Abstract
Flavin-dependent halogenases chlorinate or brominate their substrates in an environmentally friendly manner, only requiring the cofactor reduced flavin adenine dinucleotide (FADH2), oxygen, and halide salts. The tryptophan 6-halogenase Thal exhibits two flexible loops, which become ordered (substrate-binding loop) or adopt a closed conformation (FAD loop) upon substrate or cofactor binding. Here, we describe the structure of NHis-Thal-RebH5 containing an N-terminal His-tag from pET28a, which crystallized in a different space group (P21) and, surprisingly, diffracted to a higher resolution of 1.63 Å than previously deposited Thal structures (P64; ~2.2 Å) with cleaved His-tag. Interestingly, the binding of glycine in the active site can induce an ordered conformation of the substrate-binding loop.
1. Introduction
Many natural products, like chloramphenicol [1], vancomycin [2,3], and others [4,5,6], have a halide substituent, which is important for their biological activity. The unfavorable conventional chemical synthesis of these products requires toxic elemental halides and Lewis acids as catalysts and lacks substrate specificity as well as regioselectivity. Therefore, flavin-dependent halogenases (FDHs) came into focus as potential biocatalysts for aromatic compounds in organic synthesis because they halogenate regioselectively, substrate-specifically, and in an environmentally friendly manner [7,8], only requiring molecular oxygen, reduced flavin adenine dinucleotide (FADH2), and halide salts (mainly Cl− or Br−) [7]. Tryptophan halogenases are one group of FDHs which halogenate in position C5 (e.g., PyrH [9]), C6 (e.g., Thal [10]), and C7 (e.g., RebH [11,12], PrnA [13]) of the indole moiety depending on the orientation of the substrate tryptophan within the active site [9,10]. Structurally, the active site is located at the interface of the two subdomains, a conserved box-shaped and a pyramidal subdomain. The cofactor flavin adenine dinucleotide (FAD) becomes bound by the box-shaped subdomain and the halide ion is bound near the isoalloxazine ring of the cofactor [13].
Crystal structures of various tryptophan halogenases revealed two loop regions that undergo conformational changes upon binding of ligands [14]. The substrate-binding loop is disordered when the substrate-binding site (further referred to as the active site) is empty and adopts a defined structure when tryptophan is bound. The FAD loop is disordered (e.g., in PyrH [9] and BrvH [15]) or adopts an open conformation (e.g., in Thal [10]) in the apo enzyme but closes in the presence of FAD (e.g., in PrnA [13], PyrH [9], SttH [16], and Tar14 [17]). Crystal packing can prevent some of these movements; e.g., in RebH, the FAD loops of two symmetry mates form a crystal contact locking the loop in the open conformation even when FAD is bound [11,12]. In apo Xcc4156, the FAD loop in the open conformation participates in crystal contact so that attempts to soak FAD into these crystals causes their destruction [18]. Therefore, it may be of interest to obtain more than one crystal form of a single halogenase to assess the influence of crystal packing on the conformation of functionally important regions.
Thal, also known as ThdH, is a tryptophan 6-halogenase from Streptomyces albogriseolus involved in thienodolin biosynthesis [19,20]. By changing five amino acids in the active site of Thal to the corresponding residues of the related tryptophan 7-halogenase RebH, we could switch the regioselectivity almost completely. The resulting Thal variant Thal-RebH5 showed an overall conversion similar to that of wildtype Thal, but it halogenated l-Trp mainly in position 7 (95%) [10].
The recombinant Thal used for halogenation assays was produced as N-terminally His-tagged protein from a pET-28a-based expression plasmid [21]. We had expected a negative impact of the 20-residue long tag on crystallization. In a rational approach to obtain crystals of wildtype Thal (wt-Thal), we had therefore switched to a pET-M11-based plasmid that allowed efficient removal of the N-terminal His-tag by tobacco etch virus (TEV) protease [10]. With untagged wt-Thal, we had quickly obtained reproducible crystals diffracting to maximally 2.12 Å that allowed determining structures of Thal in the presence of various ligands [10,14]. During revision of the manuscript on the switch of regioselectivity in Thal [10], we wanted to obtain crystals of Thal-RebH5 as quickly as possible. In parallel to re-cloning Thal-RebH5 from pET-28a to pET-M11, we therefore purified N-terminally His-tagged Thal-RebH5 expressed in pET-28a (NHis-Thal-RebH5). While this protein did not crystallize under the conditions used for untagged wt-Thal, we obtained crystals in an unrelated condition. These crystals were less reproducible and had a less desirable habit than those of untagged Thal, but they diffracted to higher resolution. Here, we describe two structures of NHis-Thal-RebH5 obtained with these new crystals.
