Abstract
Bioproduction of vinylphenol derivatives, such as 4-vinylguaiacol (4-VG) and 4-vinylphenol (4-VP), from 4-hydroxycinnamic acids, such as ferulic acid (FA) and p-coumaric acid (pCA), employing whole cells expressing phenolic acid decarboxylases (PAD) as a biocatalyst has attracted much attention in recent years. However, the accumulation of 4-VG or 4-VP in the cell may cause high cytotoxicity to Escherichia coli (E. coli) and consequently cell death during the process. In this study, we firstly report the functional display of a phenolic acid decarboxylase (BLPAD) from Bacillus licheniformis using a GDSL autotransporter from Pseudomonas putida on the cell surface of E. coli. Expression and localization of BLPAD on E. coli were verified by SDS-PAGE and protease accessibility. The PelB signal peptide is more effective in guiding the translocation of BLPAD on the cell surface than the native signal peptide of GDSL, and the cell surface displaying BLPAD activity reached 19.72 U/OD600. The cell surface displaying BLPAD showed good reusability and retained 63% of residual activity after 7 cycles of repeated use. In contrast, the residual activity of the intracellular expressing cells was approximately 11% after 3 cycles of reuse. The molar bioconversion yields of 72.6% and 80.4% were achieved at the concentration of 300 mM of FA and pCA in a biphasic toluene/Na2HPO4–citric acid buffer system, respectively. Its good reusability and efficient catalysis suggested that the cell surface displaying BLPAD can be used as a whole-cell biocatalyst for efficient production of 4-VG and 4-VP.
1. Introduction
4-Vinylguaiacol (4-VG), also known as 2-methoxy-4-vinylphenol, is a high value-added product widely used in food, cosmetic, pharmaceutical, and chemical industries [1,2]. It was also used as a precursor for the synthesis of bio-based polymers such as oxygenated polystyrenes [3,4]. Recently, it was reported that 4-VG could be efficiently biotransformed into vanillin by the carotenoid cleavage oxygenase family enzymes [5,6,7]. 4-Vinylphenol (4-VP, 4-hydroxystyrene) is a monomer for production of polymers and a petroleum-based feedstock for resins, elastomers, and adhesives [8,9]. The bioproduction of 4-VG or 4-VP from ferulic acid (FA) or p-coumaric acid (pCA) employing whole cells harboring intracellular phenolic acid decarboxylases (PAD) as biocatalysts has attracted much attention recently due to some advantages over the free phenolic acid decarboxylases, such as unnecessary cell lysis and enzyme purification, thereby significantly reducing production cost [8,10]. However, the accumulation of 4-VG or 4-VP in the cell may cause a high cytotoxicity to Escherichia coli (E. coli) and, consequently, cell death during the process [11,12]. In addition, reactivity of whole-cell catalysts containing intracellular enzymes also hampered the barriers between enzymes and their substrates [13,14].
Cell surface display systems overcome some of the limitations associated with free enzymes or intracellular enzyme catalysis systems by exposing the heterologous enzymes on the outer membrane of the cell by fusing with anchoring proteins [15]. The enzymatic reaction takes place outside the cell membrane. So, neither substrate nor product needs to be internalized or excreted through the cell membrane; therefore, cytotoxicity of the end product to the cell is alleviated, and biocatalysis is favored [16]. These systems have a wide range of biotechnological applications in many areas such as whole-cell biocatalysis for bioconversion, biosensing, biosorption, and immobilization [17,18,19,20,21,22,23]. The GDSL autotransporter is a distinctive autotransporter that consists of a lipolytic enzyme of the GDSL family of lipases or esterases as a passenger domain at the N-terminus and a β-barrel domain at C-terminus, which is necessary for secretion of the passenger protein to the outer membrane [24]. Unlike typical autotransporters, the passenger domain in the GDSL autotransporter is not cleaved off and covalently attach to the respective C-terminal β-domain after translocation on the exterior membrane, which makes it an excellent vehicle to display various enzymes on the surface of E. coli used for whole-cell biocatalysis [25].
Herein, we firstly report the functional display of a phenolic acid decarboxylase (BLPAD) from Bacillus licheniformis (B. licheniformis) on the E. coli cell surface for efficient production of 4-VG and 4-VP. BLPAD displayed high organic solvent tolerance and outstanding catalytic activity in the bioconversion of FA and pCA into 4-VG and 4-VP, respectively [26]. The translocator domain in a GDSL autotransporter is employed as an anchoring motif to display BLPAD on the cell surface. Two signal peptides, including the native GDSL signal and pelB signal sequences, were evaluated for their effectives in guiding outer membrane translocation of the target protein on the E. coli cell surface. We demonstrated that BLPAD was targeted on the surface of E. coli by the autotransporter pathway in its active form, it was more efficiently guided by the pelB signal peptide than native signal peptide, and the cell surface displaying BLPAD was effective as a whole-cell catalyst in the bioproduction of 4-VG and 4-VP from FA and pCA, respectively.
