Light-Driven Biocatalysis in Liposomes and Polymersomes: Where Are We Now?
Abstract
1. Introduction
2. Comparison of Liposomes and Polymersomes as Artificial Compartments
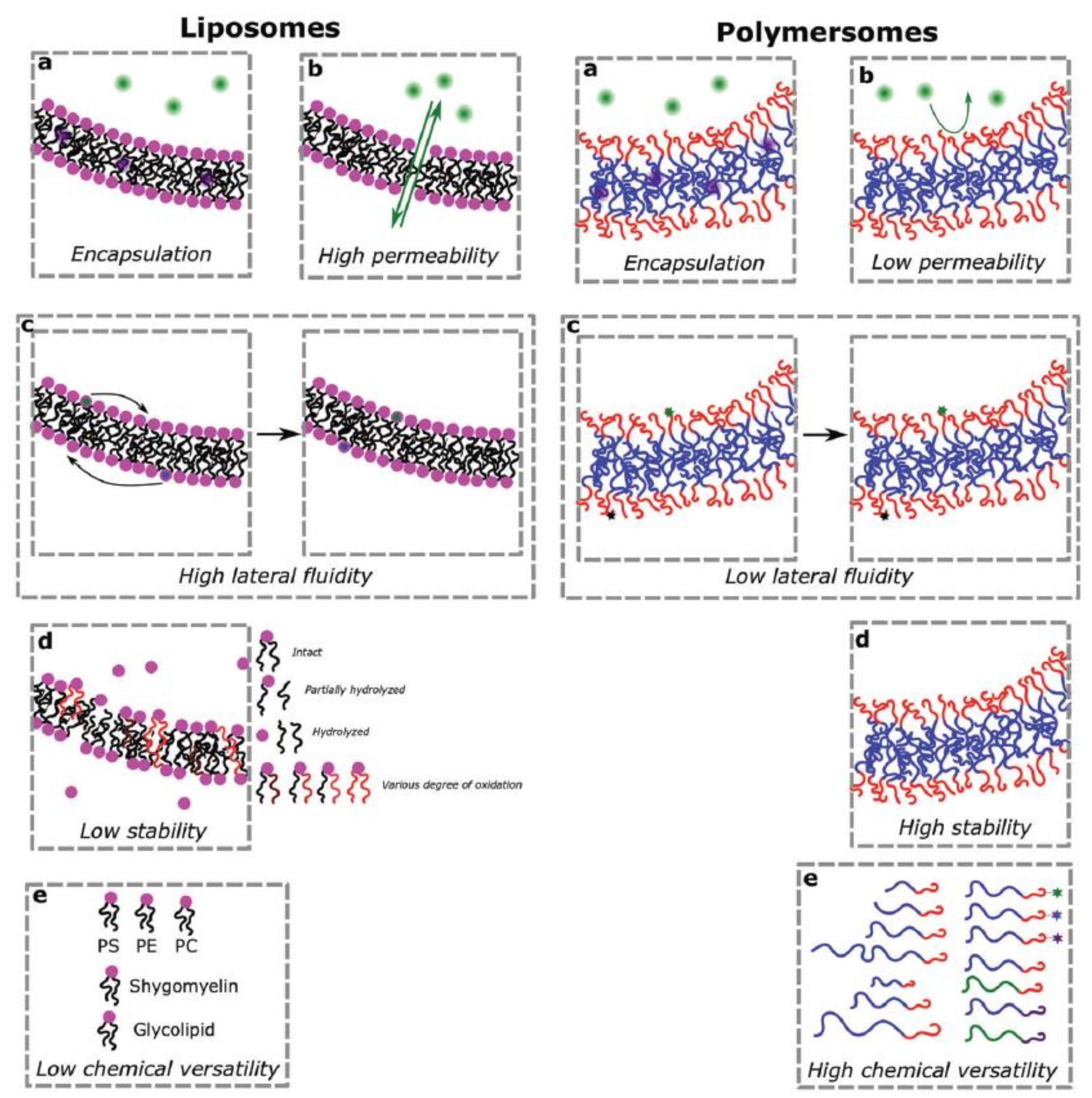
2.1. General Properties of Liposomes
2.2. Major Fields of Application of Liposomes
2.3. Functionalization of Liposomes for Light-Driven Biocatalysis
2.4. General Properties of Polymersomes
2.5. Major Fields of Application of Polymersomes
2.6. Advantages and Challenges of Using Polymersomes
2.7. Functionalization of Polymersomes for Light-Driven Biocatalysis
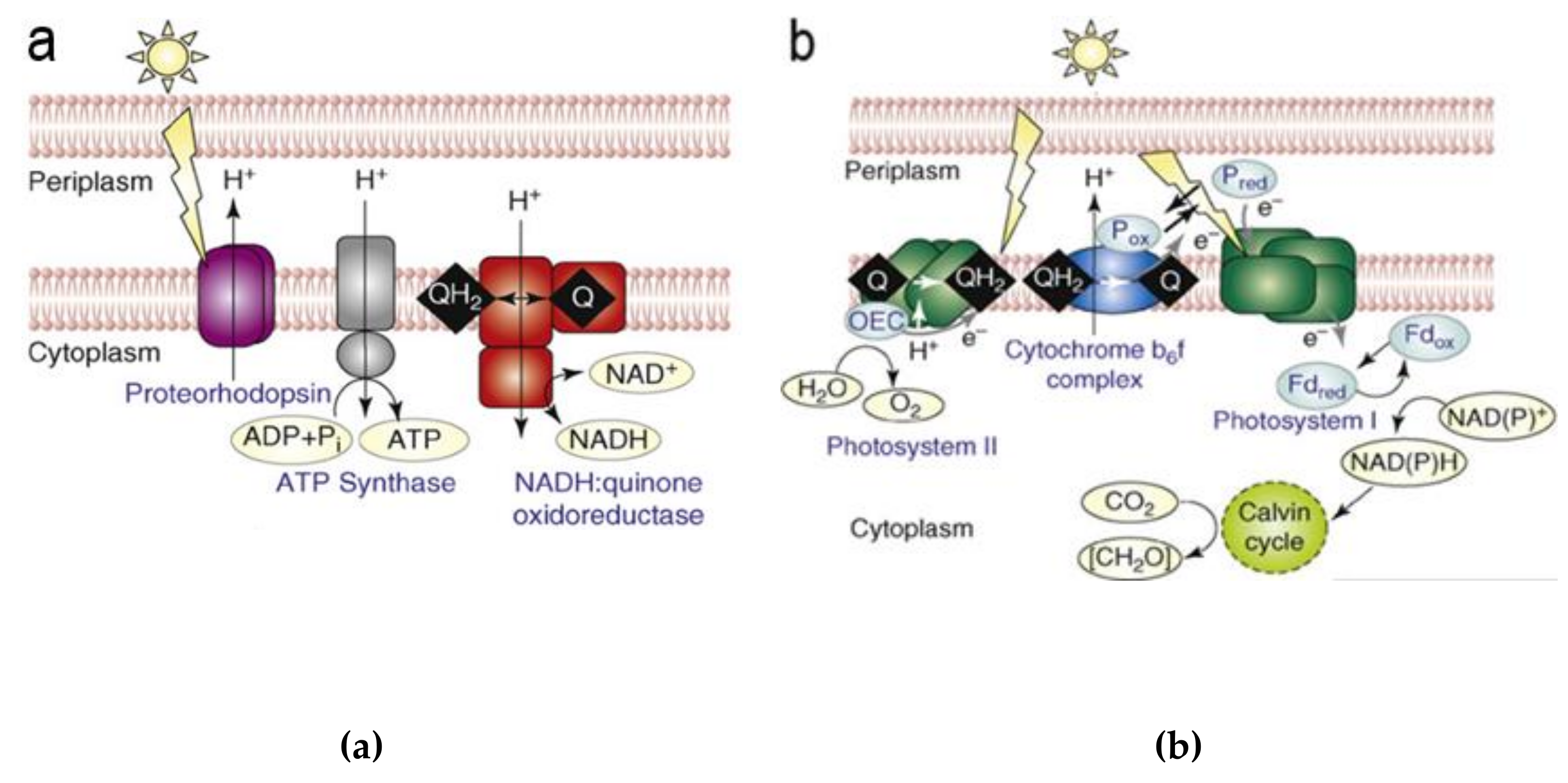
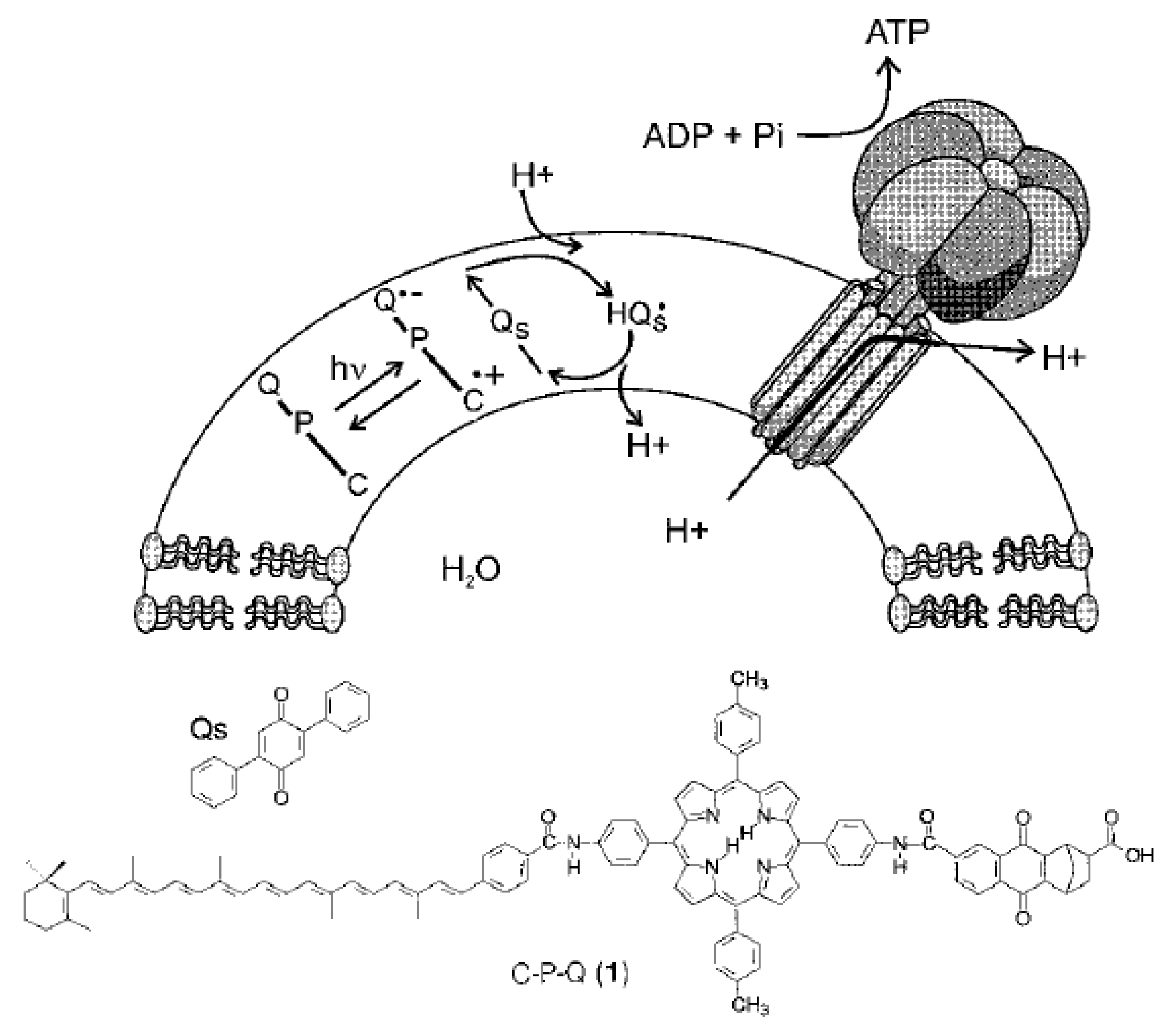
3. (Artificial) Photosynthetic Systems for Light-Driven Proton Translocation
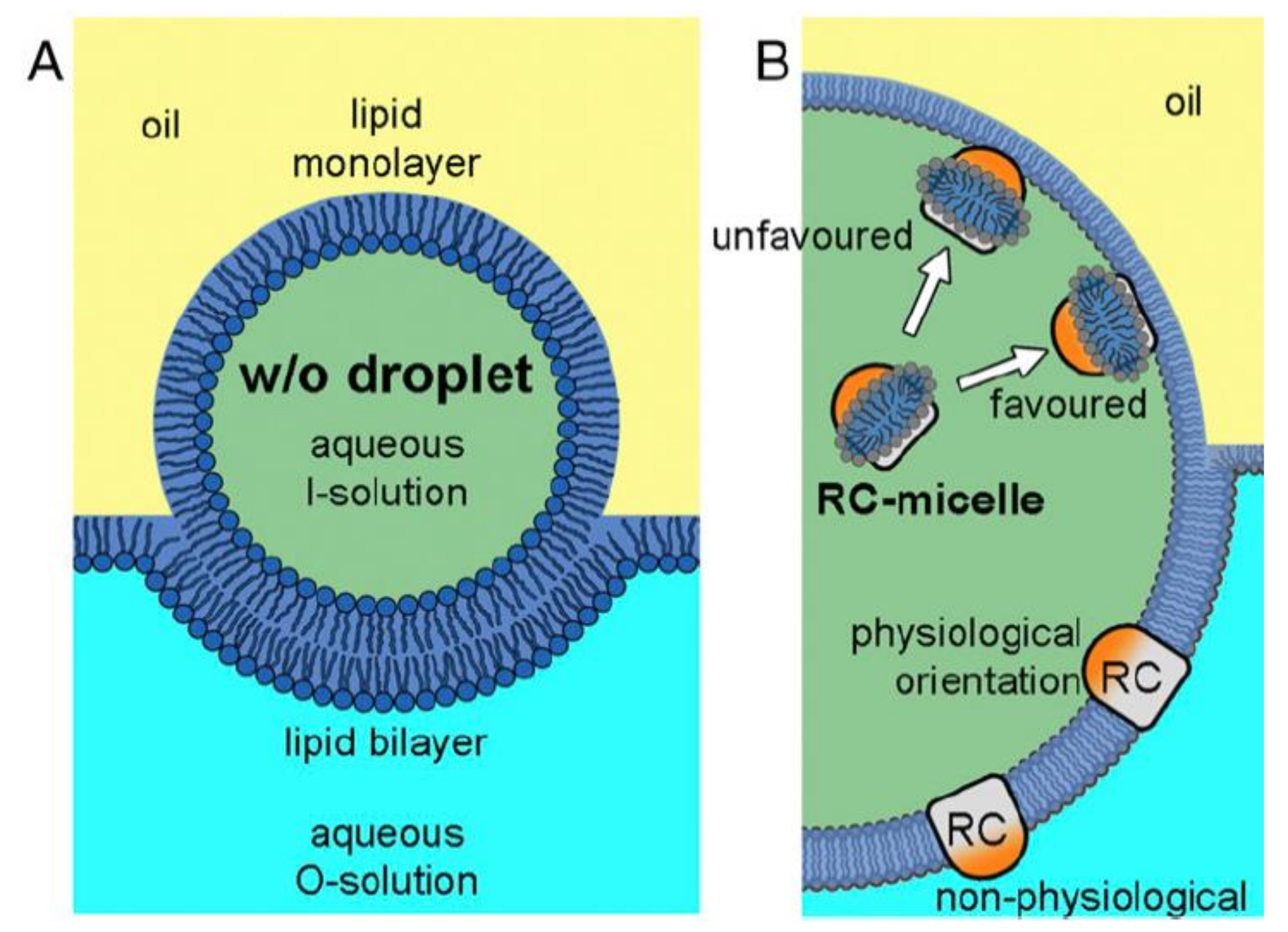
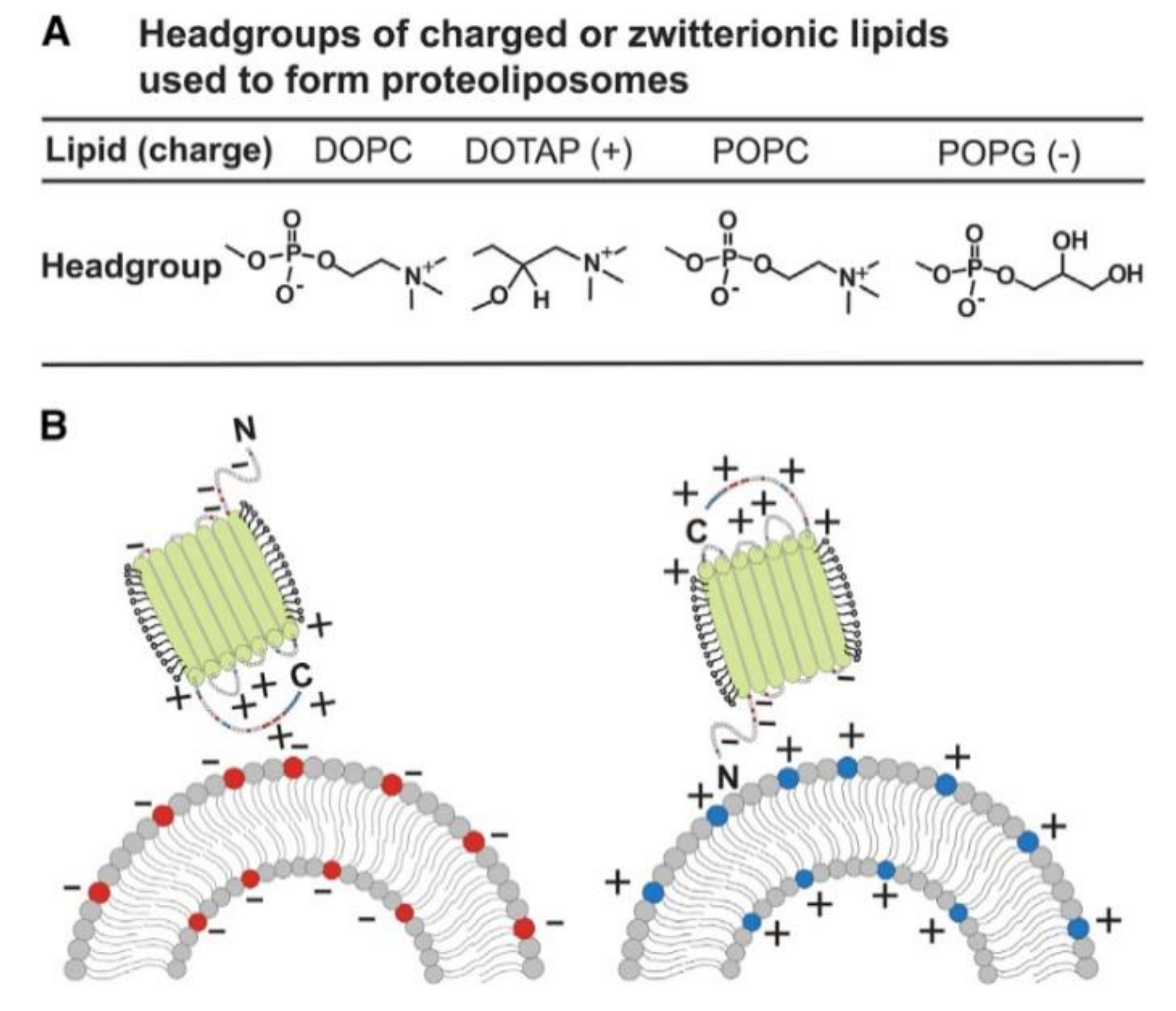
4. Insertion Orientation and Efficiency
5. Light-Driven Biocatalysis in Artificial Compartments
6. Why Is Light-Driven Biocatalysis Interesting?
7. Conclusion and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schwander, T.; Schada von Borzyskowski, L.; Burgener, S.; Cortina, N.S.; Erb, T.J. A synthetic pathway for the fixation of carbon dioxide in vitro. Science 2016, 354, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Rideau, E.; Dimova, R.; Schwille, P.; Wurm, F.R.; Landfester, K. Liposomes and polymersomes: A comparative review towards cell mimicking. Chem. Soc. Rev. 2018, 47, 8572–8610. [Google Scholar] [CrossRef] [PubMed]
