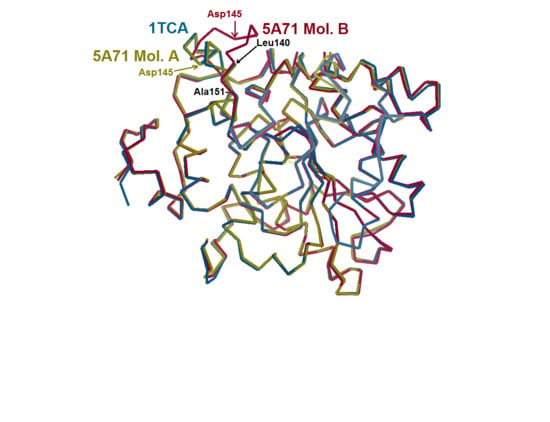
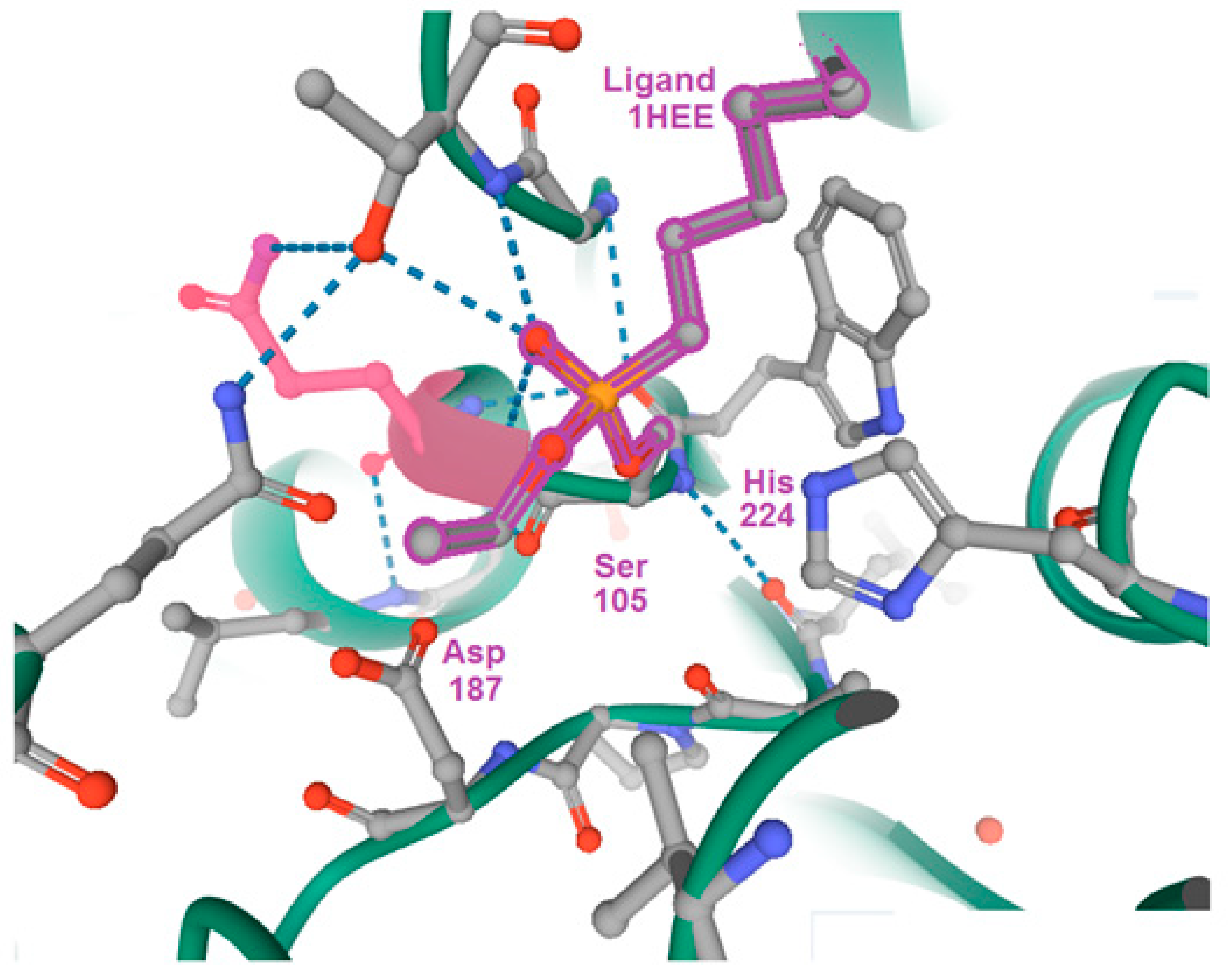
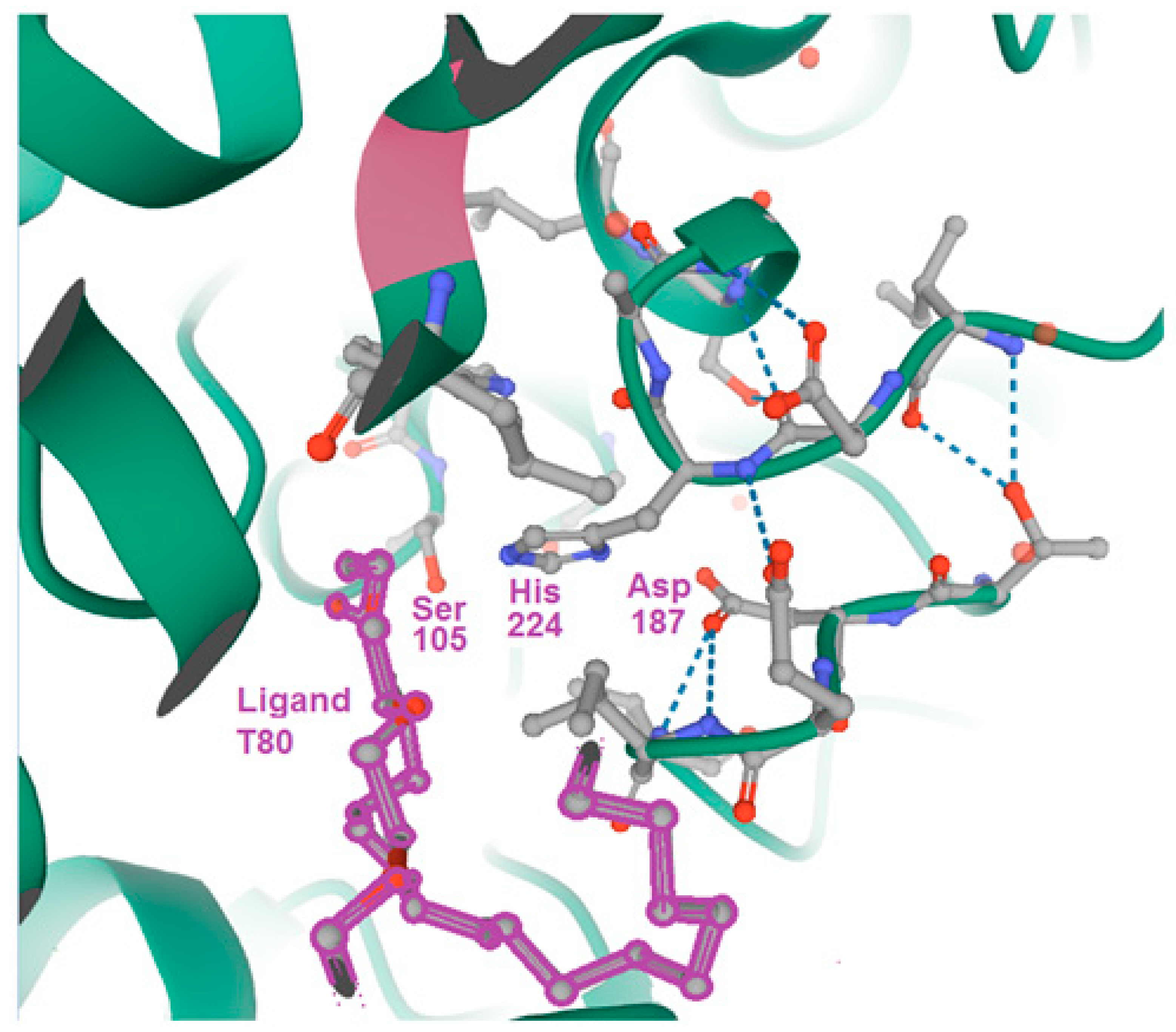
Quarter of a Century after: A Glimpse at the Conformation and Mechanism of Candida antarctica Lipase B
Abstract
Author Contributions
Funding
Conflicts of Interest
References
- Kirk, O.; Christensen, M.W. Lipases from Candida antarctica: Unique biocatalysts from a unique origin. Org. Prog. Res. Dev. 2002, 6, 446–451. [Google Scholar] [CrossRef]
- Wu, Q.; Soni, P.; Reetz, M.T. Laboratory evolution of enantiocomplementary Candida antarctica lipase B mutants with broad substrate scope. J. Am. Chem. Soc. 2013, 135, 1872–1881. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Rani, K.Y.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Kazlauskas, R.J. Hydrolases in Organic Synthesis: Regio- and Stereoselective Biotransformations; Wiley: Weinheim, Germany, 1999. [Google Scholar] [CrossRef]
- Casas-Godoy, L.; Gasteazoro, F.; Duquesne, S.; Bordes, F.; Marty, A.; Sandoval, G. Lipases: An overview. Methods Mol. Biol. 2018, 1835, 3–38. [Google Scholar] [CrossRef] [PubMed]
- Kiełbasiński, P.; Kwiatkowska, M.; Mikołajczyk, M. Chapter 10—Chiral heteroatom-containing compounds. In Future Directions in Biocatalysis, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 191–250. [Google Scholar] [CrossRef]
- Alatorre-Santamaria, S.; Gotor-Fernandez, V.; Gotor, V. Chemoenzymatic synthesis of optically active cis- and trans-2-(1H-imidazol-1-yl)cycloalkanamines. Eur. J. Org. Chem. 2011, 6, 1057–1063. [Google Scholar] [CrossRef]
- Frykman, H.; Ohrner, N.; Norin, T.; Hult, K. S-Ethyl thiooctanoate as acyl donor in lipase-catalyzed resolution of secondary alcohols. Tetrahedron Lett. 1993, 34, 1367–1370. [Google Scholar] [CrossRef]
- Patterson, L.D.; Miller, M.J. Enzymatic deprotection of the cephalosporin 3′-acetoxy group using Candida antarctica lipase B. J. Org. Chem. 2010, 75, 1289–1292. [Google Scholar] [CrossRef]
- Santaniello, E.; Casati, S.; Ciuffreda, P. Lipase-catalyzed deacylation by alcoholysis: A selective, useful transesterification reaction. Curr. Org. Chem. 2006, 10, 1095–1123. [Google Scholar] [CrossRef]