2. Materials and Methods
2.1. Plasmids and Molecular Cloning
The plasmid vector pET-28a-thal-rebH5 encoding Thal-RebH5, a quintuple mutant originating from Thal wildtype (Uniprot ID: A1E280) from Streptomyces albogriseolus, was kindly provided by Dr. Hannah Minges, Organic Chemistry III, Bielefeld University. The molecular cloning and mutation procedure were described earlier [10].
2.2. Expression and Purification of Recombinant Thal-RebH5
The expression and purification of NHis-Thal-RebH5 was performed according to the described protocol for wt-Thal [10], except that a Co-NTA (pure Cube Co-NTA agarose, Cube Biotech, Monheim am Rhein, Germany) was used instead of Ni-NTA and the cleavage of the His-tag was omitted.
2.3. Crystallization and Data Collection
Purified NHis-Thal-RebH5 (protein buffer: 10 mM Tris pH 7.4, 50 mM NaCl, 1 mM TCEP) was crystallized using the sitting drop vapor diffusion method at 20 °C with a drop ratio of 1:1 of protein solution (~15 mg·mL−1) and reservoir solution [((1) 0.1 M bicine pH 9.0, 20% (w/v) PEG 4000, 10% (v/v) glycerol, and 0.02 M amino acid mix consisting of L-Glu, L-Ala, D-Ala, Gly, L-Lys, D-Lys, L-Ser, and D-Ser) and ((2) 0.1 M bicine pH 9.0 and 10% (w/v) PEG 4000 supplemented with 5 mM tryptophan)]. Crystals appeared in different shapes, ranging from almost hexagonal prisms (see Figure 1a) to plates (Figure 1b) within four days. The crystal habit of NHis-Thal-RebH5 was inferior to that of untagged Thal (Figure 1d). Crystals of NHis-Thal-RebH5 were generally smaller, often plate-shaped instead of nicely extended into all three dimensions, and often intergrown (Figure 1c). For cryoprotection, the crystals were transferred to reservoir solution supplemented with glycerol ((1) 20% (v/v) and (2) 35% (v/v)), before being flash cooled and stored in liquid nitrogen. Crystals of His-tagged Thal were mechanically less robust than those of untagged Thal and easily broke during mounting. Data sets of NHis-Thal-RebH5 were collected at a wavelength of 0.98 Å and a temperature of 100 K at beamline P13 operated by EMBL Hamburg at the PETRA III storage ring at DESY, Hamburg, Germany [22].
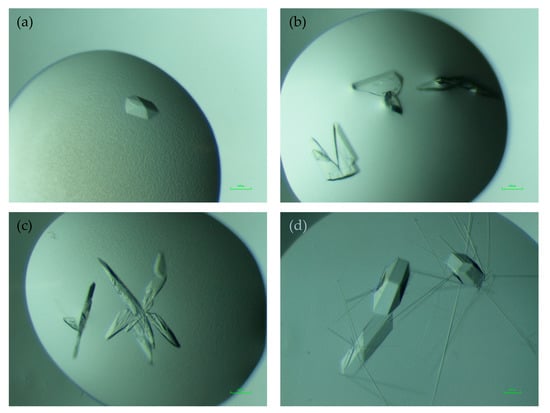
Figure 1.
Crystal morphology of NHis-Thal-RebH5. (a) Crystal of the described structure NHis-Thal-RebH5 (1) with an almost hexagonal morphology and diffraction to 1.63 Å. (b) Plate-shaped crystals of the described structure NHis-Thal-RebH5 (2), which diffracted to 1.84 Å. (c) Further examples of the various morphologies of NHis-Thal-RebH5 crystals. (d) Crystals of untagged Thal-RebH5 (6IB5). Scale bar in green corresponds to 100 µm.