2. Results and Discussion
2.1. Construction of the Vector System for Phenolic Acid Decarboxylase Autodisplay on Escherichia coli
Two plasmids, pET22b-pelblpad and pET22b-gdslblpad, containing BLPAD chimeras with a PelB signal peptide or a GDSL autotransporter signal peptide (SPGDSL) were constructed to display phenolic acid decarboxylase on the cell surface of E. coli. The wild GDSL autotransporter consists of signal peptide (SPGDSL) at the N-terminus followed by a passenger domain (esterase protein), the α-helical linker peptide (LK), and C-terminal β-barrel domain (Figure 1A). SPGDSL and the passenger domain were replaced with a PelB signal peptide sequence and BLPAD protein in the BLPAD chimera with a PelB signal peptide (Figure 1B), or, alternatively, only the passenger domain was replaced with BLPAD protein in the BLPAD chimera with a GDSL autotransporter signal peptide (Figure 1C). In this GDSL autotransporter-based cell surface display system, the translated fusion protein will be transported into the periplasmic space via Sec machinery guiding the signal peptide [27]. After the signal peptide is cleaved, the fusion protein can be translocated across the outer membrane with the help of the β-barrel domain. Finally, the N-terminal exogenous protein remains covalently attached to their respective C-terminal β-barrel domains on the cell surface [24]. Therefore, the functional displayed phenolic acid decarboxylase (BLPAD) from B. licheniformis on the E. coli cell surface was used as a whole-cell catalyst for the efficient production of 4-VG and 4-VP in this study (Figure 2).

Figure 1.
Schematic representation of the hybrid constructs for cell surface display of phenolic acid decarboxylase (BLPAD). (A) The wild GDSL autotransporter. (B) The BLPAD chimera with a PelB signal peptide sequence. (C) The BLPAD chimera with a GDSL autotransporter signal peptide sequence. SPGDSL: signal peptide of GDSL autotransporter; SPpeLB: PelB signal peptide; LK: the α-helical linker peptide; BLPAD: B. licheniformis phenolic acid decarboxylase; and β-Barrel: the translocator domain of GDSL autotransporter.
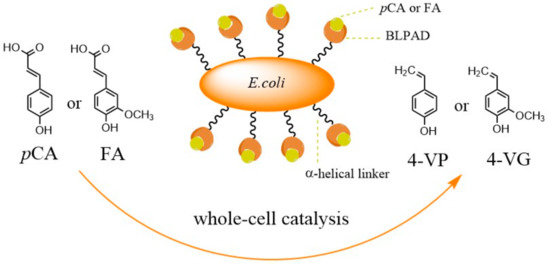
Figure 2.
Schematic diagram of the protein surface displayed on the cell and the catalysis of 4-hydroxycinnamic acid to vinyl phenol derivatives.
2.2. Expression and Localization of the Autodisplayed Phenolic Acid Decarboxylase on E. coli
The two constructed plasmids were transformed into E. coli strains BL21(DE3) and BL21(DE3) pLysS, respectively, and BLPAD was induced as described in the Materials and Methods section. To investigate localization of BLPAD in E. coli, whole-cell lysates and membrane fractions of the recombinant E. coli expressing BLPAD were analyzed by SDS-PAGE. After 4 h induction, the target protein of about 60 kDa was clearly presented in whole-cell lysates, the total membrane fraction, and the outer membrane fraction of induced cells. The molecular weight of the target protein was equal to the sum of three molecular weights of the BLPAD chimera combined with the C-terminal β-barrel domain of GDSL and linker peptide, indicating a successful expression of BLPAD as a fusion protein on the cell surface (Figure 3).

Figure 3.
Expression and membrane localization of the surface-displayed cells. (A,B) BL21(DE3) cells and BL21 (DE3) pLysS cells carrying a pET22b-pelblpad vector. (C,D) BL21(DE3) cells and BL21 (DE3) pLysS cells carrying a pET22b-gdsllpad vector, respectively. M: protein marker; Lane 1–6: whole-cell lysates of no induced cell, whole-cell lysates of induced cell, soluble fraction, total membrane fraction, inner membrane fraction, and outer membrane fraction.
To verify whether BLPAD was directed toward the external side or the periplasmic side of the outer membrane, whole cells of E. coli BL21(DE) or BL21(DE3) pLysS strains expressing BLPAD were pretreated by proteinase K. Then, the outer membrane fraction was isolated from the proteinase K treated cells. SDS-PAGE analysis showed that the corresponding protein band of BLPAD chimera on the outer membrane almost disappeared after 0.1 mg/mL proteinase K treatment (Figure 4).
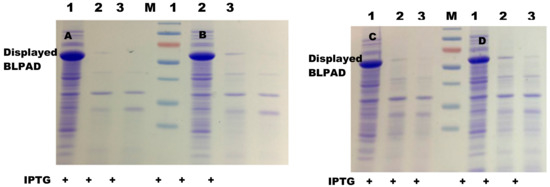
Figure 4.
