraction of the cell lysate was isolated by ultracentrifugation at 90,000× g for 1 h at 4 °C. The pellet was resuspended in a buffer containing 50 mM NaH2PO4/Na2HPO4 pH 8.0, 0.1 M NaCl and 1% n-Dodecyl β-D-maltoside (DDM) (Anatrace, Affymetrix, Maumee, OH, USA) and stirred overnight for solubilization at 4 °C. The insoluble fraction was removed by ultracentrifugation at 90,000× g for 1 h at 4 °C. The supernatant was loaded on the Ni-NTA column (Qiagen, Hilden, Germany) and KR2 was eluted in a buffer containing 50 mM NaH2PO4/Na2HPO4 pH 7.5, 0.1 M NaCl, 0.5 M imidazole and 0.1% DDM. The eluate was subjected to size-exclusion chromatography on 24 mL Superdex 200i column (GE Healthcare Life Sciences, Marlborough, MA, USA) in a buffer containing 50 mM NaH2PO4/Na2HPO4 pH 7.5, 0.1 M NaCl, 0.05% DDM. Protein-containing fractions with the minimal A280/A525 absorbance ratio were pooled and concentrated to 60 mg/mL for crystallization.
2.2. Crystallization
The crystals were grown using the in meso approach [47], similar to our previous works [35,48]. The solubilized protein in the crystallization buffer was added to the monooleoyl-formed lipidic phase (Nu-Chek Prep, Elysian, MN, USA). The best crystals were obtained using the protein concentration of 20 mg/mL. The crystals were grown using the precipitate 1.2 M sodium malonate pH 8.0 (Hampton Research, Aliso Viejo, CA, USA). Crystallization probes were set up using the NT8 robotic system (LCP version, Formulatrix, Bedford, MA, USA). The crystals were grown at 22 °C and appeared in 2–4 weeks. Before harvesting, the crystallization drop was opened and covered with 3.4 M sodium malonate solution, pH 8.0, to avoid dehydration. All crystals were harvested using micromounts (MicroLoops HT, MiTeGen, Ithaca, NY, USA) and were flash-cooled and stored in liquid nitrogen for further crystallographic analysis.
2.3. Acquisition and Treatment of Diffraction Data
The diffraction data were collected at 100 K at the Swiss Light Source (SLS, Villigen, Switzerland) beamline X06SA (PXI) equipped with an EIGER X 16M detector (Dectris, Baden-Daettwil, Baden, Switzerland). The data collection statistics are reported in Table 1. The diffraction was anisotropic as determined by decay of the CC1/2 values in 20° cones along the reciprocal cell directions. Diffraction images were processed using XDS [49] (version from 15 March 2019). XSCALE (version from 15 March 2019) was used to merge different datasets. Staraniso server [50] was used to merge, scale, assess the quality, convert intensities to structure factor amplitudes, and generate Free-R labels.

Table 1.
Crystallographic data collection and refinement statistics. R.m.s.: root mean square.
2.4. Structure Determination and Refinement
The structure was solved using molecular replacement with MOLREP [51] (version 11.7.02) and the structure of the wild type KR2 in the pentameric form (Protein Data Bank Identifier 6YC3 [41]) as a search model. The model was refined manually using Coot [52] (version 0.8.9.2) and REFMAC5 [53] (version 5.8.0258) from the CCP4 suite [54]. The refinement statistics are summarized in Table 1.
2.5. Modeling of the O-State Structures of KR2-N112X Mutants
The hypothetical models of the O-state of N112A and N112G mutants were created for illustration of the proposed mechanism of functional conversions in KR2 using PyMOL (version 2.3.1). For that, the N112 of the 6XYT model (the O-state of the WT KR2) was substituted with either Ala or G residues. In the case of N112G mutant, the putative water molecule was added at the position of the OD1 atom of N112.
3. Results and Discussion
3.1. N112A Mutation Does Not Alter the Pentameric Assembly of KR2
We have expressed and purified KR2-N112A in E. coli strain C41 cells. Following the previously reported data, the purified protein has a spectrum with the characteristic maximum absorption wavelength of 522 nm [42,44].
It was shown that KR2 forms pentamers when solubilized in the DDM at pH higher than 6.0 or reconstituted in the lipids at both acidic and neutral pH values [35,55]. KR2 is also in the pentameric form in the crystals grown using in meso approach [47] at the pH as low as 4.9 [2,35,56]. Importantly, KR2 pumps sodium, only being in the pentameric form [35,55]. Nothing is known on the functionality of the monomeric form of the protein. Therefore, the analysis of the oligomeric assembly of the protein and its functional mutants could provide valuable insights into the basis of the pentamerization of KR2 and its functional role. For instance, a recent analysis of the size-exclusion chromatography (SEC) profile of the D116N mutant of KR2 demonstrated that this mutation destabilizes the protein pentamers [41].
