Unexpected Synthesis, Single-Crystal X-ray Structure, Anticancer Activity, and Molecular Docking Studies of Certain 2–((Imidazole/Benzimidazol–2–yl)thio)–1–arylethanones
Abstract
1. Introduction
2. Results and Discussion
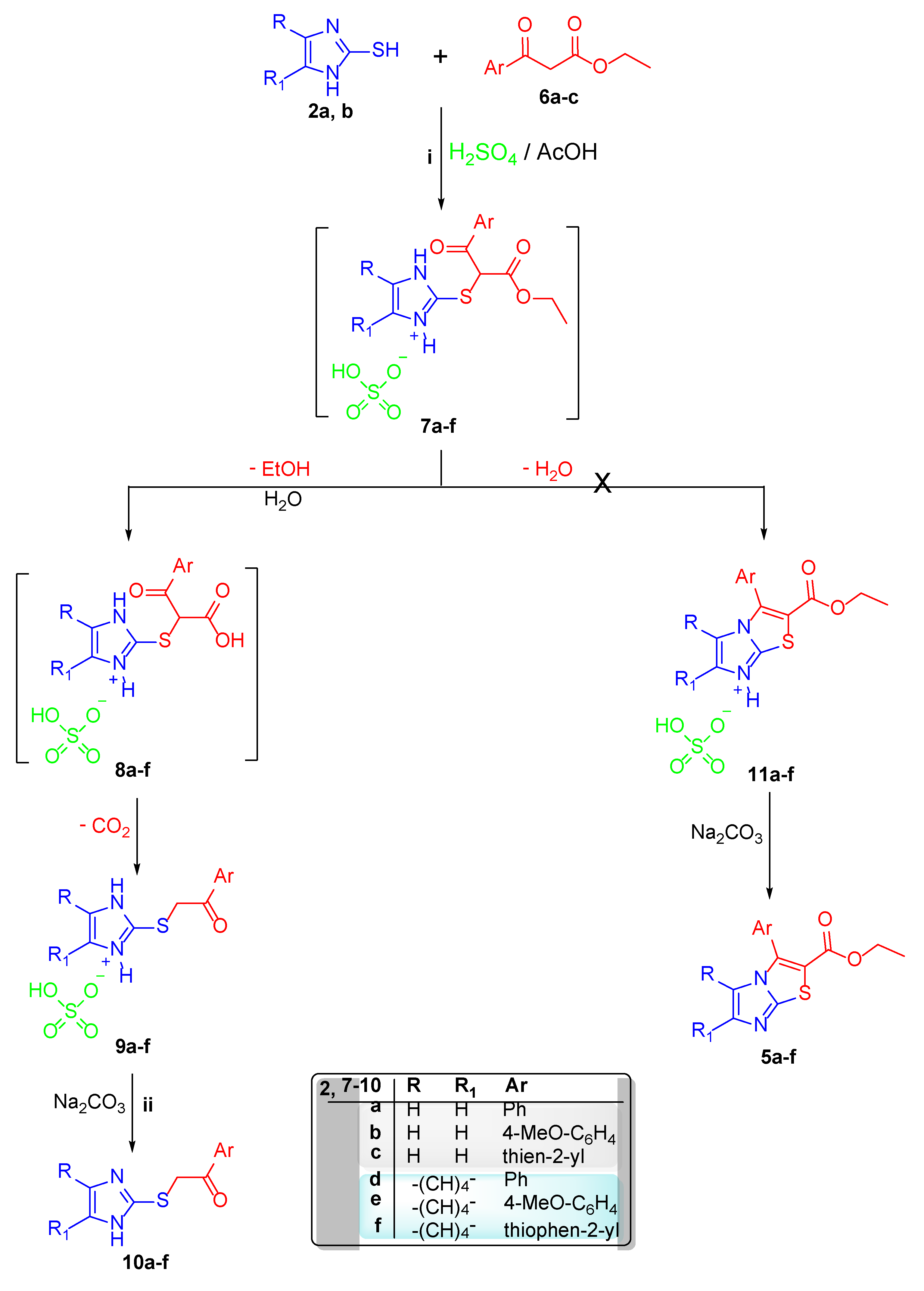
2.1. Chemistry
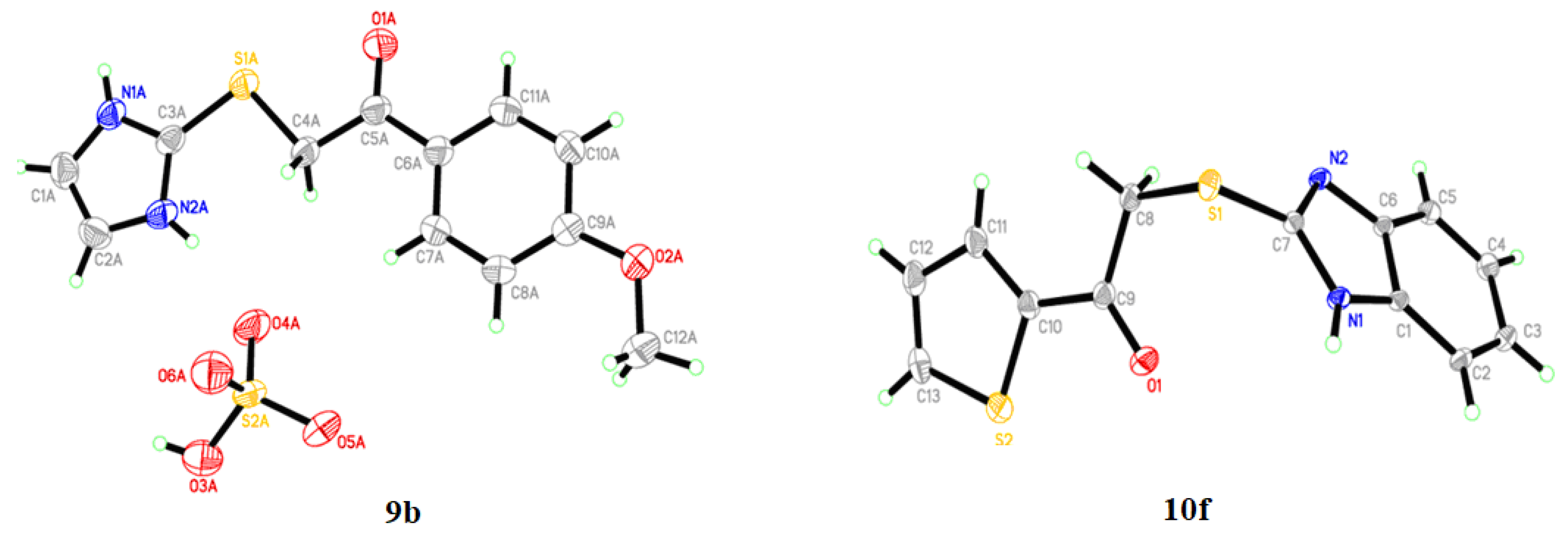
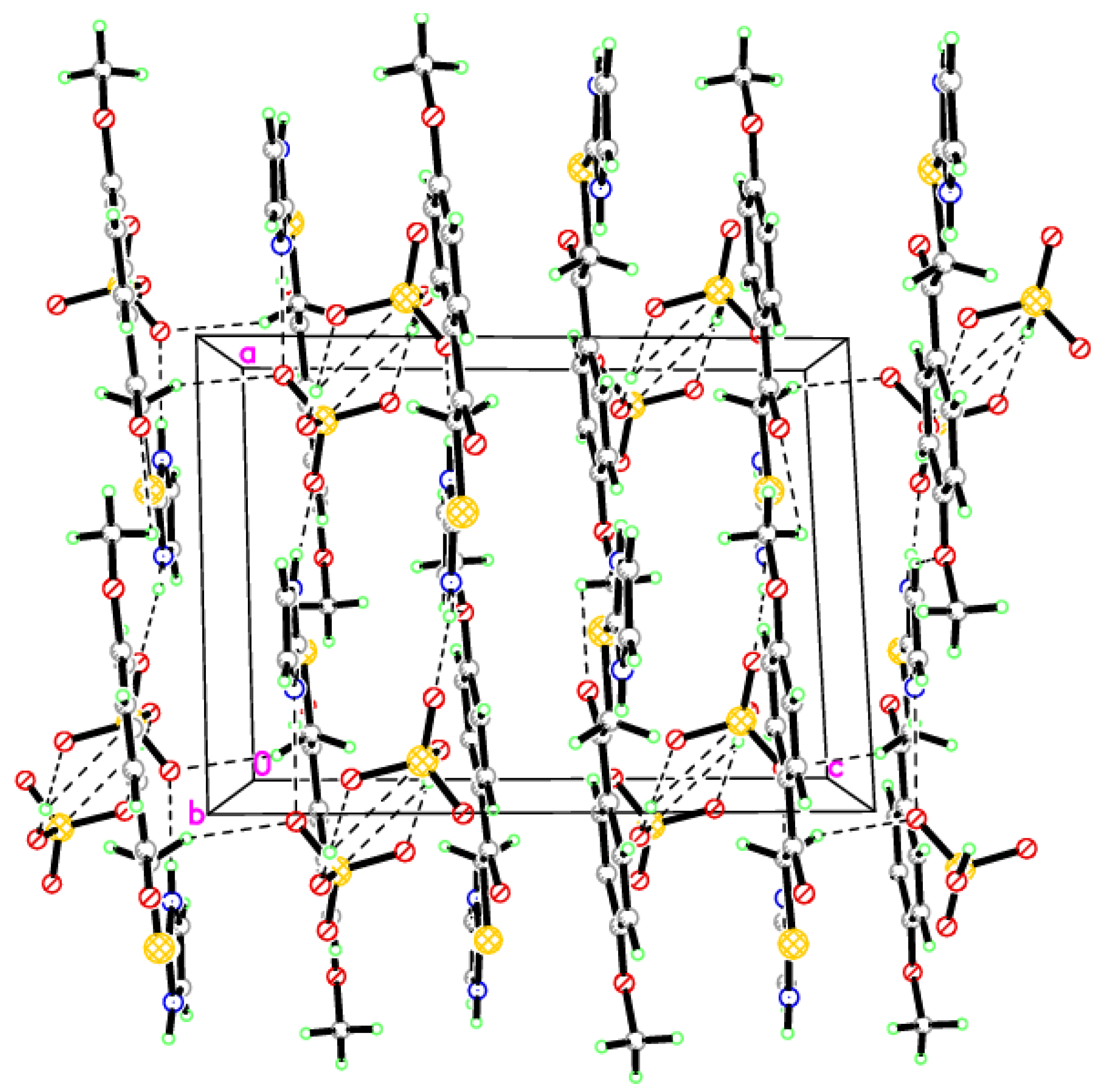
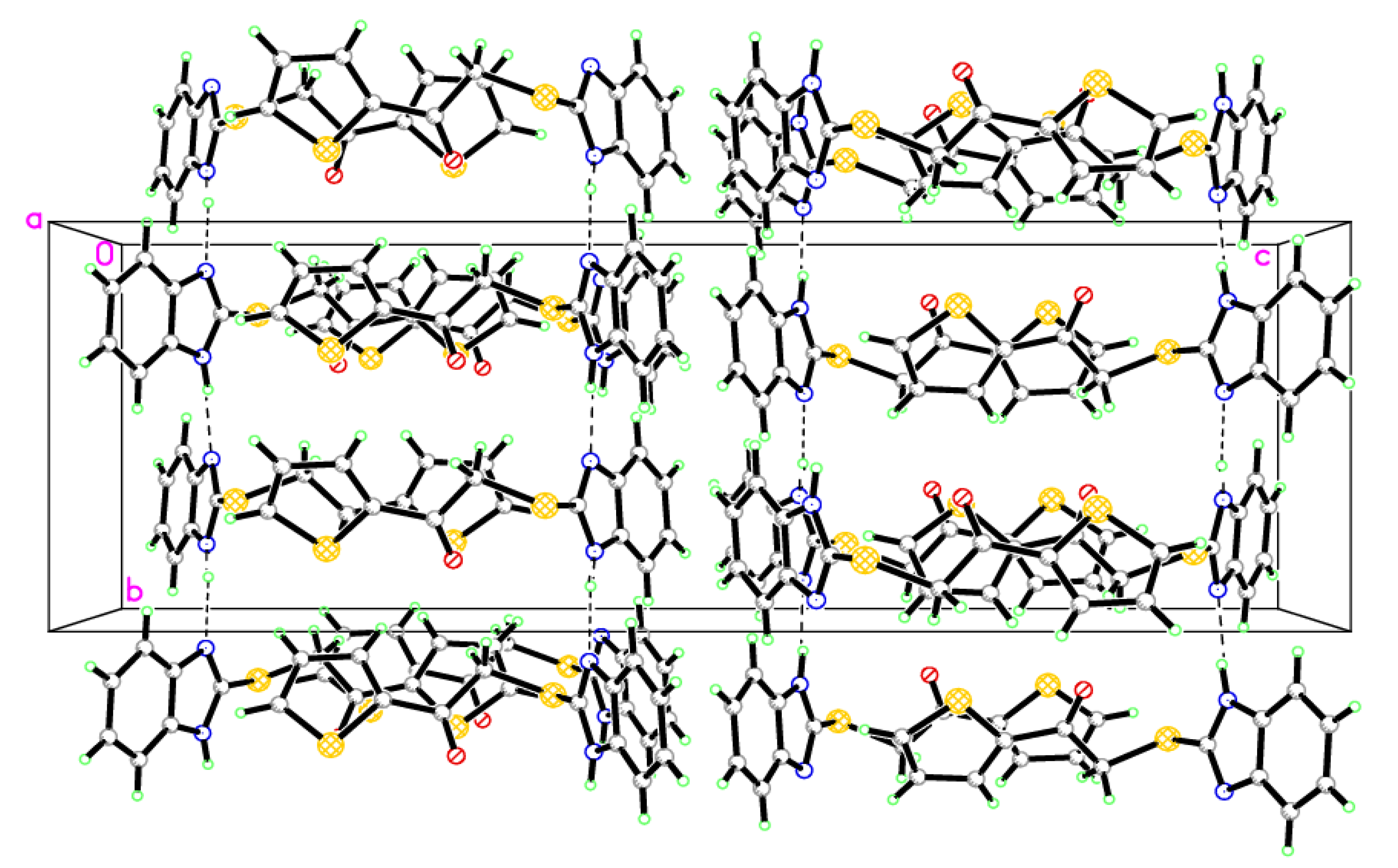
2.2. Single-Crystal X-ray Crystallographic Analysis
2.3. Biological Evaluation
Cytotoxic Action against Breast Cancer Cell Lines
2.4. CDK Inhibitory Action
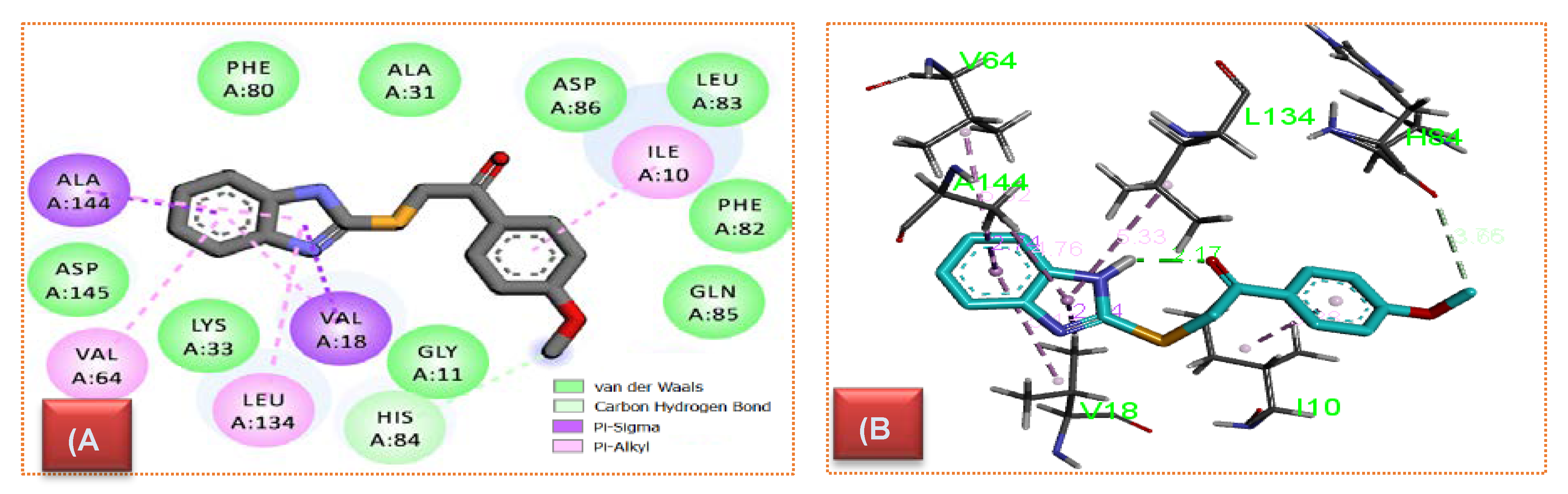
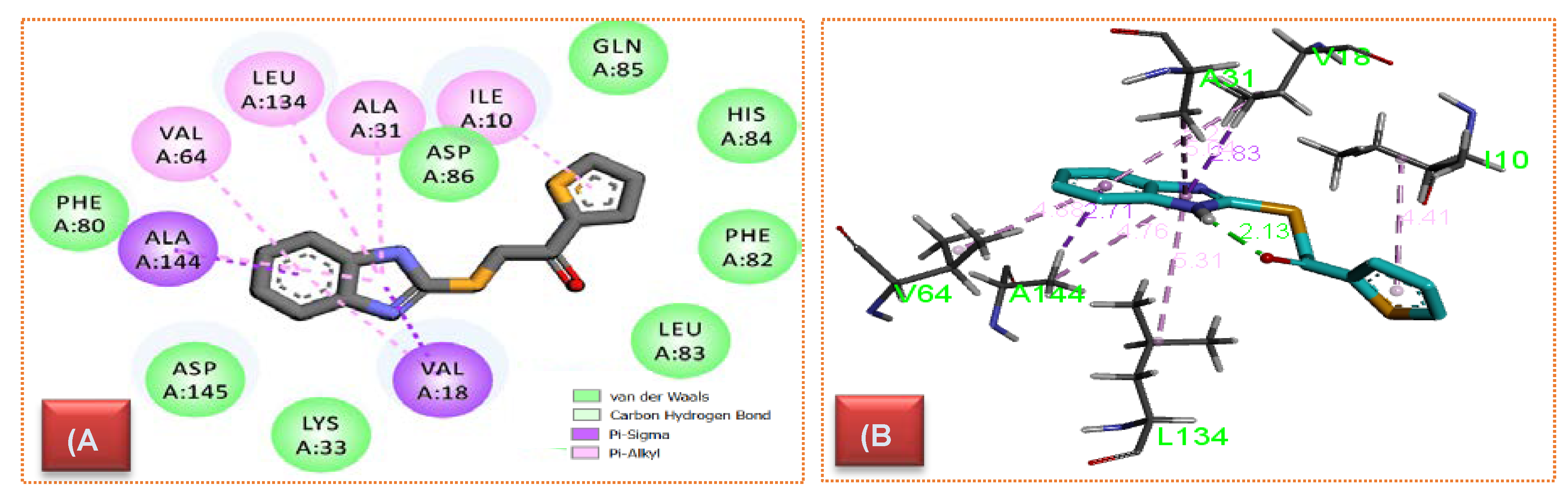
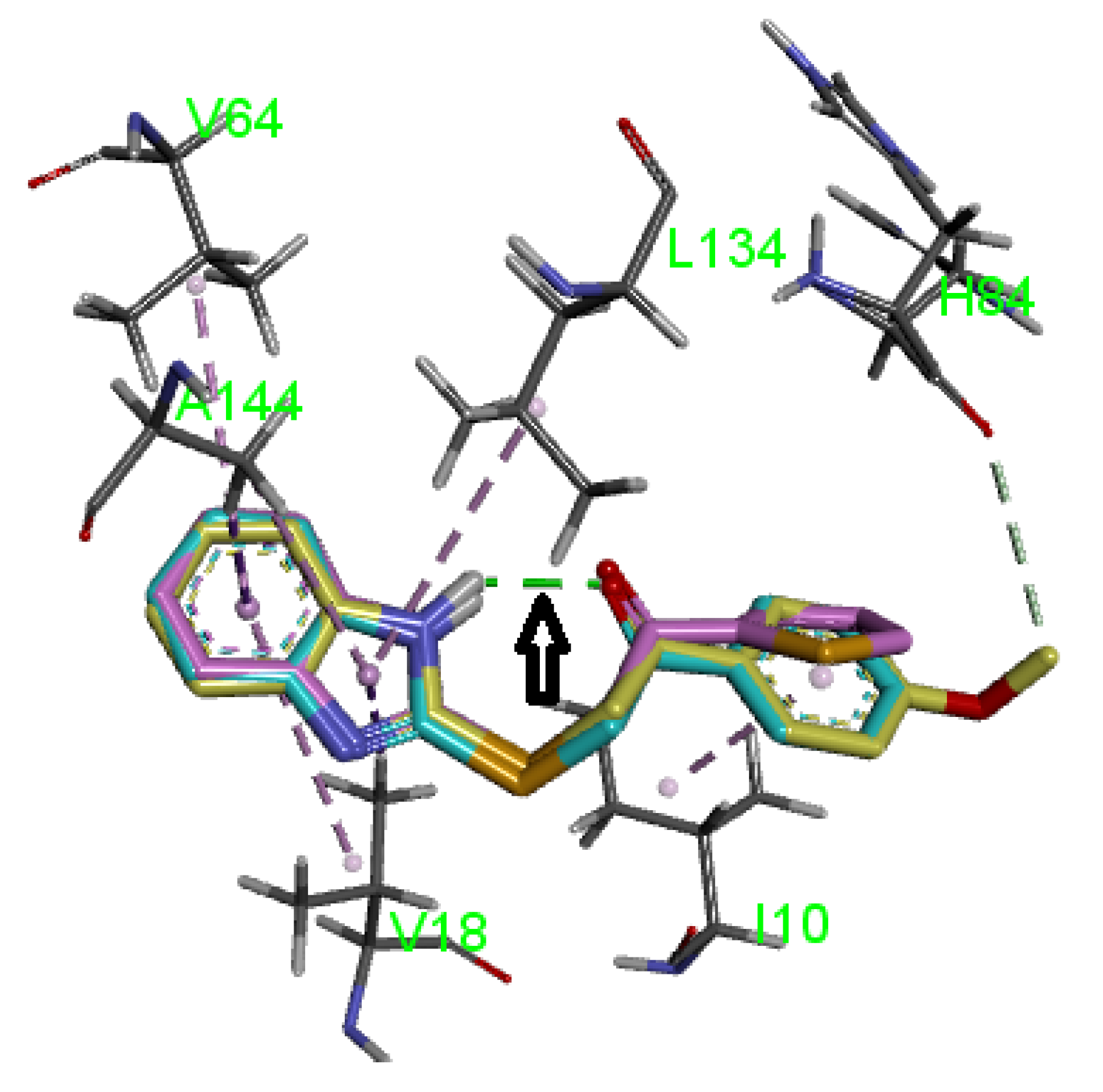
2.4.1. CDK Molecular Modeling
2.4.2. In Vitro CDK2 Assay
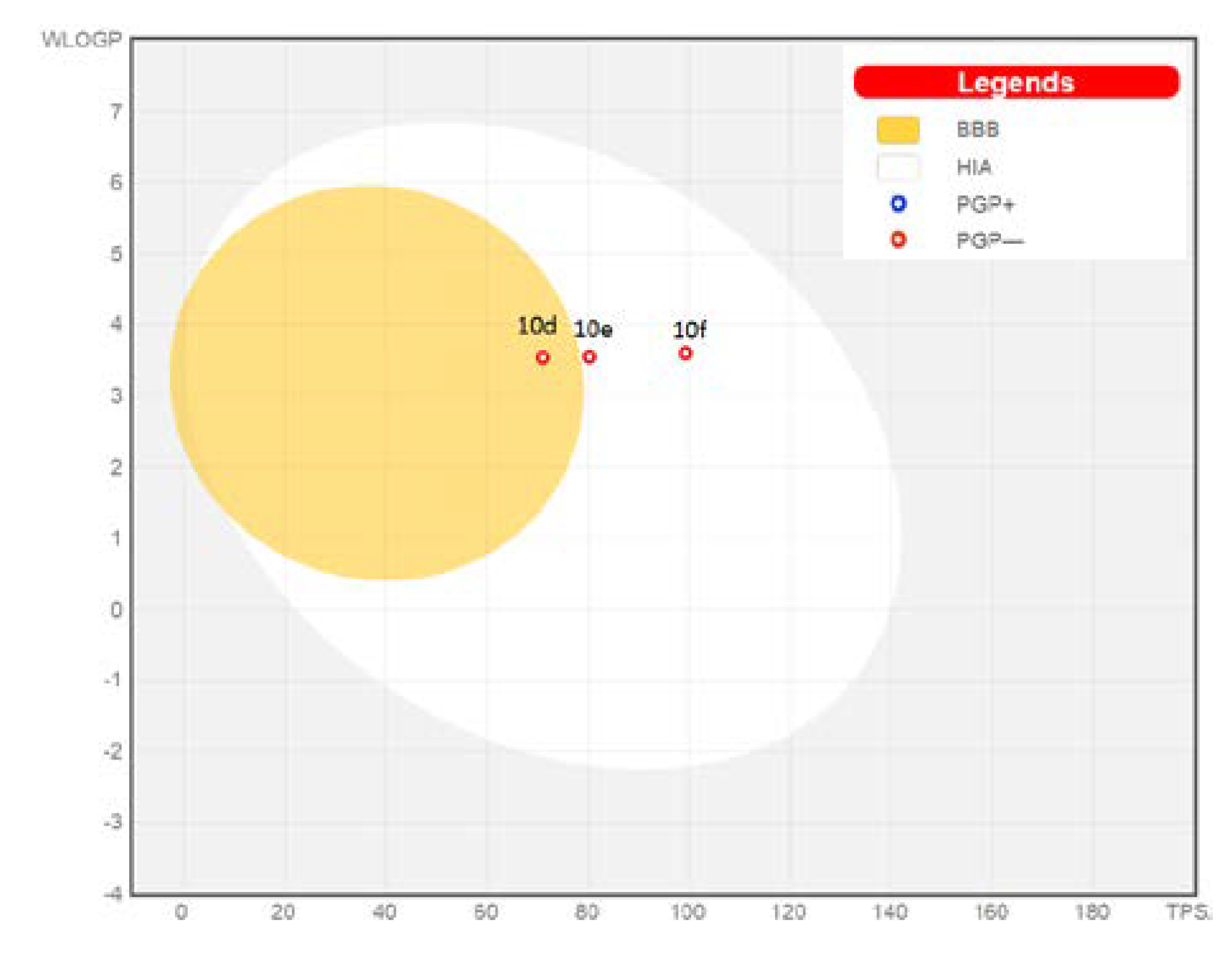
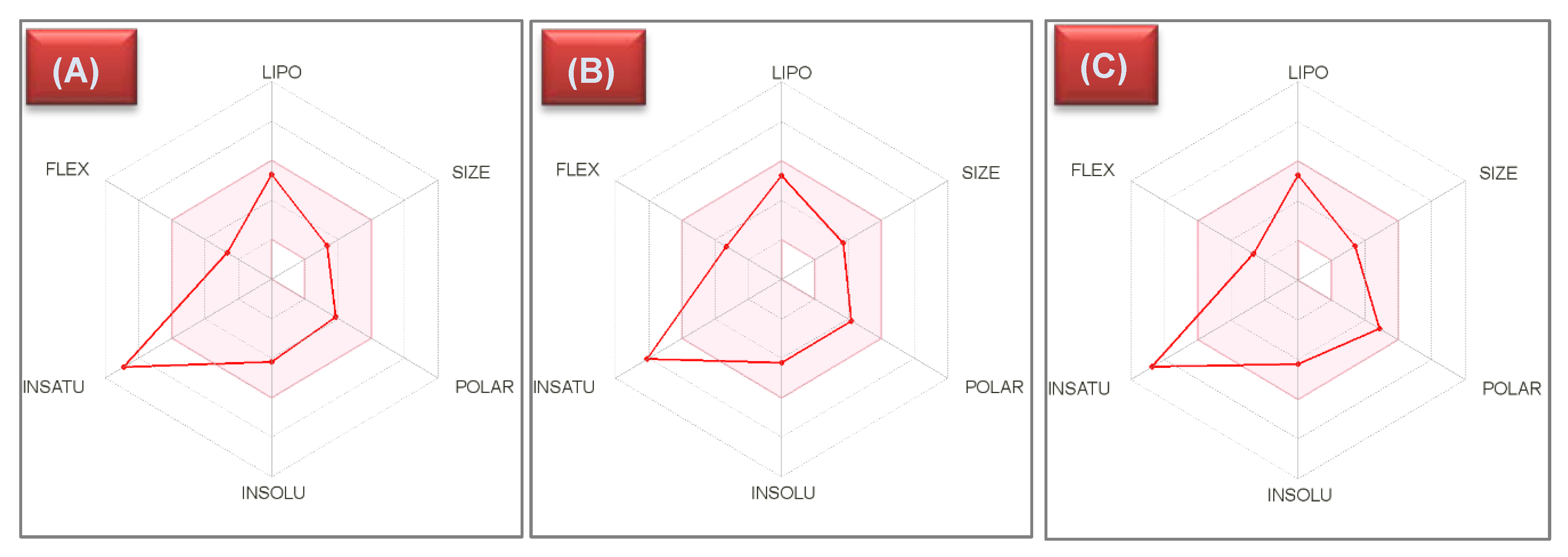
2.5. Pharmacokinetics and Druglikeness Properties’ Prediction for Benzimidazoles 10d–f
3. Materials and Methods
3.1. Chemistry
3.1.1. General
3.1.2. General Procedures for Synthesis of Salts 9a–f
2–((2–Oxo2–Phenylethyl)thio)1–H–Imidazol3–ium Hydrogen Sulfate (9a)
2–((2–(4–Methoxyphenyl)2–Oxoethyl)thio)1–H–Imidazol3–ium Hydrogen Sulfate (9b)
2–((2-Oxo2–(Thiophen2–yl)ethyl)thio)1–H-Imidazol3–Ium Hydrogen Sulfate (9c)
3.1.3. Procedures for Preparation of Imidazole/Benzimidazol-Based Derivatives 10a–f
2–((1H–Imidazol2–yl)thio)1–Phenylethan1–One (10a)
2–((1H–Imidazol2–yl)thio)1–(4–Methoxyphenyl)ethan1–One (10b)
2–((1H–Imidazol2–yl)thio)1–(Thiophen2–yl)ethan1–One (10c)
3.2. X-ray Crystallographic Analysis
3.3. Biological Evaluations
3.4. In Silico Studies
3.4.1. Molecular Modeling