- Tanner, P.; Baumann, P.; Enea, R.; Onaca, O.; Palivan, C.; Meier, W. Polymeric Vesicles: From Drug Carriers to Nanoreactors and Artificial Organelles. Acc. Chem. Res. 2011, 44, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Bangham, A.D.; Horne, R.W. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef]
- Alavi, M.; Karimi, N.; Safaei, M. Application of Various Types of Liposomes in Drug Delivery Systems. Adv. Pharm. Bull. 2017, 7, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Rahimpour, Y.; Hamishehkar, H. Liposomes in cosmeceutics. Expert Opin. Drug Delivery 2012, 9, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Azadmard-Damirchi, S.; Peighambardoust, S.H.; Valizadeh, H.; Hesari, J. Liposomes as carrier vehicles for functional compounds in food sector. J. Exp. Nanosci. 2016, 11, 737–759. [Google Scholar] [CrossRef]
- Jesorka, A.; Orwar, O. Liposomes: Technologies and Analytical Applications. Annu. Rev. Anal. Chem. 2008, 1, 801–832. [Google Scholar] [CrossRef]
- Lu, K.-Y.; Tao, S.-C.; Yang, T.-C.; Ho, Y.-H.; Lee, C.-H.; Lin, C.-C.; Juan, H.-F.; Huang, H.-C.; Yang, C.-Y.; Chen, M.-S.; et al. Profiling lipid-protein interactions using nonquenched fluorescent liposomal nanovesicles and proteome microarrays. Mol. Cell. Proteomics 2012, 11, 1177–1190. [Google Scholar] [CrossRef]