The oligomeric state of the KR2-N112A was shown to be pentameric in lipids [55]. However, there is a great penalty in terms of free energy for the protein oligomer to dissociate being inside the lipid membrane. Thus, to probe the stability of the KR2-N112A pentamers, its oligomeric state should be studied in other environments, which allow the disruption of the protein complex. The more detailed analysis of the KR2-N112A is also of specific interest, as the N112 side chain is pointed towards the oligomerization interface of KR2 and is hydrogen-bonded via a water molecule (Wat471 in the case of the chain A of the 6YC3 model [41]) to the main chain oxygen of H30 residue of the nearby protomer, additionally stabilizing the pentameric assembly of the protein (Figure 1a,b).
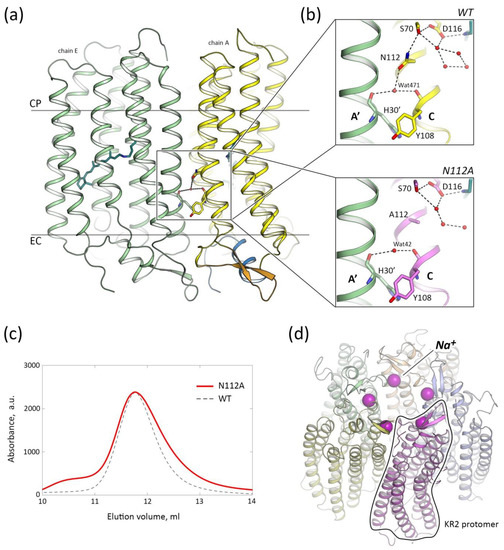
Figure 1.
Pentameric organization of KR2-N112A. (a) Side view of two neighboring protomers (chain A, yellow and chain E, green of the 6YC3 model) of KR2 pentamer in the membrane. N-terminal α-helix of chain A is colored blue. BC loop of the chain A is colored orange. Cytoplasmic (CP) and extracellular (EC) sides of the membrane are marked. (b) Detailed view of the inter-protomers contacts in the region of the N112 of the WT (top part) and A112 of the N112A mutant (bottom part). Neighboring protomer is colored green. The retinal cofactor is colored teal. Helices are marked with bold capital letters. (c) Size-exclusion chromatography (SEC) profiles of the N112A mutant (red line) and WT KR2 (black dashed line). The position of the absorption peak is the same for the WT KR2 and N112A variant (~11.8 mL when using 24 mL Superdex 200i column). The absorption was measured at 525 and 522 nm for the WT [21] and N112A [42] KR2, respectively. The small left peak of the KR2-N112A profile likely corresponds to the fraction of the aggregates. (d) KR2-N112A pentamer, view from the cytoplasmic side. Sodium ions, located at the oligomerization interface, are shown with purple spheres. One of five KR2 protomers is contoured for clarity.
In the present study, we performed SEC for the purification and analysis of the KR2-N112A. The SEC profile is similar to that of the WT protein (Figure 1c). The slight broadening of the peak in the KR2-N112A SEC profile may originate from the different viscosity of the samples, likely due to the fluctuating detergent concentration (Figure 1a). We cannot exclude that the tailing of the SEC profile peak may also come from other differing properties of the samples. Nevertheless, the KR2-N112A did not pass through the 100 kDa filter when concentrated for the crystallization, which is characteristic for the pentameric WT protein, but not for smaller oligomers of, for instance, H30K and Y154F mutants of KR2 [35]. As will be described later, KR2-N112A also forms pentamers in crystals, similar to those of the WT protein (Figure 1d).
Therefore, we conclude that the interactions of the N112 side chain are not critical for the pentamerization of KR2, and the water molecule, hydrogen-bonded to the N112 in the ground state, is likely to appear for the stabilization of the polar side chain of the residue.
3.2. Crystal Structure of the N112A Protomer
The N112A mutant was concentrated to ~60 mg/mL and crystallized using in meso approach [47]. The crystals appeared within a month and diffracted to 2.4 Å resolution. We obtained the KR2-N112A structure in its biologically relevant pentameric form at the resolution of 2.4 Å. As the diffraction of the KR2-N112A crystals was anisotropic, we merged the X-ray diffraction data from 6 crystals to improve the quality of the electron density maps and the corresponding model (Table 1, Figure S1).
The crystal packing, as well as the overall fold of the mutant, is very similar to that of the WT protein (Figure 1d). The root-mean-square deviation (RMSD) between the ground state structures of the N112A mutant and WT KR2 (PDB ID: 6YC3 [41]) is 0.15 Å. At the same time, 109–115 residues of the helix C are slightly shifted closer to the core of the protein (Figure 2a). Thus, the helix C of KR2-N112A is deflected in the same manner as it appears in the «compact» conformation of pentameric KR2 [2,56] and also in the monomeric form of the protein [2,35] (Figure 2b). However, in the case of the mutant, the shift of the helix is much smaller (Figure 2b).