3.4.2. Pharmacokinetics and Druglikeness Properties’ Prediction for Benzimidazoles 10d–f
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yadav, G.; Ganguly, S. Structure activity relationship (SAR) study of benzimidazole scaffold for different biological activities: A mini-review. Eur. J. Med. Chem. 2015, 97, 419–443. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, W.; Khan, M.F.; Verma, G.; Shaquiquzzaman, M.; Rizvi, M.A.; Mehdi, S.H.; Akhter, M.; Alam, M.M. Therapeutic evolution of benzimidazole derivatives in the last quinquennial period. Eur. J. Med. Chem. 2016, 126, 705–753. [Google Scholar] [CrossRef] [PubMed]
- Tahlan, S.; Kumar, S.; Narasimhan, B. Pharmacological significance of heterocyclic 1H-benzimidazole scaffolds: A review. BMC Chem. 2019, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Gaba, M.; Mohan, C. Development of drugs based on imidazole and benzimidazole bioactive heterocycles: Recent advances and future directions. Med. Chem. Res. 2016, 25, 173–210. [Google Scholar] [CrossRef]
- Bräse, S. Privileged Scaffolds in Medicinal Chemistry: Design, Synthesis, Evaluation; Royal Society of Chemistry: Cambridge, UK, 2015; Chapter 2; pp. 98–113. ISBN 9781−7−82620−303. [Google Scholar]
- Ali, I.; Lone, M.N.; Aboul-Enein, H.Y. Imidazoles as potential anticancer agents. MedChemComm 2017, 8, 1742–1773. [Google Scholar] [CrossRef]
- Shrivastava, N.; Naim, M.J.; Alam, M.J.; Nawaz, F.; Ahmed, S.; Alam, O. Benzimidazole scaffold as anticancer agent: Synthetic approaches and structure–activity relationship. Arch. Pharm. 2017, 350, e201700040. [Google Scholar] [CrossRef]