2.4. Data Processing, Structure Determination and Refinement
The data sets were processed with XDS [23] and scaled with XSCALE [23]. NHis-Thal-RebH5 diffracted to (1) 1.63 Å and (2) 1.84 Å, respectively, cutting at I/sig(I)~2. The crystals (see Figure 1) belonged to the space group P21 as determined by the program aimless [24] from the CCP4 package [25]. The structures of NHis-Thal-RebH5 were determined by molecular replacement using the program Phaser [26] and the structure of apo-Thal chain A (PDB ID 6H43) as the search model. Phaser placed two molecules per asymmetric unit with no clashes.
For the NHis-Thal-RebH5 structures, an extensive water search was performed using AutoBuild [27] from the PHENIX suite [28]. The structures were further improved by modeling in COOT [29] and restrained refinement in Refmac5 [30] using NCS restraints for (1) or in PHENIX using TLS refinement with one TLS group per chain and NCS restraints for (2). The structures were optimized and validated using COOT [29] and the pdb validation server [31] until R values converged to (1) Rfree of 20.2% and Rwork of 16.7% and (2) Rfree of 19.9% and Rwork of 16.6%, respectively. A summary of the data processing and refinement statistics is shown in Table 1. Figures of the final structures were generated using PyMOL (Schrödinger, New York, NY, USA). The RMSD analysis was derived from pairwise structure-based sequence alignments with the DALI server [32] (mode all against all).

Table 1.
Data collection and refinement statistics. The statistics of the deposited structure Thal-RebH5 (6IB5) lacking a His-tag [10] is listed for comparison.
2.5. Accession Numbers
The atomic coordinates and structure factors of NHis-Thal-RebH5 (1) and (2) have been deposited in the PDB as entries 7AQU and 7AQV, respectively.
3. Results
3.1. Crystallization Details and Comparison to Thal-RebH5
NHis-Thal-RebH5 crystallizes in different crystallization conditions than untagged Thal-RebH5 [10], with a different and inconsistent crystal morphology ranging from plates, which were often intergrown, to almost hexagonal structures (Figure 1). Even so, the NHis-Thal-RebH5 crystals diffracted with 1.63 Å to 2.17 Å better than untagged Thal-RebH5 (>2.28 Å in 98%) and untagged wt-Thal (>2.2 Å). The space group P21 differs from untagged Thal that crystallized in P64. However, both crystal forms have two monomers per asymmetric unit.
3.2. Comparison of the Previously Described P64 and the New P21 Crystal Forms
NHis-Thal-RebH5 crystallizes as homodimer but is a monomer in solution (size exclusion chromatography) like Thal-RebH5 (PDB ID 6IB5). Both protomers of the new structures are very similar to each other and to those of structures previously obtained in space group P64 (see Table S1 in the supplementary material). All five mutations distinguishing Thal-RebH5 from wildtype Thal (Thal → Thal-RebH5: V52I, V82I, S360T, G469S, T470N) are well defined by 2Fo-Fc density. Structures (1) and (2) of NHis-Thal-RebH5 described here are isomorphous to each other and the corresponding crystals were grown under similar conditions (same buffer, pH, and PEG) in an optimization grid screen. (1) additionally contained glycerol and an amino acid mix derived from the original crystallization condition of the Morpheus screen [33], and (2) additionally contained 5 mM l-Trp in order to obtain a substrate-bound structure. No density for Trp was present in the active sites of (2). This is consistent with our observations of crystals of untagged Thal-RebH5 where only 6 of 47 data sets of Thal-RebH5 co-crystallized or soaked with l-Trp showed at least some density for bound Trp [10]. Even upon soaking the P64 crystals with saturated l-Trp solution, the resulting difference density could not be modeled reliably [10], indicating the Thal-RebH5 binds Trp less tightly than wt-Thal. In (1), the active site is empty in chain B but contains a glycine in chain A that superposes with the position of the backbone of the native substrate tryptophan (Figure 2). Therefore, the electron density of the substrate-binding loop of chain A is better defined than for B, where amino acids (AA) 448–451 and 456 were not modeled (Figure 3), but not as good as when the substrate tryptophan is bound. This verifies previous observations that predominantly the active site of chain A binds substrate, whereas chain B remains empty or binds substrate with lower occupancy [10,14]. For structure (2) with two empty active sites, both substrate-binding loops are not defined and were not modeled (chain A: AA 450–456; chain B: AA 452–456) (Figure 3). The amino acid Tyr366 is of interest as it may couple the conformation of the substrate-binding and the FAD loop [14]. The side chain of Tyr366 adopts two very different conformations depending on the occupation of the active site with tryptophan. In both chains of (1) and (2), Tyr366 adopts the conformer typical for an empty active site. Due to the better resolution, a previously predicted H-bond between Tyr366 and Asn54 could be modeled with no clashes and a distance around 2.4 Å in both chains and structures. Phe 112 undergoes a peptide flip upon binding of the substrate tryptophan. In the new P21 crystal form, both chains of the structures (1) and (2) feature Phe 112 in the typical unbound conformation underlining the emptiness of the active sites.