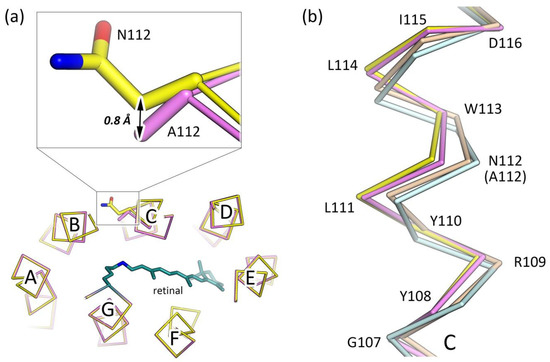
Figure 2.
Deflection of the helix C in the N112A mutant of KR2. (a) Displacement of the Cβ atoms of A112 and N112 of the N112A and WT KR2 structures, respectively. The 0.8 Å shift towards the inside of the KR2 protomer is shown with a black arrow. The retinal cofactor is colored teal. (b) Side view of the structural alignment of the helix C of the “expanded” conformation of the WT KR2 (yellow, PDB ID: 6YC3 [41]), KR2-N112A (violet, PDB ID: 6YT4, present work), “compact” conformation of the WT KR2 (light-orange, PDB ID: 4XTN, chain I [2]), and monomeric form of the WT KR2 (light-cyan, PDB ID: 4XTL [2]).
The sodium ion, found in the structures of the WT protein at the pentamerization interface, is also present in the model of KR2-N112A (Figure 1b). The organization of the binding site is the same as that in the WT protein, which is in contrast to the FTIR spectroscopy data on the N112A mutant of KR2 presented in [21]. Indeed, Inoue et al. found that KR2-N112A binds sodium in the ground state; however, the binding site is modified in the mutant [21]. Similar inconsistency of the FTIR spectroscopy data on KR2 with the high-resolution structural data was already found for the H30A mutant of the protein. Particularly, the same study [21] reported no sodium binding in the ground state of KR2-H30A. By contrast, the binding site was revealed in the 2.2 Å resolution crystal structure of the mutant in its pentameric form (PDB ID: 6YC1) [41]. In this case, similarly to the N112A mutant, the site is identical to that of the WT protein. We suggest that such inconsistency may originate from the differences of the sample preparation, or from the different lipid environments of the protein in the spectroscopy experiments and in the crystals.
3.3. The Organization of the RSB Region of KR2-N112A
The WT KR2 may form two principal conformations, which were named «expanded» and «compact» (Figure 3b,c) [35,56]. In the «expanded» conformation, there is a large water-filled cavity near the RSB, called the Schiff base cavity 1 (SBC1) [35]. In this configuration, the N112 side chain is pointed outside of the KR2 protomer and faces the oligomerization interface (Figure 1a,b). It is stabilized by the hydrogen bond with the water molecule (Wat471 for the chain A of the 6YC3 model [41]) (Figure 3b). To avoid the steric conflict, L74 side chain orients inside the protomer. On the contrary, in the «compact» conformation, the SBC1 is absent, and N112 is turned to the D116 and S70 residues and is hydrogen bonded to these residues (Figure 3c). Consequently, L74 is flipped outside the protein to allow enough space for the N112 (Figure 3c).
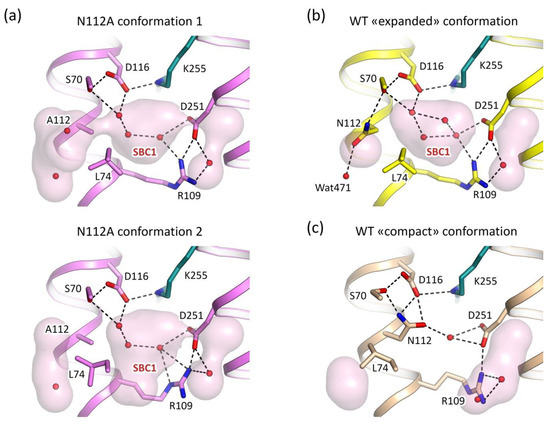
Figure 3.
Schiff base region of the N112A mutant. (a) Two conformations of the RSB region of the N112A mutant of KR2 (PDB ID: 6YT4, present work). The flip of the L74 side chain leads to the enlargement of the Schiff base cavity 1 (marked SBC1 in the Figure) in the conformation 2. (b) The “expanded” conformation of the WT KR2 (PDB ID: 6YC3 [41]). (c) The “compact” conformation of the WT KR2 (PDB ID: 4XTN, chain I [2]). Water molecules are shown with red spheres. Cavities were calculated using HOLLOW [57] and are shown with a pink surface. Hydrogen bonds are shown with black dashed lines. The side chain of the K255 residue and covalently bound retinal cofactor are colored teal.