Protease accessibility analyses of the surface-displayed cells. (A,B) BL21(DE3) cells and BL21 (DE3) pLysS cells carrying a pET22b-pelblpad vector. (C,D) BL21(DE3) cells and BL21 (DE3) pLysS cells carrying a pET22b-gdsllpad vector, respectively. M: protein marker, Lane 1–3: the outer membrane fractions from no proteinase K treated cells, with 0.05 mg/mL proteinase K treated cells, and with 0.1 mg/mL proteinase K treated cells, respectively. PrK: proteinase K; IPTG: isopropyl β-D-thiogalactoside.
As demonstrated previously, proteinase K is not able to pass the outer membrane and enter the periplasm [28]. These results clearly confirmed that the expressing BLPAD was exposed on the surface; therefore, BLPAD on the outer membrane degraded after treatment with proteinase K. After confirming successful display of BLPAD on the E. coli cell surface, whole-cell catalytic activities of four recombinant strains with or without proteinase K treatment were also assayed in this study. As shown in Figure 5, after induction, the activities of BL21(DE3) and BL21(DE3) pLysS strains carrying the pET22b-pelblpad vector reached 19.72 and 19.38 U/OD600, respectively, and the corresponding strains carrying the pET22b-gdslblpad vector reached 10.90 and 14.74 U/OD600, respectively. In contrast, the strains transformed with blank vector almost had no activity, and the activities of proteinase K treated strains were also tremendously reduced (Figure 5).
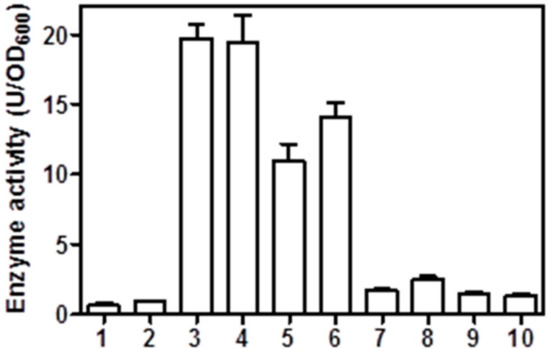
Figure 5.
Phenolic acid decarboxylase activity of the surface-displayed cells. 1 and 2: BL21(DE3) and BL21(DE3)pLysS3 cells carrying a blank pET-22b vector, respectively; 3 and 4: BL21(DE3) and BL21(DE3) pLysS cells carrying a pET22b-pelblpad vector, respectively; 5 and 6: BL21(DE3) and BL21(DE3) pLysS cells carrying a pET22b-gdslblpad vector; 7 and 8: proteinase K treated BL21(DE3) and BL21(DE3) pLysS cells carrying a pET22b-pelblpad vector, respectively; and 9 and 10: proteinase K treated BL21(DE3) and BL21(DE3) pLysS cells carrying a pET22b-gdslblpad vector, respectively. Catalytic activity was measured under standard assay conditions.
GDSL autotransporters have proved to be excellent vehicles for the E. coli surface display of various enzymes, particularly for members of the lipase and esterase families [27,29,30,31]. However, not all proteins can successfully be autodisplayed on the cell surface in active forms due to limitations of the passage domains, such as molecular weight and number of disulfide bonds [32,33]. The high catalytic activities of BLPAD autodisplayed E. coli cells demonstrated that the GDSL autotransporter system is applicable to display BLPAD with the β-barrel structural motif in an active form for application of whole-cell biocatalysts. The N-terminal signal peptide is needed by Sec machinery for translocation across the inner membrane [28]. Comparably, expressing cells carrying the pET22b-pelblpad vector with the pelB signal sequence displayed higher catalytic activity than the expressing cells carrying pET22b-gdslblpad vector with the native signal sequence, suggesting that the pelB signal sequence is more effective in guiding the transfer of BLPAD into the periplasmic space (Figure 5).