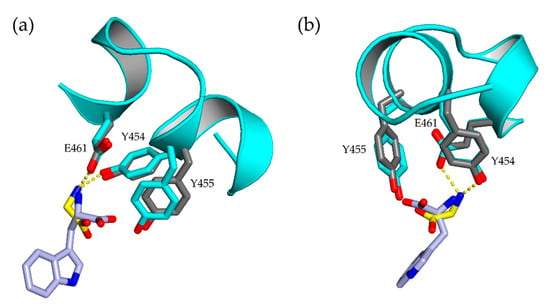
Figure 2.
Active sites of NHis-Thal-RebH5 (1) and Trp-Thal (6H44). Alignment of the bound substrate tryptophan (light blue) of Trp-Thal (6H44) (gray) and the active site of NHis-Thal-RebH5 (1) chain A (cyan) where a glycine (yellow) could be modeled, which superposes with the backbone of the tryptophan. In the figure, the structure in (a) is rotated relative to that in (b).
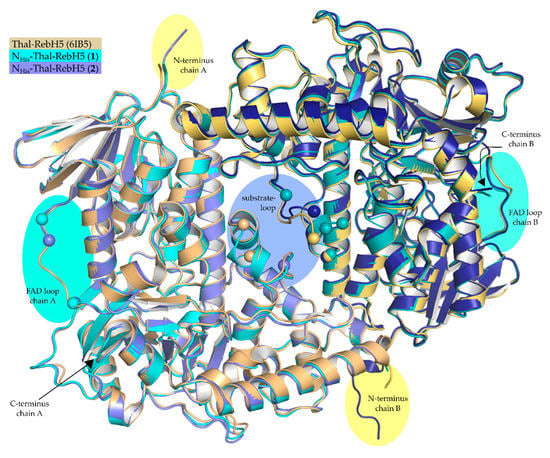
Figure 3.
Alignment of Thal-RebH5 (yellow/orange), NHis-Thal-RebH5 (1) (cyan) and (2) (light blue/blue). Overall, all three structures are very similar, only differing in loop sections and length of the N-terminus (highlighted in yellow). Disordered regions like the FAD loop (highlighted in turquoise) or substrate-binding loop (highlighted in violet) are marked structure-specific with a sphere of the last and next modeled Cα- atom.
The FAD loop (AA 40–49), which is visible in all previously determined and published structures of Thal, is partly not defined and, therefore, not modeled in chain A in (1) (AA 41–44) and (2) (AA 42–44) (Figure 3). Upon lowering the RMSD values under 0.6, density for these residues appears, suggesting that the FAD loop is not totally disordered. In chain B of (1) and (2) the FAD loop is defined and in an open conformation with a slightly different conformation than in Thal-RebH5 (PDB code 6IB5) (see Figure S1 in the supplementary material).
3.3. Crystal Contacts and Possible Impact of the His-tag
Compared to the other published Thal structures with the space group P64 NHis-Thal-RebH5 (P21) is packed more densely (see Figure S2 in the supplementary material) (Matthews coefficient: P64 3.34 Å3/Da and ~63% solvent; NHis-Thal-RebH5 2.28 Å3/Da and ~46% solvent;) and, therefore, more symmetry-related molecules are closer (nine interfaces for (1), eight interfaces for (2), and six interfaces for Thal-RebH5 (6IB5, P64)). The PISA server [34] analyzed, in total, 16 H-bonds and 15 salt bridges for (1) and 11 H-bonds and 18 salt bridges for (2), compared to 16 H-bonds and 9 salt bridges for Thal-RebH5 (6IB5, P64).