In the ground state under physiological conditions, and being in the biologically relevant pentameric state, KR2 is in the «expanded» conformation [35] (Figure 3b). The conformation, similar to the «compact» one, appears transiently in the O intermediate state of the KR2 photocycle when the sodium ion is bound in the core of the protein [41]. Thus, the conformational «expanded»-to-«compact» and «compact»-to-«expanded» switches guide the uptake and release of the sodium ion by the protein [41].
Initially, the «compact» conformation was found in the ground state of the WT KR2 at acidic pH values [35] (Figure 3c). It was also demonstrated that KR2 pumps protons at pH 4.3 [35]. Therefore, the «compact» conformation was suggested to be a determinant of the proton-pumping mode of KR2 [35]. However, the proton-pumping ability of KR2 at low pH values remains speculative due to the small ∆pH signals in the reported experiments [35]. Consequently, authors of ref [35] suggested that alternatively, at acidic pH, KR2 is in an inactive state with the protonated RSB counterion D116. If this is the case, the residual weak pumping activity might correspond to the remaining minor fraction of the protein with deprotonated D116 [35]. As N112A mutation converts KR2 into the proton pump, the study of the conformation of the ground state of KR2-N112A under physiological conditions might help to distinguish between the two proposed mechanisms of KR2 functional and structural switches upon pH decrease. It could also provide insights into the roles of the «expanded» and «compact» conformations of KR2.
Consequently, we next analyzed the organization of the RSB region of the N112A mutant of KR2. In the mutant, the asparagine side chain of 112th residue is absent, which gives more space for the L74. Following this, L74 adopts two alternative conformations, one of which is similar to that of the «expanded» form (Figure 3a, Figure S1). At the same time, the second is closer to that of the «compact» conformation of the WT protein (Figure 3a, Figure S1).
It should be noted that the R109 side chain adopts two conformations, similar to those in the monomeric model of the KR2 (PDB ID: 4XTL [2]). This is most likely caused by the minor deflection of the helix C (Figure 2b).
Although A112 is located closer to the water molecules in the SBC1 of the KR2 ground state (Cβ atom of A112 in the N112A mutant is shifted by 0.8 Å to the inside of the protein in comparison to the Cβ atom of N112 in the 6YC3 model), the positions of the water molecules in the cavity remain the same to those in the WT protein (Figure 2a and Figure 3a). This is consistent with the recent FTIR spectroscopy data on KR2 and its N112A mutant, where the authors concluded from the N-D stretching vibrations shifts that the organization of the RSB region is nearly the same in the N112A and WT proteins [45]. Hence, the SBC1 is retained in KR2-N112A and is of the same size as that in the WT protein; it is even enlarged in the second conformation of the molecule when L74 is flipped outside the protomer.
Therefore, we conclude that KR2-N112A shapes the conformation, similar to the «expanded» one of the WT protein, however, with minor alterations. In other words, N112A mutation does not result in the appearance of the «compact» conformation in the ground state of the protein, which could be associated with the conversion of KR2 into a proton pump. This allows us to speculate that either the forming of the «compact» conformation is not the determinant of the switch from the sodium- to the proton-pumping mode, or there could be other mechanisms underneath the functional conversion of the KR2-N112A.
3.4. Role of N112 in the Ion Selectivity of KR2
As it was demonstrated in the mutational study of KR2, N112 is (at least) one of the determinants of the Na+/H+ selectivity of the protein [44]. As the N112 side chain is pointed away from the active center of the protein in the ground state and interacts neither with the water molecules in the SBC1 nor with the other critical residues of the putative sodium pathway, the molecular basis of the mechanism of this selectivity remains elusive.
Recently, the crystal structure of the O intermediate state of KR2 has been reported, which showed the transient sodium binding site in the core of the KR2 protomer (PDB ID: 6XYT) [41]. It was shown that with the rise of the O-state, N112 flips towards D116 and S70 residues (Figure S2). Thus, the orientation of the N112 is similar to that in the «compact» conformation. In this configuration, the ion is coordinated by the side chains of S70, N112, and D116 residues. The geometry is adapted for the binding of sodium. Indeed, the cation is coordinated by five oxygen atoms with a mean sodium-oxygen distance of 2.3 Å (Figure S2). However, the role of N112 in the ion selectivity of KR2 was not discussed in [41].
The flipping motion of the N112 side chain could not occur in the KR2-N112A; thus, the «expanded»-to-«compact» conformational switch is also not possible in the mutant. At the same moment, it was shown that KR2-N112A does not form the O-state [42,44]; however, it forms all other intermediates, such as K, L, and M. Therefore, we suggest that N112 is perfectly optimized in KR2 (as well as the corresponding Asn in any other NaRs) for an accurate transient binding of the sodium ion in the O-state, and does not show any key structural and/or functional role in the ground state of the protein.