2.3. Bioconversion of Ferulic Acid by Whole-Cell Catalysis of Expressing E. coli
Four different BLPAD autodisplayed E. coli cells were compared to catalyze ferulic acid into 4-VG, in the monoaqueous phase or in the biphasic toluene/Na2HPO4–citric acid buffer system, in order to evaluate their bioconversion capability as a whole-cell biocatalyst. As shown in Figure 6, the molar conversion yields of the expressing BL21 (DE3) and BL21 (DE3) pLysS strains carrying a pET22b-pelblpad vector reached 62% or 40% in the monoaqueous phase, respectively, or almost 100% in the biphasic system after a 50 min reaction. Meanwhile, the corresponding values for expressing BL21 (DE3) and BL21 (DE3) pLysS strains carrying a pET22b-gdslblpad vector were 27% and 37% in the monoaqueous phase or 86% and 81% in the biphasic system, respectively. The higher conversion yields of the cells carrying a pET22b-pelblpad vector than the cells carrying pET22b-gdslblpad vector were attributed to higher catalytic activities of the whole cells carrying a pET22b-pelblpad vector as mentioned above (Figure 5). The lower yields presented in the monoaqueous system when compared to the biphasic system mainly were due to toxicity of product accumulated in the aqueous phase in the monoaqueous system [9,11]. In the biphasic system, the product could be extracted from the aqueous phase into the organic phase. Then, the residual concentration of product is lower in the aqueous phase of biphasic system compared to the aqueous phase of monophasic system. Biotransformation of hydroxycinnamic acid to 4-vinyl phenol derivatives was also feasibly affected by the toxicity of substrate, particularly as the substrate concentration increased [34]. The bioconversion efficiency of expressing BL21 (DE3) cells carrying a pET22b-pelblpad vector was further evaluated under a higher concentration of initial substrate to test the behavior of this system with different substrates. As shown in Table 1, the molar bioconversion yields of whole-cell catalysis for FA and pCA reached up to 72.6% and 80.4%, respectively, at the concentration of 300 mM in a biphasic toluene/Na2HPO4–citric acid buffer system. In contrast, the molar bioconversion yields of whole-cell catalysis for caffeic acid (CA) and sinapic acid (SA) were only 7.24% and 4.48%, respectively (Table 1). Whole-cell biocatalysis by using E. coli cells overexpressing the intracellular BLPAD protein or containing blank plasmid, as described previously [26], was also performed in the same reaction conditions. Molar bioconversion yields of E. coli cells overexpressing intracellular BLPAD reached 94.9% ± 4.8% and 99.8% ± 5.3% for FA and pCA at the concentration of 300 mM under same conditions, respectively. Additionally, the corresponding values for caffeic acid (CA) and sinapic acid (SA) were 25.8% ± 3.7% and 8.9% ± 2.5%, respectively. No product was detected when using E. coli cells containing a blank plasmid in the same reaction conditions. Comparatively, the bioconversion efficiencies of hydroxycinnamic acid in high concentration conditions were relatively lower than intracellular expressing BLPAD E. coli, but they were much higher than in previous literature using recombinant E. coli harboring intracellular PADs [8,9]. These results indicated that the displayed BLPAD had a high potential as a whole-cell biocatalyst for 4-VG and 4-VP bioproduction.
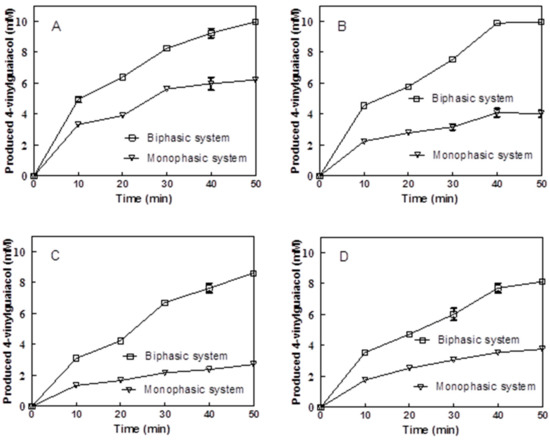
Figure 6.
Bioconversion of ferulic acid (FA) in monophasic and biphasic systems with four different surface-displayed cells. (A,B) BL21(DE3) cells and BL21 (DE3) pLysS cells carrying a pET22b-pelblpad vector, respectively. (C,D) BL21(DE3) cells and BL21 (DE3) pLysS cells carrying a pET22b-gdsllpad vector, respectively.

Table 1.
Bioconversion of 4-hydroxycinnamic acid by Escherichia coli BL21 (DE3) cells carrying a pET22b-pelblpad vector after one hour of incubation. pCA: p-coumaric acid; CA: caffeic acid; and SA: sinapic acid.
Recyclability of biocatalysts is often a desired feature for the economics of industrial biocatalysis, which may reduce the high costs of biocatalysts in general [34]. However, the toxicity effect of vinylphenol derivatives could cause serious damage to the intracellular decarboxylase-containing cells. Ben-Bassat et al. reported that after a single 4-VP production cycle, the intracellular decarboxylase-containing cells were not viable when streaked on agar plates, which limited cell recycling [9]. The autodisplayed BLPAD showed good recyclability in this study. As shown in Figure 7, the BLPAD displayed cells were successfully recycled for 7 cycles with a residual activity of 63% (The activity at the first run was taken as 100%). In contrast, the recyclability of the intracellular expressing cells was rather low, and their residual activity reduced to approximately 11% after three reuse cycles. This result indicated that BLPAD anchored on the cell surface appeared to be more stable compared to free intracellular enzyme.
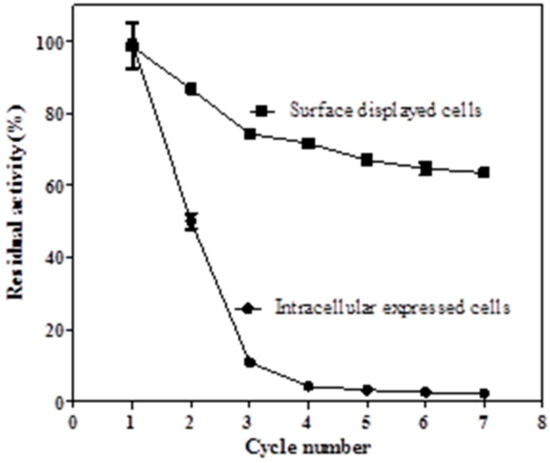
Figure 7.
Reusability of the surface-displayed and intracellular expressed cells.