In structure (1), the N-terminal His-tag and the linker are not defined by electron density. In structure (2), however, the linker could be partly modeled for chain B to Val-5. The modeled linker extends out of the dimer (Figure 3) in between two symmetry relatives. Superposition with Thal-RebH5 (6IB5) revealed almost the same amount of space for a possible linker in the P64 crystals of untagged Thal-RebH5. The surrounding of the N-terminus differs for the protomers in the new crystal form. The residue D2 is the first amino acid defined in both chains of (1). In chain A, D2 is closest to a symmetry-related A K267 (see Figure S3c in the supplementary material). In chain B, D2 is closest to a symmetry-related B E424 (see Figure S3d in the supplementary material). In both chains of (1) and in chain A of (2), D2 forms an intramolecular salt bridge with R4. In chain B of (2) the D2 side chain adopts another conformation and interacts with the partly modeled linker and therefore stabilizes it. The defined linker in chain B of (2) forms an extended crystal contact to a symmetry-related chain B, e.g., involving hydrogen bonds of G-2 with the side chains of B R420 and B E424 (see Figure S3d in the supplementary material).
4. Discussion
Comparison of Thal-RebH5 structures with His-tag ((1) and (2)) and without His-tag (6IB5) revealed a high similarity of the structures themselves and their functional regions like the FAD loop, the substrate-binding loop, the active site, the possible peptide flip of Phe112, and the switch residue Tyr366. The extended N-terminus with the partly modeled linker of the His-tag (structure (2) chain B) (Figure 3) makes an additional crystal contact. The crystals looked less promising and were mechanically less robust than previously obtained crystals of Thal constructs with cleaved His-tag.
4.1. Differing Functional Regions
One observation was the presence of a glycine in the active site of chain A of NHis-Thal-RebH5 (1) and, therefore, a more ordered substrate-binding loop of chain A compared to chain B with an empty active site. This phenomenon was already observed in apo-Thal (6H43) [10], where unmodeled 2Fo-Fc density within the active site and a defined substrate-binding loop was detected in chain A, but not in chain B. This principle was further analyzed and described by a statistical analysis of a negatively cooperative binding of the substrate tryptophan and cofactor FAD [14]. When the substrate tryptophan is bound, the substrate-binding loop becomes ordered, and when both substrate and cofactor are bound, the substrate is always bound to chain A, whereas FAD is bound in chain B.
Another observation was the partly undefined FAD loop of chain A of (1) and (2). One potential reason for this could be that residue I42 of chain A would clash strongly with B P262 of a symmetry-related molecule. In chain B, I42 does not clash and even makes stabilizing van der Waals interactions with a neighboring molecule (see Figure S3a,b in the supplementary material). In addition, there is a crystal contact of B R44 and a symmetry-related A D307, whereas chain A makes no such crystal contact.
In all other Thal structures, the FAD loop is not stabilized by a crystal contact but is always defined by electron density [10,14]. The open conformation of the FAD loop seen in the P64 structures might be the energetically lowest possible conformation in the absence of FAD and might even be present in solution.
4.2. Impact of VM on Diffraction
The denser packing of P21 compared to P64 crystals, deducible from the lower Matthews coefficient of 2.28 for P21 (NHis-Thal-RebH5), compared to 3.33 for P64 (apo-Thal, 6H43), is another possible explanation for the better diffraction. In 1968, Matthews estimated solvent contents of 27–65%, which correlates to VM values of 2.15–2.61 Å3/Dalton, for protein crystals by analyzing 116 structures [35]. Hence, the untagged P64 structures of Thal are at the high end of the VM distribution. Kantardjieff and Rupp [36] reanalyzed in 2002 the VM distribution of 10,471 non-redundant protein crystal forms and observed a broader range with a mode of 2.34 Å3/Dalton corresponding to a solvent content of ~47%. They also analyzed the VM distribution along the resolution and observed lower VM values at higher resolution, indicating that crystals diffract better when they are packed tightly.