In this approximation, the fact that N112D/S/T/G mutants retain sodium-pumping activity [44] could be easily explained. Indeed, Asp often coordinates sodium ions in proteins and is similar to Asn in terms of geometry and size. Ser and Thr are also able to coordinate cations due to the presence of the oxygen atoms in their side chains. However, their size is smaller than that of the asparagine group, which may explain significantly lowered sodium-pumping activity of the N112S/T mutants. In the case of KR2-N112G, the retaining of the sodium pumping activity is surprising. At the same time, the transiently bound sodium ion in N112G mutant is likely coordinated by the additional water molecule, which may be located at the position of the N112 OD1 atom of the O-state of KR2 (PDB ID: 6XYT [41]) (See Section 2.5 of Materials and Methods, Figure S2). The appearance of the water molecule at this position is constricted in the case of N112A, due to the steric conflict with the Ala side chain, which may explain the absence of the sodium-pumping activity of the KR2-N112A, but not of KR2-N112G (Figure S2).
On the other hand, similar to N112A, N112C/P/V/E/Q/L/I/M/F/W mutations convert KR2 into a proton pump. This fact is easily understandable since these mutations cannot support efficient sodium binding inside the protein. Therefore, they also cannot provide the formation of the O-state, which is characteristic of the sodium-pumping mode of KR2. Particularly, nonpolar side chains of Pro, Val, Leu, Ile, Met, Phe, and Trp amino acid residues are not consistent with the sodium coordination. Cys was demonstrated to coordinate metal ions like manganese, iron, copper, cobalt, nickel, and zinc [58]. However, it does not participate in the binding of sodium, potassium, magnesium, and calcium [58]. At the same time, polar Glu and Gln residues are supposedly too bulky to allow enough space for sodium binding inside the KR2 protomer.
4. Conclusions
In this work, we described the crystal structure of functionally important N112A mutant of the light-driven sodium pump KR2. The mutation converts the protein to an outward proton pump. N112 is a part of the NDQ motif, characteristic for all known NaRs (Figure 4). Our study provides a deeper understanding of the structural rearrangements, which occur in response to the variations of the critical functional residues of the protein. Although the overall fold and the retinal Schiff base region of the mutant are similar to the «expanded» conformation of the WT KR2, the SBC1 is enlarged in the KR2-N112A. The helix C is also slightly deflected in the mutant. The N112A mutation also does not affect the functionally important pentameric assembly of the protein; thus, the involvement of the N112 into the interprotomer interactions in the ground state of KR2 is not crucial for the stability of the oligomer.
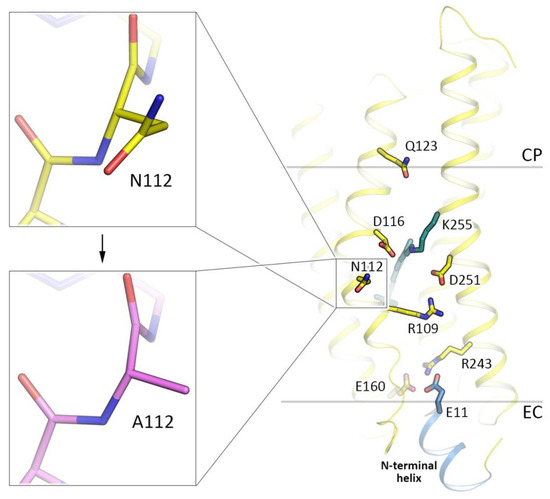
Figure 4.
Location of the N112A substitution site (yellow, the WT of KR2, PDB ID: 6YC3 [41]; magenta, the N112A variant of KR2, present work, PDB ID: 6YT4). Hydrophobic/hydrophilic membrane core boundaries were calculated with PPM server [59] and are shown with gray lines. Cytoplasmic (CP) and extracellular (EC) sides of the membrane are marked. N-terminal α-helix of KR2 is colored blue. Helices A, B, and BC loop are hidden for clarity.
Hence, the reported here crystal structure of the KR2-N112A, together with the available functional and spectroscopy data, allows us to conclude that the N112 does not show any important structural and/or functional roles in the ground state of the protein. Therefore, our study shows that the analysis of the ground state of N112A cannot solely explain the Na+-to-H+ pump conversion of KR2. However, N112 is vital for the organization of the transient sodium binding site in the O-state of the KR2 photocycle. Thus, we speculate that the major role of N112 as a key functional amino acid residue is in the organization of the site. For the complete understanding of the functional conversions of NaRs, and the principles of the ion selectivity of the proteins, further investigations on KR2 are required. Particularly, the structures of the intermediate states of KR2 mutants are essential.