- Andualema, B.; Gessesse, A. Microbial lipases and their industrial applications: Review. Biotechnology 2012, 11, 100–118. [Google Scholar] [CrossRef]
- Naik, S.; Basu, A.; Saikia, R.; Madan, B.; Paul, P.; Chaterjee, R.; Brask, J.; Svendsen, A. Lipases for use in industrial biocatalysis: Specificity of selected structural groups of lipases. J. Mol. Catal. B Enzym. 2010, 65, 18–23. [Google Scholar] [CrossRef]
- Sharma, D.; Sharma, B.; Shukla, A.K. Biotechnological approach of microbial lipase: A review. Biotechnology 2011, 10, 3–40. [Google Scholar] [CrossRef]
- Carboni-Oerlemans, C.; Dominguez de Maria, P.; Tuin, B.; Bargeman, G.; van der Meer, A.; van Gemert, R. Hydrolase-catalysed synthesis of peroxycarboxylic acids: Biocatalytic promiscuity for practical applications. J. Biotechnol. 2006, 126, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Carlqvist, P.; Svedendahl, M.; Branneby, C.; Hult, K.; Brinck, T.; Berglund, P. Exploring the active-site of a rationally redesigned lipase for catalysis of Michael-type additions. ChemBioChem 2005, 6, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Hult, K.; Berglund, P. Enzyme promiscuity: Mechanism and applications. Trends Biotechnol. 2007, 25, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Madalińska, L.; Kwiatkowska, M.; Cierpiał, T.; Kiełbasiński, P. Investigations on enzyme catalytic promiscuity: The first attempts at a hydrolytic enzyme-promoted conjugate addition of nucleophiles to α,β-unsaturated sulfinyl acceptors. J. Mol. Catal. B. Enzym 2012, 81, 25–30. [Google Scholar] [CrossRef]
- Sharma, U.K.; Sharma, N.; Kumar, R.; Kumar, R.; Sinha, A.K. Biocatalytic promiscuity of lipase in chemoselective oxidation of aryl alcohols/acetates: A anique synergism of CAL-B and Br for the metal-free H2O2 activation. Org. Lett. 2009, 11, 4846–4848. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, B.-K.; Lin, X.-F. Enzymatic promiscuity for organic synthesis and cascade process. Curr. Org. Chem. 2010, 14, 1966–1988. [Google Scholar] [CrossRef]