- Zhu, H.; Bilgin, M.; Bangham, R.; Hall, D.; Casamayor, A.; Bertone, P.; Lan, N.; Jansen, R.; Bidlingmaier, S.; Houfek, T.; et al. Global Analysis of Protein Activities Using Proteome Chips. Science 2001, 293, 2101. [Google Scholar] [CrossRef]
- Jørgensen, I.L.; Kemmer, G.C.; Pomorski, T.G. Membrane protein reconstitution into giant unilamellar vesicles: A review on current techniques. Eur. Biophys. J. 2017, 46, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Racker, E.; Stoeckenius, W. Reconstitution of Purple Membrane Vesicles Catalyzing Light-driven Proton Uptake and Adenosine Triphosphate Formation. J. Biol. Chem. 1974, 249, 662–663. [Google Scholar] [PubMed]
- Richard, P.; Rigaud, J.-L.; GrÄBer, P. Reconstitution of CF0F1 into liposomes using a new reconstitution procedure. Eur. J. Biochem. 1990, 193, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Pitard, B.; Richard, P.; Duñach, M.; Rigaud, J.-L. ATP Synthesis by the F0F1 ATP Synthase from Thermophilic Bacillus PS3 Reconstituted into Liposomes with Bacteriorhodopsin. Eur. J. Biochem. 1996, 235, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Steinberg-Yfrach, G.; Rigaud, J.-L.; Durantini, E.N.; Moore, A.L.; Gust, D.; Moore, T.A. Light-driven production of ATP catalysed by F0F1-ATP synthase in an artificial photosynthetic membrane. Nature 1998, 392, 479. [Google Scholar] [CrossRef] [PubMed]