- Tahlan, S.; Kumar, S.; Kakkar, S.; Narasimhan, B. Benzimidazole scaffolds as promising antiproliferative agents: A review. BMC Chem. 2019, 13, 66. [Google Scholar] [CrossRef]
- Yadav, S.; Narasimhan, B. Perspectives of benzimidazole derivatives as anticancer agents in the new era. Anticancer Agents Med. Chem. 2016, 16, 1403–1425. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Yar, M.S.; Sharma, V.K.; Khan, A.A.; Ali, Z.; Haider, M.R.; Pathak, A. Recent Progress of Benzimidazole Hybrids for Anticancer Potential. Curr. Med. Chem. 2019. [Google Scholar] [CrossRef]
- Aniket, P.; Shantanu, D.S.; Anagha, O.B.; Ajinkya, P.S. Iodine catalyzed convenient synthesis of 2-aryl1–-arylmethyl1– H-benzimidazoles in aqueous media. Int. J. ChemTech Res. 2015, 8, 496–500. [Google Scholar]
- Wan, J.P.; Gan, S.F.; Wu, J.M.; Pan, Y. Water mediated chemoselective synthesis of 1,2-disubstituted benzimidazoles using o-phenylenediamine and the extended synthesis of quinoxalines. Green Chem. 2009, 11, 1633–1637. [Google Scholar] [CrossRef]
- Di Gioia, M.L.; Cassano, R.; Costanzo, P.; Herrera Cano, N.; Maiuolo, L.; Nardi, M.; Nicoletta, F.P.; Oliverio, M.; Procopio, A. Green synthesis of privileged benzimidazole scaffolds using active deep eutectic solvent. Molecules 2019, 24, 2885. [Google Scholar] [CrossRef]
- Herrera Cano, N.; Uranga, J.G.; Nardi, M.; Procopio, A.; Wunderlin, D.A.; Santiago, A.N. Selective and eco-friendly procedures for the synthesis of benzimidazole derivatives. The role of the Er(OTf)3 catalyst in the reaction selectivity. Beilstein J. Org. Chem. 2016, 12, 2410–2419. [Google Scholar] [CrossRef] [PubMed]