3. Materials and Methods
3.1. Bacterial Strains, Vectors, and Chemicals
E. coli strains BL21 (DE3) and BL21 (DE3) pLysS (TransGene, Beijing, China) were used for expression and whole cell activity assays of the surface-displayed BLPAD. pET-22b (Novagen, Copenhagen, Denmark) was used as an expression vector. pET28b-blpad and pET-29a-GDSL, harboring the genes of BLPAD and GDSL autotransporter, respectively, were used as templates for amplifying the gene fragments of BLPAD and the translocator domain, respectively [25,26]. Lysozyme and proteinase K were obtained from Sangon Biotech (Shanghai, China). FA, pCA, CA, and SA were purchased from Sinopharm Group (Beijing, China). 4-VG and 4-VP were obtained from Sigma-Aldrich (St. Louis, MO, USA). FastPfu DNA polymerase and restriction enzymes were all obtained from Takara (Dalian, China).
3.2. Construction of Plasmids for Cell Surface Display of Phenolic Acid Decarboxylase (BLPAD)
A schematic representation of the constructs for cell surface display of BLPAD is shown in Figure 1. The translocator domain in the GDSL autotransporter (Genbank No. JQ855507) from Pseudomonas putida was used as an anchoring motif to display BLPAD on the cell surface. The gene of GDSL autotransporter encodes a preprotein of 627 amino acids, which comprises a 24 amino acid N-terminal signal peptide, a 283 amino acid GDSL esterase passenger domain (P25-Q307), a 51 amino acid linker region, and a 272 amino acids C-terminal located autotransporter β-barrel domain. Two chimeras with different signal sequences were constructed as follows. For translocation via the signal sequence of pelB, the mature sequence of BLPAD was PCR amplified using pET28b-blpad as the template and blpad-EF/blpad-ER as primers (Table 2). PCR was performed under the following conditions: one cycle at 94 °C for 5 min, 55 °C for 30 s, and 72 °C for 6 min; 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 6 min; and a final extension at 72 °C for 10 min. The restrictive sites of BamH1 and NheI were introduced into the PCR fragment at 5′ and 3′ ends, respectively. The fragment encoding for the translocator domain of the GDSL autotransporter was also PCR amplified using pET-29a-GDSL as the template and EstA-EF/EstA-ER as primers (Table 2). The restrictive sites NheI and XhoI were introduced into the PCR fragment at 5′ and 3′ ends, respectively. These two resulting PCR fragments were digested with BamH1/NheI and NheI/XhoI, respectively, and ligated to pET-22b to give pET22b-pelblpad. For translocation via the signal sequence of the GDSL autotransporter, the mature sequence of BLPAD and the signal sequence of the GDSL autotransporter at the N-terminus were amplified by primer extension PCR using pET28b-blpad as the template and blpad-F1 to F4/blpad-ER as primers (Table 2). The restrictive sites of Nde I and NheI were introduced into the PCR fragment at 5′ and 3′ ends, respectively. This PCR fragment was digested with Nde1/NheI and ligated, similar to digested pET22b-pelblpad, to give pET22b-gdslblpad.

Table 2.
Oligonucleotide primers used in this study.
3.3. Expression and Membrane Fraction Isolation
Constructed plasmids pET22b-pelblpad and pET22b-gdslblpad were transformed into E. coli BL21(DE3) and E. coli BL21(DE3) pLysS strains, respectively. The recombinant strains were cultured in 50 mL of Luria–Bertani (LB) medium supplemented with ampicillin (100 μg/mL) at 37 °C with shaking at 180 rpm. After OD600 reached 0.6, the temperature was reduced to 25 °C, and the expression of recombinant BLPAD was induced with 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 5 h.
Membrane fraction isolation was performed as previously described [27]. After induction, the expressed cells were harvested by centrifugation (6000× g, 30 min, 4 °C). The pellets were resuspended in 5 mL lysis buffer (50 mM Tris–HCl, 5mM EDTA, 20% sucrose, and 0.2 mg/mL lysozyme, pH 8.0) and were disrupted by sonication in an ice bath. The cell lysate was centrifuged at 6000× g for 30 min at 4 °C. The resulting supernatant was ultracentrifuged at 100,000× g at 4 °C for 1 h. The supernatant after ultracentrifugation was kept as a soluble fraction for future analysis. The pellet (total membrane fraction) was resuspended and incubated in 5 mL solution containing 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, and 1% TritonX-100 at room temperature for 20 min. The suspension was ultracentrifuged again as above. The resulting pellet was resuspended in 50 mM Tris–HCl (pH 8.0) as the outer membrane fraction. The supernatant was kept as an inner membrane fraction for future analysis. Protein concentrations were determined using a BCA (Bicinchoninic acid) protein assay kit (Sangon, Shanghai, China) and BSA (Bovine Serum Albumin) as a standard. Equivalent amounts of the protein fractions were analyzed by protein electrophoresis using 12% separating gel.