- Girard, P.; Pécréaux, J.; Lenoir, G.; Falson, P.; Rigaud, J.-L.; Bassereau, P. A New Method for the Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles. Biophys. J. 2004, 87, 419–429. [Google Scholar] [CrossRef]
- Milano, F.; Trotta, M.; Dorogi, M.; Fischer, B.; Giotta, L.; Agostiano, A.; Maróti, P.; Kálmán, L.; Nagy, L. Light induced transmembrane proton gradient in artificial lipid vesicles reconstituted with photosynthetic reaction centers. J. Bioenerg. Biomembr. 2012, 44, 373–384. [Google Scholar] [CrossRef]
- Dezi, M.; Di Cicco, A.; Bassereau, P.; Lévy, D. Detergent-mediated incorporation of transmembrane proteins in giant unilamellar vesicles with controlled physiological contents. Proc. Natl. Acad. Sci. USA 2013, 110, 7276–7281. [Google Scholar] [CrossRef]
- Feng, X.; Jia, Y.; Cai, P.; Fei, J.; Li, J. Coassembly of Photosystem II and ATPase as Artificial Chloroplast for Light-Driven ATP Synthesis. ACS Nano 2016, 10, 556–561. [Google Scholar] [CrossRef]
- Lee, K.Y.; Park, S.J.; Lee, K.A.; Kim, S.H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.H.; Ahn, T.K.; Parker, K.K.; et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef]
- Peyret, A.; Ibarboure, E.; Le Meins, J.-F.; Lecommandoux, S. Asymmetric Hybrid Polymer–Lipid Giant Vesicles as Cell Membrane Mimics. Adv. Sci. 2017, 5, 1700453. [Google Scholar] [CrossRef] [PubMed]