- Sapkal, S.B.; Shelke, K.F.; Sonar, S.S.; Shingate, B.B.; Shingare, M.S. Acidic ionic liquid catalyzed environmentally friendly synthesis of benzimidazole derivatives. Bull. Catal. Soc. India 2009, 2, 78–83. [Google Scholar]
- Sharma, J.; Soni, P.K.; Bansal, R.; Halve, A.K. Synthetic Approaches Towards Benzimidazoles by the Reaction of o-Phenylenediamine with Aldehydes Using a Variety of Catalysts: A Review. Curr. Org. Chem. 2018, 22, 2280–2299. [Google Scholar] [CrossRef]
- Brain, C.T.; Brunton, S.A. An intramolecular palladium-catalysed aryl amination reaction to produce benzimidazoles. Tetrahedron Lett. 2002, 43, 1893–1895. [Google Scholar] [CrossRef]
- Nale, D.B.; Bhanage, B.M. The use of various o-phenylenediamines and N-substituted formamides as C 1 sources in a zinc-catalyzed cyclization in the presence of poly (methylhydrosiloxane) provides benzimidazoles in good yields. Benzoxazole and benzothiazole derivates can also be synthesized. Synlett 2015, 26, 2831–2834. [Google Scholar]
- Mahesh, D.; Sadhu, P.; Punniyamurthy, T. Copper (II)-catalyzed oxidative cross-coupling of anilines, primary alkyl amines, and sodium azide using TBHP: A route to 2-substituted benzimidazoles. J. Org. Chem. 2016, 81, 3227–3234. [Google Scholar] [CrossRef]
- Abdel-Aziz, H.A.; Ghabbour, H.A.; Eldehna, W.M.; Al-Rashood, S.T.; Al-Rashood, K.A.; Fun, H.K.; Al-Tahhan, M.; Al-Dhfyan, A. 2-((Benzimidazol-2-yl) thio)-1-arylethan-1-ones: Synthesis, crystal study and cancer stem cells CD133 targeting potential. Eur. J. Med. Chem. 2015, 104, 1–10. [Google Scholar] [CrossRef]