Finally, the engineering of KR2 variants with modified or optimized pumping efficiency and ion selectivity is of high importance for the development of KR2-based optogenetic tools. However, the crystal structure of the ground state of KR2-N112A is a beautiful demonstration of the fact that the rational design of such variants should be supported by the structures of the intermediate state’s models of not only WT but functional mutants of the protein.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4352/10/6/496/s1, Figure S1: Examples of the electron density maps of the KR2-N112A structure., Figure S2: Transient sodium binding site in the O-state of the KR2 photocycle.
Author Contributions
Conceptualization, K.K. and V.G.; investigation, N.M., K.K., C.B., T.B., R.A., A.R., N.I., A.A., M.R., V.G.; writing—original draft preparation, K.K. and N.M.; writing—review and editing, K.K. and V.G.; visualization, K.K., and N.M.; supervision, V.G.; project administration, N.M., K.K. and V.G.; funding acquisition, V.G, V.C., D.D., and M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Foundation for Basic Research (grants 17-00-00164, 17-00-00165, 17-00-00166, and 17-00-00167 komfi).
Acknowledgments
We acknowledge the Paul Scherrer Institut, Villigen, Switzerland, for the provision of synchrotron radiation beamtime at beamline X06SA (PXI) of the SLS and would like to thank Takashi Tomizaki for assistance. Atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank under the accession code 6YT4.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| WT | Wild type |
| KR2 | Krokinobacter eikastus rhodopsin 2 |
| SEC | Size exclusion chromatography |
| RSB | Retinal Schiff base |
| SBC1 | Schiff base cavity 1 |
References
- Gushchin, I.; Gordeliy, V. Microbial Rhodopsins. In Membrane Protein Complexes: Structure and Function; Springer: Singapore, 2018; pp. 19–56. [Google Scholar]
- Gushchin, I.; Shevchenko, V.; Polovinkin, V.; Kovalev, K.; Alekseev, A.; Round, E.; Borshchevskiy, V.; Balandin, T.; Popov, A.; Gensch, T.; et al. Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 2015, 22, 390–396. [Google Scholar] [CrossRef]
- Gushchin, I.; Chervakov, P.; Kuzmichev, P.; Popov, A.N.; Round, E.; Borshchevskiy, V.; Ishchenko, A.; Petrovskaya, L.; Chupin, V.; Dolgikh, D.A.; et al. Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria. Proc. Natl. Acad. Sci. USA 2013, 110, 12631–12636. [Google Scholar] [CrossRef]
- Luecke, H.; Schobert, B.; Richter, H.T.; Cartailler, J.P.; Lanyi, J.K. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 1999, 291, 899–911. [Google Scholar] [CrossRef]
- Kovalev, K.; Volkov, D.; Astashkin, R.; Alekseev, A.; Gushchin, I.; Haro-Moreno, J.M.; Chizhov, I.; Siletsky, S.; Mamedov, M.; Rogachev, A.; et al. High-resolution structural insights into the heliorhodopsin family. Proc. Natl. Acad. Sci. USA 2020, 117, 4131–4141. [Google Scholar] [CrossRef]
- Zabelskii, D.; Alekseev, A.; Kovalev, K.; Oliviera, A.-S.; Balandin, T.; Soloviov, D.; Bratanov, D.; Volkov, D.; Vaganova, S.; Astashkin, R.; et al. Viral channelrhodopsins: Calcium-dependent Na+/K+ selective light-gated channels. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shihoya, W.; Inoue, K.; Singh, M.; Konno, M.; Hososhima, S.; Yamashita, K.; Ikeda, K.; Higuchi, A.; Izume, T.; Okazaki, S.; et al. Crystal structure of heliorhodopsin. Nature 2019, 574, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Needham, D.M.; Yoshizawa, S.; Hosaka, T.; Poirier, C.; Choi, C.J.; Hehenberger, E.; Irwin, N.A.T.; Wilken, S.; Yung, C.-M.; Bachy, C.; et al. A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc. Natl. Acad. Sci. USA 2019, 116, 20574–20583. [Google Scholar] [CrossRef] [PubMed]
- Oesterhelt, D.; Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 1971, 233, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Matsuno-Yagi, A.; Mukohata, Y. ATP synthesis linked to light-dependent proton uptake in a red mutant strain of Halobacterium lacking bacteriorhodopsin. Arch. Biochem. Biophys. 1980, 199, 297–303. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Li, H.; Spudich, J.L. Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications. Annu. Rev. Biochem. 2017, 86, 845–872. [Google Scholar] [CrossRef]