- Devaux, P.F. Static and dynamic lipid asymmetry in cell membranes. Biochemistry 1991, 30, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- S, B.M. Phospholipids: More about Less. Nature 1972, 236, 11. [Google Scholar] [CrossRef]
- Heijne, G.; Gavel, Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 1988, 174, 671–678. [Google Scholar] [CrossRef]
- Lin, Q.; London, E. The Influence of Natural Lipid Asymmetry upon the Conformation of a Membrane-inserted Protein (Perfringolysin O). J. Biol. Chem. 2014, 289, 5467–5478. [Google Scholar] [CrossRef] [PubMed]
- LoPresti, C.; Lomas, H.; Massignani, M.; Smart, T.; Battaglia, G. Polymersomes: nature inspired nanometer sized compartments. J. Mater. Chem. 2009, 19, 3576–3590. [Google Scholar] [CrossRef]
- Winzen, S.; Bernhardt, M.; Schaeffel, D.; Koch, A.; Kappl, M.; Koynov, K.; Landfester, K.; Kroeger, A. Submicron hybrid vesicles consisting of polymer–lipid and polymer–cholesterol blends. Soft Matter 2013, 9, 5883–5890. [Google Scholar] [CrossRef]
- Discher, B.M.; Won, Y.-Y.; Ege, D.S.; Lee, J.C.M.; Bates, F.S.; Discher, D.E.; Hammer, D.A. Polymersomes: Tough Vesicles Made from Diblock Copolymers. Science 1999, 284, 1143. [Google Scholar] [CrossRef]
- Palivan, C.G.; Goers, R.; Najer, A.; Zhang, X.Y.; Car, A.; Meier, W. Bioinspired polymer vesicles and membranes for biological and medical applications. Chem. Soc. Rev. 2016, 45, 377–411. [Google Scholar] [CrossRef]