- Kundys, A.; Białecka-Florjańczyk, E.; Fabiszewska, A.; Małajowicz, J. Candida antarctica lipase B as catalyst for cyclic esters synthesis, their polymerization and degradation of aliphatic polyesters. J. Polym. Environ. 2018, 26, 396–407. [Google Scholar] [CrossRef]
- Xie, Y.; An, J.; Yang, G.; Wu, G.; Zhang, Y.; Cui, L.; Feng, Y. Enhanced enzyme kinetic stability by increasing rigidity within the active site. J. Biol. Chem. 2014, 289, 7994–8006. [Google Scholar] [CrossRef]
- Kim, S.-K.; Park, Y.-C.; Lee, H.-H.; Jeon, S.-T.; Min, W.-K.; Seo, J.-H. Simple amino acid tags improve both expression and secretion of Candida antarctica lipase B in recombinant Escherichia coli. Biotechnol. Bioeng. 2015, 112, 346–355. [Google Scholar] [CrossRef]
- Qian, Z.; Horton, J.R.; Cheng, X.; Lutz, S. Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation. J. Mol. Biol. 2009, 393, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Kiełbasiński, P.; Mikołajczyk, M. Chiral heteroatom-containing compounds. In Future Directions in Biocatalysis; Matsuda, T., Ed.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 159–203. [Google Scholar] [CrossRef]
- Krasiński, G.; Cypryk, M.; Kwiatkowska, M.; Mikołajczyk, M.; Kiełbasiński, P. Molecular modeling of the lipase-catalyzed hydrolysis of acetoxymethyl(i-propoxy) phenylphosphine oxide and its P-borane analogue. J. Mol. Graph. Model. 2012, 38, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, M.; Krasiński, G.; Cypryk, M.; Cierpiał, T.; Kiełbasiński, P. Lipase-mediated stereoselective transformations of chiral organophosphorus P-boranes revisited: Revision of the absolute configuration of alkoxy(hydroxymethyl)phenylphosphine P-boranes. Tetrahedron Asymmetry 2011, 22, 1581–1590. [Google Scholar] [CrossRef]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Strzelczyk, P.; Bujacz, G.D.; PKiełbasiński, P.; Błaszczyk, J. Crystal and molecular structure of hexagonal form of lipase B from Candida antarctica. Acta Biochim. Pol. 2016, 63, 103–109. [Google Scholar] [CrossRef]
- Uppenberg, J.; Oehrner, N.; Norin, M.; Hult, K.; Kleywegt, G.J.; Patkar, S.; Waagen, V.; Anthonsen, T.; Jones, T.A. Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols. Biochemistry 1995, 34, 16838–16851. [Google Scholar] [CrossRef]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef]