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef] [PubMed]
- Schobert, B.; Lanyi, J.K. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 1982, 257, 10306–10313. [Google Scholar] [CrossRef] [PubMed]
- Bogomolni, R.A.; Spudich, J.L. The photochemical reactions of bacterial sensory rhodopsin-I. Flash photolysis study in the one microsecond to eight second time window. Biophys. J. 1987, 52, 1071–1075. [Google Scholar] [CrossRef]
- Béjà, O.; Spudich, E.N.; Spudich, J.L.; Leclerc, M.; DeLong, E.F. Proteorhodopsin phototrophy in the ocean. Nature 2001, 411, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef]
- Nagel, G. Channelrhodopsin-1: A Light-Gated Proton Channel in Green Algae. Science 2002, 296, 2395–2398. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Spudich, J.L. Structurally Distinct Cation Channelrhodopsins from Cryptophyte Algae. Biophys. J. 2016, 110, 2302–2304. [Google Scholar] [CrossRef]
- Oppermann, J.; Fischer, P.; Silapetere, A.; Liepe, B.; Rodriguez-Rozada, S.; Flores-Uribe, J.; Peter, E.; Keidel, A.; Vierock, J.; Kaufmann, J.; et al. MerMAIDs: A family of metagenomically discovered marine anion-conducting and intensely desensitizing channelrhodopsins. Nat. Commun. 2019, 10, 3315. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Janz, R.; Liu, X.; Spudich, J.L. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 2015, 349, 647–650. [Google Scholar] [CrossRef]
- Inoue, K.; Ono, H.; Abe-Yoshizumi, R.; Yoshizawa, S.; Ito, H.; Kogure, K.; Kandori, H. A light-driven sodium ion pump in marine bacteria. Nat. Commun. 2013, 4, 1678. [Google Scholar] [CrossRef]
- Kim, K.; Kwon, S.-K.; Jun, S.-H.; Cha, J.S.; Kim, H.; Lee, W.; Kim, J.F.; Cho, H.-S. Crystal structure and functional characterization of a light-driven chloride pump having an NTQ motif. Nat. Commun. 2016, 7, 12677. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Ito, S.; Kato, Y.; Nomura, Y.; Shibata, M.; Uchihashi, T.; Tsunoda, S.P.; Kandori, H. A natural light-driven inward proton pump. Nat. Commun. 2016, 7, 13415. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, V.; Mager, T.; Kovalev, K.; Polovinkin, V.; Alekseev, A.; Juettner, J.; Chizhov, I.; Bamann, C.; Vavourakis, C.; Ghai, R.; et al. Inward H+ pump xenorhodopsin: Mechanism and alternative optogenetic approach. Sci. Adv. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Tsunoda, S.P.; Singh, M.; Tomida, S.; Hososhima, S.; Konno, M.; Nakamura, R.; Watanabe, H.; Bulzu, P.-A.; Banciu, H.L.; et al. Schizorhodopsins: A family of rhodopsins from Asgard archaea that function as light-driven inward H+ pumps. Sci. Adv. 2020, 6, eaaz2441. [Google Scholar] [CrossRef]
- Bulzu, P.-A.; Andrei, A.-Ş.; Salcher, M.M.; Mehrshad, M.; Inoue, K.; Kandori, H.; Beja, O.; Ghai, R.; Banciu, H.L. Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nat. Microbiol. 2019, 4, 1129–1137. [Google Scholar] [CrossRef]
- Pushkarev, A.; Inoue, K.; Larom, S.; Flores-Uribe, J.; Singh, M.; Konno, M.; Tomida, S.; Ito, S.; Nakamura, R.; Tsunoda, S.P.; et al. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 2018, 558, 595–599. [Google Scholar] [CrossRef]
- Yutin, N.; Koonin, E.V. Proteorhodopsin genes in giant viruses. Biol. Direct 2012, 7, 34. [Google Scholar] [CrossRef]
- Bratanov, D.; Kovalev, K.; Machtens, J.-P.; Astashkin, R.; Chizhov, I.; Soloviov, D.; Volkov, D.; Polovinkin, V.; Zabelskii, D.; Mager, T.; et al. Unique structure and function of viral rhodopsins. Nat. Commun. 2019, 10, 4939. [Google Scholar] [CrossRef]
- Luck, M.; Mathes, T.; Bruun, S.; Fudim, R.; Hagedorn, R.; Tran Nguyen, T.M.; Kateriya, S.; Kennis, J.T.M.; Hildebrandt, P.; Hegemann, P. A Photochromic Histidine Kinase Rhodopsin (HKR1) That Is Bimodally Switched by Ultraviolet and Blue Light. J. Biol. Chem. 2012, 287, 40083–40090. [Google Scholar] [CrossRef]
- Yoshida, K.; Tsunoda, S.P.; Brown, L.S.; Kandori, H. A unique choanoflagellate enzyme rhodopsin exhibits light-dependent cyclic nucleotide phosphodiesterase activity. J. Biol. Chem. 2017, 292, 7531–7541. [Google Scholar] [CrossRef]