- Baumann, P.; Tanner, P.; Onaca, O.; Palivan, C.G. Bio-Decorated Polymer Membranes: A New Approach in Diagnostics and Therapeutics. Polymers 2011, 3, 173–192. [Google Scholar] [CrossRef]
- Elani, Y.; Law, R.V.; Ces, O. Vesicle-based artificial cells as chemical microreactors with spatially segregated reaction pathways. Nat. Commun. 2014, 5, 5305. [Google Scholar] [CrossRef] [PubMed]
- Schoonen, L.; van Hest, J.C.M. Compartmentalization Approaches in Soft Matter Science: From Nanoreactor Development to Organelle Mimics. Adv. Mater. 2016, 28, 1109–1128. [Google Scholar] [CrossRef] [PubMed]
- Gaitzsch, J.; Huang, X.; Voit, B. Engineering Functional Polymer Capsules toward Smart Nanoreactors. Chem. Rev. 2016, 116, 1053–1093. [Google Scholar] [CrossRef] [PubMed]
- Klermund, L.; Poschenrieder, S.T.; Castiglione, K. Biocatalysis in Polymersomes: Improving Multienzyme Cascades with Incompatible Reaction Steps by Compartmentalization. Acs Catal. 2017, 7, 3900–3904. [Google Scholar] [CrossRef]
- Bermudez, H.; Brannan, A.K.; Hammer, D.A.; Bates, F.S.; Discher, D.E. Molecular weight dependence of polymersome membrane structure, elasticity, and stability. Macromolecules 2002, 35, 8203–8208. [Google Scholar] [CrossRef]
- Lorenceau, E.; Utada, A.S.; Link, D.R.; Cristobal, G.; Joanicot, M.; Weitz, D.A. Generation of Polymerosomes from Double-Emulsions. Langmuir 2005, 21, 9183–9186. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Eisenberg, A. Polymer vesicles. Science 2002, 297, 967–973. [Google Scholar] [CrossRef]