- Cygler, M.; Schrag, J.D.; Ergan, F. Advances in structural understanding of lipases. Biotechnol. Genet. Eng. Rev. 1992, 10, 143–184. [Google Scholar] [CrossRef][Green Version]
- Palekar, A.A.; Vasudevan, P.T.; Yan, S. Purification of lipase: A review. Biocatal. Biotransform. 2009, 18, 177–200. [Google Scholar] [CrossRef]
- Brämer, C.; Schreiber, S.; Scheper, T.; Beutel, S. Continuous purification of Candida antarctica lipase B using 3-membrane adsorber periodic counter-current chromatography. Eng. Life Sci. 2018, 18, 414–424. [Google Scholar] [CrossRef]
- Van Tassel, L.; Moilanen, A.; Ruddock, L.W. Efficient production of wild-type lipase B from Candida antarctica in the cytoplasm of Escherichia coli. Protein Expr. Purif. 2020, 165, 105498. [Google Scholar] [CrossRef] [PubMed]
- Potrzebowski, M.J.; Grossmann, G.; Błaszczyk, J.; Wieczorek, M.W.; Sieler, J.; Knopik, P.; Komber, H. X-ray and solid state NMR studies of bis(5,5-dimethyl-2-thioxo-l,3,2-dioxaphosphorinan-2-yl) disulfide and diselenide. Inorg. Chem. 1994, 33, 4688–4695. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Rose, A.S.; Kovca, J.; Burley, S.K.; Velankar, S. Mol*: Towards a common library and tools for web molecular graphics. MolVA/EuroVis Proceedings, 2018. MolVA/EuroVis Proc. 2018. [Google Scholar] [CrossRef]
- Benson, S.P.; Pleiss, J. Self-assembly nanostructures of triglyceride-water interfaces determine functional conformations of Candida antarctica lipase B. Langmuir 2017, 33, 3151–3159. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Cryst. 2004, D60, 2126–2132. [Google Scholar] [CrossRef]
- Skjot, M.; De Maria, L.; Chatterjee, R.; Svendsen, A.; Patkar, S.A.; Ostergaard, P.R.; Brask, J. Understanding the plasticity of the α/β hydrolase fold: Lid swapping on the Candida antarctica lipase B results in chimeras with interesting biocatalytic properties. ChemBioChem 2009, 10, 520–527. [Google Scholar] [CrossRef]
- Xu, J.; Cen, Y.; Singh, W.; Fan, J.; Wu, L.; Lin, X.; Zhou, J.; Huang, M.; Reetz, M.T.; Wu, Q. Stereodivergent protein engineering of a lipase to access all possible stereoisomers of chiral esters with two stereocenters. J. Am. Chem. Soc. 2019, 141, 7934–7945. [Google Scholar] [CrossRef]