4.3. Impact of His-tag on Crystallization
Changing the position of the His-tag (N- or C-terminal) may already have effects on the protein [37], such as differing activity, inhibition, or conformation [38,39]. There are contradictory publications on the impact of His-tags on crystallization. Smits et al. [39] report only the construct with His-tag yielded crystals, whereas Plavša et al. [40] could improve the diffraction quality in terms of resolution, spot shape, and spot size by adding thrombin in situ immediately before crystallization to cleave the His-tag (using a pET-28a vector). This also resulted in a different crystal morphology like we observed for tagged and untagged Thal-RebH5 (see Figure 1). The His-tag can change the properties of the protein [41,42] and, therefore, it is possible that the tagged and untagged protein do not crystallize in the same crystallization conditions [43], just like with NHis-Thal-RebH5 and untagged Thal-RebH5.
Bucher et al. [44] chose a more systematic approach. They analyzed the crystallization habit of the protein PfuMBP with various short tags and could detect an influence of the tag amino acid sequence on the formation of crystals and their ability to diffract. On the other hand, they observed no impact of the His-tag on the crystallization of PfuMBP and concluded that the His-tag is “not intrinsically detrimental to the formation of protein crystals” [44] (p. 397) in context of the numerous deposited His-tagged structures [44].
Carson et al. [45] statistically analyzed, in 2007, the impact of His-tags on the crystallization of 1142 tagged in comparison to 11906 untagged protein structures deposited at the PDB and detected that “the resolution and R-factors were consistently better for the his-tagged structures but the differences are not statistically significant” [45] (p. 301). Only small structural differences between tagged and untagged protein structures were detected, which agrees with our results of high similarity of the structures themselves and their functional regions. Moreover, the majority of the tags (90%) were disordered or involved in crystal contacts, as well as, on average, ten residues long [43,45]. This again coincides with our results of reliable density only for five residues of the linker and an additional crystal contact involving the N-terminus of chain B. In addition, most of the His-tags protruded outside of the protein just like NHis-Thal-RebH5 (2) [43,45]. In total, “structures with well-defined His-tags are rare” [41] (p. 300) and “have only marginally different values” [41] (p. 299) of R-factors, VM and B-factors [45].
In summary, it is not entirely possible to predict the impact of His-tags on the crystallization of one’s protein, but there are so many His-tagged structures deposited in the PDB that there is no mandatory need to cleave the His-tag. Tagged and untagged Thal-RebH5 is a great example, showing that it is worth a try to test a different construct and a different crystal morphology, even when the crystals do not look promising. Furthermore, the binding of a glycine in the active site of the tryptophan 6-halogeanse Thal can induce an ordered conformation of the substrate-binding loop.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4352/10/12/1135/s1, Figure S1: Differing FAD loop of NHis-Thal-RebH5 (1), (2) and Thal-RebH5, Figure S2: Crystal packing of NHis-Thal-RebH5 (P21), Figure S3: Crystal contacts of NHis-Thal-RebH5 (1) and (2), Table S1: Structural RMSD analysis of untagged and tagged tryptophan halogenase Thal variants.
Author Contributions
Conceptualization, H.H.N.; formal analysis, A.-C.M. and H.H.N.; investigation, A.-C.M., T.P. and H.H.N.; data curation, A.-C.M. and H.H.N.; writing—original draft preparation, A.-C.M.; writing—review and editing, A.-C.M. and H.H.N.; visualization, A.-C.M.; supervision and project administration, H.H.N. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge support for the publication costs by the Deutsche Forschungsgemeinschaft and the Open Access Publication Fund of Bielefeld University.