- Abdel-Aziz, H.A.; Eldehna, W.M.; Ghabbour, H.; Al-Ansary, G.H.; Assaf, A.M.; Al-Dhfyan, A. Synthesis, crystal study, and anti-proliferative activity of some 2-benzimidazolylthioacetophenones towards triple-negative breast cancer MDA-MB4–68 cells as apoptosis-inducing agents. Int. J. Mol. Sci. 2016, 17, 1221. [Google Scholar] [CrossRef]
- Al-Ansary, G.H.; Eldehna, W.M.; Ghabbour, H.A.; Al-Rashood, S.T.; Al-Rashood, K.A.; Eladwy, R.A.; Al-Dhfyan, A.; Kabil, M.M.; Abdel-Aziz, H.A. Cancer stem cells CD133 inhibition and cytotoxicity of certain 3-phenylthiazolo [3,2-a]benzimidazoles: Design, direct synthesis, crystal study and in vitro biological evaluation. J. Enzym. Inhib. Med. Chem. 2017, 32, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of bond lengths determined by X-ray and neutron diffraction. Part 1. Bond lengths in organic compounds. J. Chem. Soc. Perkins Trans. 2 1987, 12, S1–S19. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D. New colorimetric cyto-toxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, P.; Andres, D.; Kwiatkowski, D.A.; Kolbe, C.C.; Lienau, P.; Siemeister, G.; Lucking, U.; Stegmann, C.M. Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9. ACS Chem. Biol. 2016, 11, 1710–1719. [Google Scholar] [CrossRef]
- Chen, P.; Lee, N.V.; Hu, W.; Xu, M.; Ferre, R.A.; Lam, H.; Bergqvist, S.; Solowiej, J.; Diehl, W.; He, Y.A.; et al. Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance. Mol. Cancer Ther. 2016, 15, 2273–2281. [Google Scholar] [CrossRef]
- Lolli, G.; Lowe, E.D.; Brown, N.R.; Johnson, L.N. The Crystal Structure of Human CDK7 and Its Protein Recognition Properties. Structure 2004, 12, 2067–2079. [Google Scholar] [CrossRef]