3.4. Protease Accessibility
The expressed cells were washed twice using phosphate buffered saline (PBS) and resuspended in 50 mM Tris–HCl buffer (pH 8.0) containing 5 mM CaCl2 and 0.1 mg/mL proteinase K. The cell suspension was incubated at 37 °C and 150 rpm for 30 min. The reaction was stopped by addition of PMSF (Phenylmethanesulfonyl fluoride) solution (up to 5 mM). The cells were pelleted by centrifugation and washed with PBS again. The outer membrane fraction was isolated from the obtained pellet as described above and analyzed by SDS-PAGE.
3.5. Bioconversion of Ferulic Acid by Whole-Cell Catalysis of Expressing E. coli
Comparisons of bioconversion capabilities of four different BLPAD autodisplayed E. coli cells were performed in a 2 mL glass tube with Teflon seals at 37 °C and 400 rpm shaking for set times. The mixture for the monoaqueous phase consisted of 0.9 mL of 200 mM Na2HPO4–citric acid buffer (pH 6.0) containing 10 mM FA and 0.1 mL of BLPAD autodisplayed cells (OD600 = 1). For a biphasic reaction system, an equal volume of toluene was added into above mixture. Triplicate samples were withdrawn at intervals (10 min), and the reaction was terminated by adding 50 μL of 50% trichloroacetic acid. Bioconversion of different substrates with higher initial concentrations by BLPAD autodisplayed E. coli cells carrying a pET22b-pelblpad vector was carried out in a biphasic reaction system, which consisted of 10 mL of toluene and 10 mL of Na2HPO4–citric acid buffer containing different initial concentrations of substrates and E. coli cells (2 OD600 in final), at 200 rpm and 37 °C for 1 h [35]. Whole-cell catalysis by using E. coli cells overexpressing intracellular BLPAD protein or containing a blank plasmid as described previously [26] was also performed in the same reaction conditions. The substrate and product formed in aqueous and organic phases were quantified by high-performance liquid chromatography (HPLC) as previously described [26].
Reusability of the surface-displayed BLPAD was determined by measuring the activity of recombinant cells at a concentration of 50 mM FA in a monophasic system as described above. Recombinant E. coli cells with autodisplayed BLPAD were induced as above. Induction of recombinant E. coli cells expressing intracellular BLPAD was performed as previously described [26]. After induction, cells from the 50 mL culture were harvested by centrifugation at 6500 rpm and 4 °C for 10 min, washed twice with Na2HPO4–citric acid (200 mM, pH 6.0) buffer, and were resuspended in the same buffer. The reaction was carried out in 15 mL of reaction mixture at 200 rpm and 37 °C for 15 min. After each reaction, the cells were collected by centrifugation and washed with the same buffer twice. Then, the buffer containing 50 mL of FA was added to start a new batch of catalysis. The generated product was quantified by HPLC as previously described [26]. The residual activity was calculated by defining the activity of the first reaction as 100%.
3.6. Phenolic Acid Decarboxylase Activity Assay
The whole cell activity of BLPAD autodisplayed E. coli was assayed in the mixture containing 0.8 mL of 200 mM Na2HPO4–citric acid buffer (pH 6.0), 0.1 mL of 50 mM FA, and 0.1mL of diluted cell suspension in a 10 mL glass tube with Teflon seals at 37 °C for 10 min. The BLPAD autodisplayed E. coli BL21 (DE3) cells were cultivated and induced as described above. One milliliter of methanol was added into the mixture to terminate the reaction. The generated product was quantified by HPLC as previously described [26]. One unit (U) of BLPAD activity was defined as the amount of enzyme that generated 1 μmol 4-VG per minute under the conditions described above. The value was presented as unit per milliliter of cells with one OD600 density (U/OD600) in order to compare the activity of different surface-displayed cells on the basis of the same number of cells used.
4. Conclusions
In this study, a phenolic acid decarboxylase from B. licheniformis was functionally autodisplayed on the E. coli cell surface using a GDSL autotransporter. Expression and localization of BLAPD on the cell surface of E. coli was verified by SDS-PAGE and protease accessibility. The pelB signal peptide is more effective in guiding the translocation of BLAPD on the cell surface than the native signal peptide of GDSL. Molar bioconversion yields of 72.6% and 80.4% were achieved at a concentration of 300 mM of FA and pCA in a biphasic system, respectively. Furthermore, the cell surface displayed BLPAD showed good reusability and retained 63% of residual activity after 7 cycles of repeated use, indicating that the cell surface displayed BLPAD can be used as a whole-cell biocatalyst for efficient production of 4-VG and 4-VP.
Author Contributions
L.L. (Lulu Li) contributed to the study design, performed the bulk of the experimental work, data analysis and interpretation, and drafted the manuscript. X.W. contributed to vector construction and characterization of the activity assay. L.L. (Liangkun Long) and S.D. contributed to the study design, data analysis and interpretation, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Forestry Science and Technology Promotion Project [(2016)41], the Key Research and Development Program of Jiangsu (BE2015759).
Acknowledgments
The Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) supported this study.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
FA: ferulic acid; 4-VG: 4-vinyl guaiacol; pCA: p-coumaric acid; CA: caffeic acid; SA: sinapic acid; BLPAD: Bacillus licheniformis phenolic acid decarboxylases; OD600: optical density at 600 nm; PBS: phosphate buffered saline; PCR: polymerase chain reaction; IPTG: isopropyl-β-D-thiogalactopyranoside; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; LB medium: Luria-Bertani medium; HPLC: high-performance liquid chromatography.