- Le Meins, J.F.; Sandre, O.; Lecommandoux, S. Recent trends in the tuning of polymersomes’ membrane properties. Eur. Phys. J. E. Soft Matter 2011, 34. [Google Scholar] [CrossRef]
- Andersen, O.S.; Koeppe, R.E. Bilayer Thickness and Membrane Protein Function: An Energetic Perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef]
- Garni, M.; Thamboo, S.; Schoenenberger, C.-A.; Palivan, C.G. Biopores/membrane proteins in synthetic polymer membranes. Biochim. Biophys. Acta Biomembr. 2017, 1859, 619–638. [Google Scholar] [CrossRef]
- Itel, F.; Najer, A.; Palivan, C.G.; Meier, W. Dynamics of Membrane Proteins within Synthetic Polymer Membranes with Large Hydrophobic Mismatch. Nano Lett. 2015, 15, 3871–3878. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, T.S.; Klermund, L.; Wang, G.; Castiglione, K. Membrane functionalization of polymersomes: alleviating mass transport limitations by integrating multiple selective membrane transporters for the diffusion of chemically diverse molecules. Nanotechnology 2018, 29, 44LT01. [Google Scholar] [CrossRef] [PubMed]
- Nallani, M.; Benito, S.; Onaca, O.; Graff, A.; Lindemann, M.; Winterhalter, M.; Meier, W.; Schwaneberg, U. A nanocompartment system (synthosome) designed for biotechnological applications. J. Biotechnol. 