- Bettayeb, K.; Baunbak, D.; Delehouze, C.; Loaec, N.; Hole, A.J.; Baumli, S.; Endicott, J.A.; Douc-Rasy, S.; Benard, J.; Oumata, N.; et al. CDK Inhibitors Roscovitine and CR8 Trigger Mcl1– Down-Regulation and Apoptotic Cell Death in Neuroblastoma Cells. Genes Cancer 2010, 1, 369–380. [Google Scholar] [CrossRef]
- Wei, Y.; Li, J.; Qing, J.; Huang, M.; Wu, M.; Gao, F.; Lin, J. Discovery of novel hepatitis C virus NS5B polymerase inhibitors by combining random forest, multiple e-pharmacophore modeling and docking. PLoS ONE 2016, 11, e0148181. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXTL-PC (Version 5.1); Siemens Analytical Instruments, Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Sabt, A.; Abdelhafez, O.M.; El-Haggar, R.S.; Madkour, H.M.; Eldehna, W.M.; El-Khrisy, E.E.D.A.; Abdel-Rahman, M.A.; Rashed, L.A. Novel coumarin-6-sulfonamides as apoptotic anti-proliferative agents: Synthesis, in vitro biological evaluation, and QSAR studies. J. Enzym. Inhib. Med. Chem. 2018, 33, 1095–1107. [Google Scholar] [CrossRef]
- Said, M.A.; Eldehna, W.M.; Nocentini, A.; Fahim, S.H.; Bonardi, A.; Elgazar, A.A.; Kryštof, V.; Soliman, D.H.; Abdel-Aziz, H.A.; Gratteri, P.; et al. Sulfonamide-based ring-fused analogues for CAN508 as novel carbonic anhydrase inhibitors endowed with antitumor activity: Design, synthesis, and in vitro biological evaluation. Eur. J. Med. Chem. 2020, 189, 112019. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comp. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Available online: https://3dsbiovia.com/resource-center/downloads/ (accessed on 6 March 2020).