- Gómez-Consarnau, L.; Raven, J.A.; Levine, N.M.; Cutter, L.S.; Wang, D.; Seegers, B.; Arístegui, J.; Fuhrman, J.A.; Gasol, J.M.; Sañudo-Wilhelmy, S.A. Microbial rhodopsins are major contributors to the solar energy captured in the sea. Sci. Adv. 2019, 5, eaaw8855. [Google Scholar] [CrossRef]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 2015, 18, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, K.; Polovinkin, V.; Gushchin, I.; Alekseev, A.; Shevchenko, V.; Borshchevskiy, V.; Astashkin, R.; Balandin, T.; Bratanov, D.; Vaganova, S.; et al. Structure and mechanisms of sodium-pumping KR2 rhodopsin. Sci. Adv. 2019, 5, eaav2671. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Inoue, K.; Kandori, H. Kinetic Analysis of H+–Na+ Selectivity in a Light-Driven Na+ -Pumping Rhodopsin. J. Phys. Chem. Lett. 2015, 6, 5111–5115. [Google Scholar] [CrossRef]
- Kato, H.E.; Inoue, K.; Abe-Yoshizumi, R.; Kato, Y.; Ono, H.; Konno, M.; Hososhima, S.; Ishizuka, T.; Hoque, M.R.; Kunitomo, H.; et al. Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 2015, 521, 48–53. [Google Scholar] [CrossRef]
- Balashov, S.P.; Imasheva, E.S.; Dioumaev, A.K.; Wang, J.M.; Jung, K.-H.; Lanyi, J.K. Light-Driven Na+ Pump from Gillisia limnaea: A High-Affinity Na+ Binding Site Is Formed Transiently in the Photocycle. Biochemistry 2014, 53, 7549–7561. [Google Scholar] [CrossRef]
- Gerwert, K.; Freier, E.; Wolf, S. The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 606–613. [Google Scholar] [CrossRef]
- Nishimura, N.; Mizuno, M.; Kandori, H.; Mizutani, Y. Distortion and a Strong Hydrogen Bond in the Retinal Chromophore Enable Sodium-Ion Transport by the Sodium-Ion Pump KR2. J. Phys. Chem. B 2019, 123, 3430–3440. [Google Scholar] [CrossRef]
- Kovalev, K.; Astashkin, R.; Gushchin, I.; Orekhov, P.; Volkov, D.; Zinovev, E.; Marin, E.; Rulev, M.; Alekseev, A.; Royant, A.; et al. Molecular mechanism of light-driven sodium pumping. Nat. Commun. 2020, 11, 2137. [Google Scholar] [CrossRef]
- Inoue, K.; Konno, M.; Abe-Yoshizumi, R.; Kandori, H. The Role of the NDQ Motif in Sodium-Pumping Rhodopsins. Angew. Chem. Int. Ed. 2015, 54, 11536–11539. [Google Scholar] [CrossRef]
- Tomida, S.; Ito, S.; Inoue, K.; Kandori, H. Hydrogen-bonding network at the cytoplasmic region of a light-driven sodium pump rhodopsin KR2. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Abe-Yoshizumi, R.; Inoue, K.; Kato, H.E.; Nureki, O.; Kandori, H. Role of Asn112 in a Light-Driven Sodium Ion-Pumping Rhodopsin. Biochemistry 2016, 55, 5790–5797. [Google Scholar] [CrossRef]
- Tomida, S.; Ito, S.; Mato, T.; Furutani, Y.; Inoue, K.; Kandori, H. Infrared spectroscopic analysis on structural changes around the protonated Schiff base upon retinal isomerization in light-driven sodium pump KR2. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148190. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Landau, E.M.; Rosenbusch, J.P. Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 14532–14535. [Google Scholar] [CrossRef] [PubMed]
- Gordeliy, V.I.; Labahn, J.; Moukhametzianov, R.; Efremov, R.; Granzin, J.; Schlesinger, R.; Büldt, G.; Savopol, T.; Scheidig, A.J.; Klare, J.P.; et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 2002, 419, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Tickle, I.J.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. STARANISO; Global Phasing Ltd.: Cambridge, UK, 2018. [Google Scholar]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP 4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Inoue, K.; Ikeda, K.; Konno, M.; Singh, M.; Kataoka, C.; Abe-Yoshizumi, R.; Kandori, H.; Uchihashi, T. Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 2018, 8, 8262. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, I.; Shevchenko, V.; Polovinkin, V.; Borshchevskiy, V.; Buslaev, P.; Bamberg, E.; Gordeliy, V. Structure of the light-driven sodium pump KR2 and its implications for optogenetics. FEBS J. 2016, 283, 1232–1238. [Google Scholar] [CrossRef]
- Ho, B.K.; Gruswitz, F. HOLLOW: Generating Accurate Representations of Channel and Interior Surfaces in Molecular Structures. BMC Struct. Biol. 2008, 8, 49. [Google Scholar] [CrossRef]
- Harding, M.M.; Nowicki, M.W.; Walkinshaw, M.D. Metals in protein structures: A review of their principal features. Crystallogr. Rev. 2010, 16, 247–302. [Google Scholar] [CrossRef]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).