- Chen, H.; Meng, X.; Xu, X.; Liu, W.; Li, S. The molecular basis for lipase stereoselectivity. Appl. Microbiol. Biotechnol. 2018, 102, 3487–3495. [Google Scholar] [CrossRef]
- Cen, Y.; Singh, W.; Arkin, M.; Moody, T.S.; Huang, M.; Zhou, J.; Wu, Q.; Reetz, M.T. Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Park, C.-G.; Kwon, M.-A.; Song, J.-K.; Kim, D.-M. Cell-free synthesis and multifold screening of Candida antarctica lipase B (CalB) variants after combinatorial mutagenesis of hot spots. Biotechnol. Prog. 2011, 27, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Kim, S.; Park, J.; Joe, S.; Min, B.; Oh, J.; Song, J.; Park, S.-Y.; Park, S.; Lee, H. Structural and experimental evidence for the enantiomeric recognition toward a bulky sec-alcohol by Candida antarctica lipase B. ACS Catal. 2016, 6, 7458–7465. [Google Scholar] [CrossRef]
- Kazlauskas, R.J.; Weissfloch, A.N.; Rappaport, A.T.; Cuccia, L.A. A rule to predict which enantiomer of a secondary alcohol reacts faster in reactions catalyzed by cholesterol esterase, lipase from Pseudomonas cepacia, and lipase from Candida rugosa. J. Org. Chem. 1991, 56, 2656–2665. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Błaszczyk, J.; Kiełbasiński, P. Quarter of a Century after: A Glimpse at the Conformation and Mechanism of Candida antarctica Lipase B. Crystals 2020, 10, 404. https://doi.org/10.3390/cryst10050404
Błaszczyk J, Kiełbasiński P. Quarter of a Century after: A Glimpse at the Conformation and Mechanism of Candida antarctica Lipase B. Crystals. 2020; 10(5):404. https://doi.org/10.3390/cryst10050404
Chicago/Turabian StyleBłaszczyk, Jarosław, and Piotr Kiełbasiński. 2020. "Quarter of a Century after: A Glimpse at the Conformation and Mechanism of Candida antarctica Lipase B" Crystals 10, no. 5: 404. https://doi.org/10.3390/cryst10050404
APA StyleBłaszczyk, J., & Kiełbasiński, P. (2020). Quarter of a Century after: A Glimpse at the Conformation and Mechanism of Candida antarctica Lipase B. Crystals, 10(5), 404. https://doi.org/10.3390/cryst10050404