References
- Mishra, S.; Sachan, A.; Vidyarthi, A.S.; Sachan, S.G. Transformation of ferulic acid to 4-vinyl guaiacol as a major metabolite: A microbial approach. Rev. Environ. Sci. Biotechnol. 2014, 13, 377–385. [Google Scholar] [CrossRef]
- Tinikul, R.; Chenprakhon, P.; Maenpuen, S.; Chaiyen, P. Biotransformation of plant-derived phenolic acids. Biotechnol. J. 2018, 13, 1700632. [Google Scholar] [CrossRef] [PubMed]
- Kodaira, K.; Onishi, Y.; Ito, K. An oligomerization of 2-methoxy- 4-vinylphenol. Makromol. Chem. Rapid. Commun. 1980, 1, 427–431. [Google Scholar] [CrossRef]
- Takeshima, H.; Satoh, K.; Karnigaito, M. Bio-based functional styrene monomers derived from naturally occurring ferulic acid for poly (vinylcatechol) and poly (vinylguaiacol) via controlled radical polymerization. Macromolecules 2017, 50, 4206–4216. [Google Scholar] [CrossRef]
- Fufuya, T.; Miura, M.; Kino, K. A coenzyme-independent decarboxylase/oxygenase cascade for the efficient synthesis of vanillin. ChemBioChem 2014, 15, 2248. [Google Scholar]
- Tang, J.; Shi, L.; Li, L.; Long, L.; Ding, S. Expression and characterization of a 9-cis-epoxycarotenoid dioxygenase from Serratia sp. ATCC 39006 capable of biotransforming isoeugenol and 4-vinylguaiacol to vanillin. Biotechnol. Rep. 2018, 18, e00253. [Google Scholar] [CrossRef]
- Ni, J.; Wu, Y.T.; Tao, F.; Peng, Y.; Xu, P. A coenzyme-free biocatalyst for the value-added utilization of lignin-derived aromatics. J. Am. Chem. Soc. 2018, 140, 16001–16005. [Google Scholar] [CrossRef]
- Yang, J.; Wang, S.; Lorrain, M.J.; Rho, D.; Abokitse, K.; Lau, P.C. Bioproduction of lauryl lactone and 4-vinyl guaiacol as value added chemicals in two-phase biotransformation systems. Appl. Microbiol. Biotechnol. 2009, 84, 867–876. [Google Scholar] [CrossRef]
- Ben-Bassat, A.; Breinig, S.; Crum, G.A.; Huang, L.; Altenbaugh, A.B.; Rizzo, N.; Trotman, RJ.; Vannelli, T.; Sariaslani, F.S.; Haynie, S.L. Preparation of 4-vinylphenol using pHCA decarboxylase in a two solvent medium. Org. Process. Res. Dev. 2007, 11, 278–285. [Google Scholar] [CrossRef]
- Salgado, J.M.; Rodriguez-Solana, R.; Curiel, J.A.; de las Rivas, B.; Munoz, R.; Dominguez, J.M. Production of vinyl derivatives from alkaline hydrolysates of corn cobs by recombinant Escherichia coli containing the phenolic acid decarboxylase from Lactobacillus plantarum CECT 748T. Bioresour. Technol. 2012, 117, 274–285. [Google Scholar] [CrossRef]
- Jung, D.H.; Choi, W.; Choi, K.Y.; Jung, E.; Yun, H.; Kazlauskas, R.J.; Kim, B.G. Bioconversion of p-coumaric acid to p-hydroxystyrene using phenolic acid decarboxylase from B. amyloliquefaciens in biphasic reaction system. Appl. Microbiol. Biotechnol. 2013, 97, 1501–1511. [Google Scholar] [CrossRef] [PubMed]
- Licandro-Seraut, H.; Roussel, C.; Perpetuini, G.; Gervais, P.; Cavin, J.F. Sensitivity to vinyl phenol derivatives produced by phenolic acid decarboxylase activity in Escherichia coli and several food-borne Gram-negative species. Appl. Microbiol. Biotechnol. 2013, 97, 7853–7864. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Chen, R.R. Accelerating whole-cell biocatalysis by reducing outer membrane permeability barrier. Biotechnol. Bioeng. 2004, 87, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Biocatalysis in organic chemistry and biotechnology: Past, present, and future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Khera, E.; Wen, F. Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res. 2015, 54, 4021–4032. [Google Scholar] [CrossRef] [PubMed]
- Schuurmann, J.; Quehl, P.; Festel, G.; Jose, J. Bacterial whole-cell biocatalysts by surface display of enzymes: Toward industrial application. Appl. Microbiol. Biotechnol. 2014, 98, 8031–8046. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kondo, A. Cell surface engineering of industrial microorganisms for biorefining applications. Biotechnol. Adv. 2015, 33, 403–1411. [Google Scholar] [CrossRef] [PubMed]
- He, W.W.; Jiang, B.; Mu, W.M.; Zhang, T. Production of D-Allulose with D-Psicose 3-epimerase expressed and displayed on the surface of Bacillus subtilis spores. J. Agr. Food. Chem. 2016, 64, 7201–7207. [Google Scholar] [CrossRef]