| 9b | |||
| Bond | Å | Bond | Å |
| S1A—C3A | 1.732(2) | O2A—C12A | 1.428(3) |
| S1A—C4A | 1.807(2) | O2A—C9A | 1.354(3) |
| S1B—C4B | 1.805(2) | O1B—C5B | 1.208(3) |
| S1B—C3B | 1.729(2) | O2B—C12B | 1.433(3) |
| S2A—O4A | 1.4343(19) | O2B—C9B | 1.357(3) |
| S2A—O3A | 1.5235(19) | N1A—C3A | 1.324(3) |
| S2A—O5A | 1.4346(18) | N1A—C1A | 1.376(3) |
| S2A—O6A | 1.4764(18) | N2A—C2A | 1.370(3) |
| S2B—O4B | 1.5283(17) | N2A—C3A | 1.332(3) |
| S2B—O5B | 1.4782(18) | N1B—C3B | 1.326(3) |
| S2B—O6B | 1.4330(18) | N1B—C1B | 1.379(3) |
| S2B—O3B | 1.4346(17) | N2B—C2B | 1.372(3) |
| O1A—C5A | 1.212(3) | N2B—C3B | 1.329(3) |
| Angle | ° | Angle | ° |
| C3A—S1A—C4A | 99.79(10) | N1A—C1A—C2A | 106.5(2) |
| C3B—S1B—C4B | 99.33(10) | N2A—C2A—C1A | 107.5(2) |
| O4A—S2A—O5A | 112.49(11) | N1A—C3A—N2A | 107.11(19) |
| O4A—S2A—O6A | 111.10(11) | S1A—C3A—N1A | 122.50(16) |
| O3A—S2A—O6A | 106.18(10) | S1A—C3A—N2A | 130.35(16) |
| O5A—S2A—O6A | 111.22(11) | S1A—C4A—C5A | 106.53(14) |
| O3A—S2A—O4A | 109.48(11) | O1A—C5A—C6A | 122.4(2) |
| O3A—S2A—O5A | 106.04(11) | O1A—C5A—C4A | 119.07(19) |
| O2A—C9A—C8A | 125.06(18) | O2A—C9A—C10A | 115.36(19) |
| 10f | |||
| Bond | Å | Bond | Å |
| S1—C7 | 1.7515(12) | N1—C1 | 1.3752(16) |
| S1—C8 | 1.8058(13) | N1—C7 | 1.3554(15) |
| S2—C10 | 1.7236(13) | N2—C6 | 1.3865(15) |
| S2—C13 | 1.7082(16) | N2—C7 | 1.3224(15) |
| O1—C9 | 1.2162(18) | ||
| Angle | ° | Angle | ° |
| C7—S1—C8 | 100.47(6) | N1—C7—N2 | 113.39(10) |
| C10—S2—C13 | 91.62(7) | S1—C7—N1 | 121.61(8) |
| C1—N1—C7 | 106.66(9) | S1—C8—C9 | 113.44(9) |
| C6—N2—C7 | 104.82(9) | O1—C9—C10 | 121.54(12) |
| N1—C1—C2 | 131.98(11) | O1—C9—C8 | 122.19(11) |
| N1—C1—C6 | 105.79(10) | S2—C10—C11 | 111.21(10) |
| N2—C6—C1 | 109.33(10) | S2—C10—C9 | 119.21(10) |
| N2—C6—C5 | 130.19(10) | S2—C13—C12 | 112.46(11) |
| S1—C7—N2 | 124.83(9) | ||
| 9b | ||||
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3A—H3AA···O5B | 0.8200 | 1.8300 | 2.648(3) | 180.00 |
| O4B—H4BC···O6A | 0.8200 | 1.8200 | 2.635(3) | 179.00 |
| N2B—H2NB···O6Bi | 0.77(3) | 2.04(3) | 2.805(3) | 172(2) |
| N2A—H2NA···O4A | 0.80(3) | 1.98(3) | 2.762(3) | 168(2) |
| N1A—H1NA···O5Ai | 0.81(3) | 1.96(3) | 2.769(3) | 173(3) |
| N1B—H1NB···O3B | 0.83(3) | 1.94(3) | 2.768(2) | 173(2) |
| C1A—H1AA···O2Aii | 0.9300 | 2.5400 | 3.259(3) | 134.00 |
| C1B—H1BA···O2Biii | 0.9300 | 2.5400 | 3.274(3) | 136.00 |
| C4B—H4BA···O6Biv | 0.9700 | 2.5000 | 3.423(3) | 158.00 |
| Symmetry codes: (i) x + 1, y, z; (ii) x + 1, y + 1, z; (iii) x − 1, y − 1, z; (iv) −x + 1, −y + 1, −z + 2; (v) x − 1, y, z. | ||||
| 10f | ||||
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N1···N2i | 0.906(19) | 1.88(2) | 2.7858(14) | 173.7(16) |
| Symmetry code: (i) −x + 1/2, y + 1/2, z. | ||||

| Cpd. | Ar | IC50 (µM) a | |
|---|---|---|---|
| T–47D | MCF–7 | ||
| 10a | C6H5 | 57.68 ± 0.25 | 42.61 ± 3.08 |
| 10b | 4–CH3O–C6H4 | 39.32 ± 0.08 | 31.25 ± 2.36 |
| 10c | thien2–yl | NA b | 72.64 ± 4.05 |
| 10d | C6H5 | 8.04 ± 0.32 | 12.90 ± 1.04 |
| 10e | 4–CH3O–C6H4 | 11.17 ± 0.04 | 4.53 ± 0.33 |
| 10f | thien–2–yl | 34.94 ± 0.37 | 23.07 ± 2.19 |
| Staurosporine | – | 7.19 ± 0.43 | 6.67 ± 0.28 |
| Cpd. | Energy Score (S) kcal/mol | |||
|---|---|---|---|---|
| CDK2 | CDK4/6 | CDK7 | CDk9 | |
| 10d | −8.7 | −7.2 | −6.6 | −7.3 |
| 10e | −8.9 | −7.2 | −6.7 | −7.5 |
| 10f | −8.0 | −6.7 | −6.5 | −7.0 |
| Cpd. | Bond Type | Involved Amino Acids | Distance |
|---|---|---|---|
| 10d | Pi–Alkyl | Ile10 | 3.83 |
| Pi–Sigma | Ala144 | 2.70 | |
| Pi–Alkyl | Ala144 | 4.76 | |
| Pi–Alkyl | Leu134 | 5.42 | |
| Pi–Alkyl | Val64 | 4.92 | |
| Pi–Alkyl | Val18 | 5.20 | |
| Pi–Sigma | Val18 | 2.85 | |
| 10e | Pi–Alkyl | Ile10 | 3.88 |
| Pi–Alkyl | Ala144 | 4.76 | |
| Pi–Sigma | Ala144 | 2.47 | |