Acknowledgments
The synchrotron MX data were collected at beamline P13 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We thank Johanna Hakanpää for assistance in using the beamline. We thank Hannah Minges for providing the plasmid vector pET-28a-thal-rebH5.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Podzelinska, K.; Latimer, R.; Bhattacharya, A.; Vining, L.C.; Zechel, D.L.; Jia, Z. Chloramphenicol Biosynthesis: The Structure of CmlS, a Flavin-Dependent Halogenase Showing a Covalent Flavin-Aspartate Bond. J. Mol. Biol. 2010, 397, 316–331. [Google Scholar] [CrossRef]
- Williams, D.H.; Bardsley, B. The vancomycin group of antibiotics and the fight against resistant bacteria. Angew. Chem. Int. Ed. 1999, 38, 1172–1193. [Google Scholar] [CrossRef]
- Bischoff, D.; Pelzer, S.; Bister, B.; Nicholson, G.J.; Stockert, S.; Schirle, M.; Wohlleben, W.; Jung, G.; Süssmuth, R.D. The biosynthesis of vancomycin-type glycopeptide antibiotics-The order of the cyclization steps. Angew. Chem. Int. Ed. 2001, 40, 4688–4691. [Google Scholar] [CrossRef]
- Weichold, V.; Milbredt, D.; van Pée, K.-H. Specific Enzymatic Halogenation-From the Discovery of Halogenated Enzymes to Their Applications In Vitro and In Vivo. Angew. Chem. Int. Ed. 2016, 55, 6374–6389. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, L.C.; Drennan, C.L. Structural perspective on enzymatic halogenation. Acc. Chem. Res. 2009, 42, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.R.; Belin, L.; Sancelme, M.; Prudhomme, M.; Ollier, M.; Rapp, M.; Sevère, D.; Riou, J.F.; Fabbro, D.; Meyer, T. Structure-activity relationships in a series of substituted indolocarbazoles: Topoisomerase I and protein kinase C inhibition and antitumoral and antimicrobial properties. J. Med. Chem. 1996, 39, 4471–4477. [Google Scholar] [CrossRef] [PubMed]
- Frese, M.; Guzowska, P.H.; Voß, H.; Sewald, N. Regioselective Enzymatic Halogenation of Substituted Tryptophan Derivatives using the FAD-Dependent Halogenase RebH. ChemCatChem 2014, 6, 1270–1276. [Google Scholar] [CrossRef]
- Schnepel, C.; Sewald, N. Enzymatic Halogenation: A Timely Strategy for Regioselective C−H Activation. Chem. A Eur. J. 2017, 23, 12064–12086. [Google Scholar] [CrossRef]
- Zhu, X.; De Laurentis, W.; Leang, K.; Herrmann, J.; Ihlefeld, K.; van Pée, K.H.; Naismith, J.H. Structural Insights into Regioselectivity in the Enzymatic Chlorination of Tryptophan. J. Mol. Biol. 2009, 391, 74–85. [Google Scholar] [CrossRef]
- Moritzer, A.C.; Minges, H.; Prior, T.; Frese, M.; Sewald, N.; Niemann, H.H. Structure-based switch of regioselectivity in the flavin-dependent tryptophan 6-halogenase Thal. J. Biol. Chem. 2019, 294, 2529–2542. [Google Scholar] [CrossRef]
- Bitto, E.; Huang, Y.; Bingman, C.A.; Singh, S.; Thorson, J.S.; Phillips, G.N. The structure of Flavin-dependent tryptophan 7-halogenase RebH. Proteins Struct. Funct. Bioinform. 2007, 70, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.; Blasiak, L.C.; Koglin, A.; Drennan, C.L.; Walsh, C.T. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry 2007, 46, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Flecks, S.; Unversucht, S.; Haupt, C.; Van Pée, K.H.; Naismith, J.H. Structural biology: Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science 2005, 309, 2216–2219. [Google Scholar] [CrossRef] [PubMed]
- Moritzer, A.; Niemann, H.H. Binding of FAD and tryptophan to the tryptophan 6-halogenase Thal is negatively coupled. Protein Sci. 2019, 28, 2112–2118. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, P.R.; Widmann, C.; Wibberg, D.; Schröder, L.; Frese, M.; Kottke, T.; Kalinowski, J.; Niemann, H.H.; Sewald, N. A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Shepherd, S.A.; Menon, B.R.K.; Fisk, H.; Struck, A.-W.; Levy, C.; Leys, D.; Micklefield, J. A Structure-Guided Switch in the Regioselectivity of a Tryptophan Halogenase. ChemBioChem 2016, 17, 821–824. [Google Scholar] [CrossRef]
- Luhavaya, H.; Sigrist, R.; Chekan, J.R.; McKinnie, S.M.K.; Moore, B.S. Biosynthesis of L-4-Chlorokynurenine, an Antidepressant Prodrug and a Non-Proteinogenic Amino Acid Found in Lipopeptide Antibiotics. Angew. Chem. Int. Ed. 2019, 58, 8394–8399. [Google Scholar] [CrossRef]