- Tozakidis, I.E.P.; Brossette, T.; Lenz, F.; Maas, R.M.; Jose, J. Proof of concept for the simplified breakdown of cellulose by combining Pseudomonas putida strains with surface displayed thermophilic endocellulase, exocellulase and beta-glucosidase. Microb. Cell Fact. 2016, 15, 103. [Google Scholar] [CrossRef]
- Park, D.M.; Brewer, A.; Reed, D.W.; Lammers, L.N.; Jiao, Y.Q. Recovery of rare earth elements from low-grade feedstock leachates using engineered bacteria. Environ. Sci. Technol. 2017, 51, 13471–13480. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, J.; Fan, J.; Wang, Z.Y.; Li, L. Detection of catechol using an electrochemical biosensor based on engineered Escherichia coli cells that surface-display laccase. Anal. Chim. Acta. 2018, 1009, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Maruthamuthu, M.K.; Hong, J.; Arulsamy, K.; Somasundaram, S.; Hong, S.; Choe, W.S.; Yoo, I.K. Development of bisphenol A-removing recombinant Escherichia coli by monomeric and dimeric surface display of bisphenol A-binding peptide. Bioproc. Biosyst. Eng. 2018, 41, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, H.; Ding, N.; Ohara, Y.; Hori, K. Immobilization of Enterobacter aerogenes by a trimeric autotransporter adhesin, AtaA, and its application to biohydrogen production. Catalysts 2018, 8, 159. [Google Scholar] [CrossRef]
- Wilhelm, S.; Rosenau, F.; Kolmar, H.; Jaeger, K.E. Autotransporters with GDSL passenger domains: Molecular physiology and biotechnological applications. ChemBioChem 2011, 12, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, T.; Vanderleyden, J.; Spaepen, S. Autotransporter-based cell surface display in Gram-negative bacteria. Crit. Rev. Microbiol. 2015, 41, 109–123. [Google Scholar] [CrossRef]
- Hu, H.F.; Li, L.L.; Ding, S.J. An organic solvent-tolerant phenolic acid decarboxylase from Bacillus licheniformis for the efficient bioconversion of hydroxycinnamic acids to vinyl phenol derivatives. Appl. Microbiol. Biotechnol. 2015, 99, 5071–5081. [Google Scholar] [CrossRef] [PubMed]
- Petrovskaya, L.E.; Novototskaya-Vlasova, K.A.; Kryukova, E.A.; Rivkina, E.M.; Dolgikh, D.A.; Kirpichnikov, M.P. Cell surface display of cold-active esterase EstPc with the use of a new autotransporter from Psychrobacter cryohalolentis K5T. Extremophiles 2015, 19, 161–170. [Google Scholar] [CrossRef]
- Natale, P.; Bruser, T.; Driessen, A.J.M. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane: Distinct translocases and mechanisms. BBA-Rev. Biomembr. 2008, 1778, 1735–1756. [Google Scholar] [CrossRef]
- Schultheiss, E.; Paar, C.; Schwab, H.; Jose, J. Functional esterase surface display by the autotransporter pathway in Escherichia coli. J. Mol. Catal. B-Enzym. 2002, 18, 89–97. [Google Scholar] [CrossRef]
- Kim, S.J.; Song, J.K.; Kim, H.K. Cell surface display of Staphylococcus haemolyticus L62 lipase in Escherichia coli and its application as a whole cell biocatalyst for biodiesel production. J. Mol. Catal. B-Enzym. 2013, 97, 54–61. [Google Scholar] [CrossRef]
- Kranen, E.; Detzel, C.; Weber, T.; Jose, J. Autodisplay for the co-expression of lipase and foldase on the surface of E coli: Washing with designer bugs. Microb. Cell Fact. 2014, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Oliver, D.C.; Huang, G.; Fernandez, R.C. Identification of secretion determinants of the Bordetella pertussis BrkA autotransporter. J. Bacteriol. 2003, 185, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Nicolay, T.; Lemoine, L.; Lievens, E.; Balzarini, S.; Vanderleyden, J.; Spaepen, S. Probing the applicability of autotransporter based surface display with the EstA autotransporter of Pseudomonas stutzeri A15. Microb. Cell Fact. 2012, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Kaleem, I.; He, D.M.; Liu, G.Y.; Li, C. Efficient production of glycyrrhetic acid 3-O-mono-β-D-glucuronide by whole-cell biocatalysis in an ionic liquid/buffer biphasic system. Process. Biochem. 2012, 47, 908–913. [Google Scholar] [CrossRef]
- Chen, Y.H.; Li, L.L.; Long, L.K.; Ding, S.J. High cell-density cultivation of phenolic acid decarboxylase-expressing Escherichia coli and 4-vinylguaiacol bioproduction from ferulic acid by whole-cell catalysis. J. Chem. Technol. Biot. 2018, 93, 2415–2421. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).