| Pi–Alkyl | Leu134 | 5.33 | |
| None Classical hydrogen bond | His84 | 3.66 | |
| Pi–Alkyl | Val64 | 5.02 | |
| Pi–Alkyl | Val18 | 5.13 | |
| Pi–Sigma | Val18 | 2.84 | |
| 10f | Pi–Alkyl | Ile10 | 4.41 |
| Pi–Alkyl | Val64 | 4.88 | |
| Pi–Alkyl | Ala144 | 4.76 | |
| Pi–Sigma | Ala144 | 2.71 | |
| Pi–Alkyl | Leu134 | 5.31 | |
| Pi–Alkyl | Ala31 | 5.04 | |
| Pi–Alkyl | Val18 | 5.21 | |
| Pi–Sigma | Val18 | 2.88 |
| Cpd. | Ar | IC50 (µM) |
|---|---|---|
| CDK2 | ||
| 10d | C6H5 | 0.89 |
| 10e | 4–CH3O–C6H4 | 0.62 |
| 10f | thien–2–yl | 1.15 |
| Cpd. | BBB | GIA | P–gPsubstrate | CYP2C9 Inhibitor | CYP2C19 Inhibitor | CYP1A2 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor | PAINS Alerts | Veber Violations | Lipinski Violations |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10d | Yes | High | No | Yes | Yes | Yes | Yes | Yes | NO | 0 | 0 |
| 10e | No | High | No | Yes | Yes | Yes | Yes | Yes | NO | 0 | 0 |
| 10f | No | High | No | Yes | Yes | Yes | No | Yes | NO | 0 | 0 |
| Crystal Data | 9b | 10f | ||
|---|---|---|---|---|
| Chemical formula | C12H14N2O6S2 | C13H10N2OS2 | ||
| Mr | 346.37 | 274.35 | ||
| Temperature (K) | 296 | 293 | ||
| Crystal system, space group | Triclinic, P1− | Orthorhombic, Pbca | ||
| a, b, c (Å) | 9.6682 (2), 11.9413 (3), 13.1649 (3) | 8.6619 (2), 9.8244 (2), 29.7158 (7) | ||
| V (Å3) | 1489.42 (6) | 2528.75 (10) | ||
| Z | 4 | 8 | ||
| µ (mm−1) | 3.54 | 0.41 | ||
| Radiation type | Cu Kα | Mo Kα | ||
| Crystal size (mm) | 0.60 × 0.16 × 0.12 | 0.48 × 0.20 × 0.11 | ||
| Data Collection | ||||
| Diffractometer | Bruker APEX-II D8 venture | |||
| Absorption correction | Multi-scan, SADABS Bruker 2014 | |||
| Tmin, Tmax | 0.226, 0.674 | 0.89, 0.95 | ||
| Rint | 0.055 | 0.038 | ||
| No. of measured, independent and observed [I > 2σ(I)] reflections | 16277, 4617, 4132 | 60453, 4406, 3778 | ||
| Refinement | ||||
| R[F2 > 2σ(F2)], wR(F2), S | 0.038, 0.105, 1.05 | 0.039, 0.111, 0.99 | ||
| Reflections Number | 4617 | 4406 | ||
| Restraints Number | 0 | 0 | ||
| Parameters Number | 416 | 167 | ||
| H atoms treatment | H-atoms were treated by independent and constrained refinement mixture | |||
| Δ ρmax, Δ ρmin (e Å−3) | 0.31–0.32 | 0.52–0.39 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Warhi, T.; Said, M.A.; El Hassab, M.A.; Aljaeed, N.; Ghabour, H.A.; Almahli, H.; Eldehna, W.M.; Abdel-Aziz, H.A. Unexpected Synthesis, Single-Crystal X-ray Structure, Anticancer Activity, and Molecular Docking Studies of Certain 2–((Imidazole/Benzimidazol–2–yl)thio)–1–arylethanones. Crystals 2020, 10, 446. https://doi.org/10.3390/cryst10060446
Al-Warhi T, Said MA, El Hassab MA, Aljaeed N, Ghabour HA, Almahli H, Eldehna WM, Abdel-Aziz HA. Unexpected Synthesis, Single-Crystal X-ray Structure, Anticancer Activity, and Molecular Docking Studies of Certain 2–((Imidazole/Benzimidazol–2–yl)thio)–1–arylethanones. Crystals. 2020; 10(6):446. https://doi.org/10.3390/cryst10060446
Chicago/Turabian StyleAl-Warhi, Tarfah, Mohamed A. Said, Mahmoud A. El Hassab, Nada Aljaeed, Hazem A. Ghabour, Hadia Almahli, Wagdy M. Eldehna, and Hatem A. Abdel-Aziz. 2020. "Unexpected Synthesis, Single-Crystal X-ray Structure, Anticancer Activity, and Molecular Docking Studies of Certain 2–((Imidazole/Benzimidazol–2–yl)thio)–1–arylethanones" Crystals 10, no. 6: 446. https://doi.org/10.3390/cryst10060446
APA StyleAl-Warhi, T., Said, M. A., El Hassab, M. A., Aljaeed, N., Ghabour, H. A., Almahli, H., Eldehna, W. M., & Abdel-Aziz, H. A. (2020). Unexpected Synthesis, Single-Crystal X-ray Structure, Anticancer Activity, and Molecular Docking Studies of Certain 2–((Imidazole/Benzimidazol–2–yl)thio)–1–arylethanones. Crystals, 10(6), 446. https://doi.org/10.3390/cryst10060446