- Widmann, C.; Ismail, M.; Sewald, N.; Niemann, H.H. Structure of apo flavin-dependent halogenase Xcc4156 hints at a reason for cofactor-soaking difficulties. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 687–697. [Google Scholar] [CrossRef]
- Milbredt, D.; Patallo, E.P.; van Pée, K.-H. A Tryptophan 6-Halogenase and an Amidotransferase Are Involved in Thienodolin Biosynthesis. ChemBioChem 2014, 15, 1011–1020. [Google Scholar] [CrossRef]
- Seibold, C.; Schnerr, H.; Rumpf, J.; Kunzendorf, A.; Hatscher, C.; Wage, T.; Ernyei, A.J.; Dong, C.; Naismith, J.H.; Van Pée, K.-H. A flavin-dependent tryptophan 6-halogenase and its use in modification of pyrrolnitrin biosynthesis. Biocatal. Biotransform. 2006, 24, 401–408. [Google Scholar] [CrossRef]
- Schnepel, C.; Minges, H.; Frese, M.; Sewald, N. A High-Throughput Fluorescence Assay to Determine the Activity of Tryptophan Halogenases. Angew. Chem. Int. Ed. 2016, 55, 14159–14163. [Google Scholar] [CrossRef] [PubMed]
- Cianci, M.; Bourenkov, G.; Pompidor, G.; Karpics, I.; Kallio, J.; Bento, I.; Roessle, M.; Cipriani, F.; Fiedler, S.; Schneider, T.R. P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Rad. 2017, 24, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.W.; Read, R.J.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 64, 61–69. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Kovalevskiy, O.; Nicholls, R.A.; Long, F.; Carlon, A.; Murshudov, G.N. Overview of refinement procedures within REFMAC 5: Utilizing data from different sources. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 215–227. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Laakso, L.M. Dali server update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef] [PubMed]
- Gorrec, F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 2009, 42, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Kantardjieff, K.A.; Rupp, B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003, 12, 1865–1871. [Google Scholar] [CrossRef]
- Zhao, D.; Huang, Z. Effect of His-tag on Expression, Purification, and Structure of Zinc Finger Protein, ZNF191(243-368). Bioinorg. Chem. Appl. 2016, 2016. [Google Scholar] [CrossRef]
- Munadziroh, E.; Ulfa, E.U.; Labiqah, A.; Asmarani, O.; Puspaningsih, N.N.T. Effect of Poly-Histidine Tag Position toward Inhibition Activity of Secretory Leukocyte Protease Inhibitor as Candidate for Material Wound Healing. Avicenna J. Med. Biotechnol. 2020, 12, 32–36. [Google Scholar]
- Smits, S.H.J.; Mueller, A.; Grieshaber, M.K.; Schmitt, L. Coenzyme- and His-tag-induced crystallization of octopine dehydrogenase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 836–839. [Google Scholar] [CrossRef]
- Plavša, J.J.; Řezáčová, P.; Kugler, M.; Pachl, P.; Brynda, J.; Voburka, Z.; Celic, A.S.S.; Petri, E.T.; Škerlová, J. In situ proteolysis of an N-terminal His tag with thrombin improves the diffraction quality of human aldo-keto reductase 1C3 crystals. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 300–306. [Google Scholar] [CrossRef]
- Aslantas, Y.; Surmeli, N.B. Effects of N-Terminal and C-Terminal Polyhistidine Tag on the Stability and Function of the Thermophilic P450 CYP119. Bioinorg. Chem. Appl. 2019, 2019, 8080697. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Su, S.; Liu, F.; Xu, T.; Wang, X.; Huang, Y.; Sun, X.; Ge, X.; Chen, T.; Liu, H.; et al. Removal of the tag from His-tagged ILYd4, a human CD59 inhibitor, significantly improves its physical properties and its activity. Curr. Pharm. Des. 2012, 18, 4187–4196. [Google Scholar] [CrossRef] [PubMed]
- Tajika, Y.; Sakai, N.; Tamura, T.; Yao, M.; Watanabe, N.; Tanaka, I. Crystal structure of hypothetical protein PH0828 fromPyrococcus horikoshii. Proteins Struct. Funct. Bioinform. 2004, 57, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Bucher, M.H.; Evdokimov, A.G.; Waugh, D.S. Differential effects of short affinity tags on the crystallization of Pyrococcus furiosus maltodextrin-binding protein. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Carson, M.; Johnson, D.H.; McDonald, H.; Brouillette, C.; DeLucas, L.J. His-tag impact on structure. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 295–301. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).