2006, 123, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Siti, W.; de Hoog, H.-P.M.; Fischer, O.; Shan, W.Y.; Tomczak, N.; Nallani, M.; Liedberg, B. An intercompartmental enzymatic cascade reaction in channel-equipped polymersome-in-polymersome architectures. J. Mater. Chem. B 2014, 2, 2733–2737. [Google Scholar] [CrossRef]
- Schmitt, C.; Lippert, A.H.; Bonakdar, N.; Sandoghdar, V.; Voll, L.M. Compartmentalization and Transport in Synthetic Vesicles. Front. Bioeng. Biotechnol. 2016, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Nardin, C.; Thoeni, S.; Widmer, J.; Winterhalter, M.; Meier, W. Nanoreactors based on (polymerized) ABA-triblock copolymer vesicles. Chem. Commun. 2000, 1433–1434. [Google Scholar] [CrossRef]
- Ranquin, A.; Versees, W.; Meier, W.; Steyaert, J.; Van Gelder, P. Therapeutic nanoreactors: combining chemistry and biology in a novel triblock copolymer drug delivery system. Nano Lett. 2005, 5, 2220–2224. [Google Scholar] [CrossRef]
- Kumar, M.; Grzelakowski, M.; Zilles, J.; Clark, M.; Meier, W. Highly permeable polymeric membranes based on the incorporation of the functional water channel protein Aquaporin Z. Proc. Natl. Acad. Sci. USA 2007, 104, 20719–20724. [Google Scholar] [CrossRef]
- Graff, A.; Fraysse-Ailhas, C.; Palivan, C.G.; Grzelakowski, M.; Friedrich, T.; Vebert, C.; Gescheidt, G.; Meier, W. Amphiphilic Copolymer Membranes Promote NADH:Ubiquinone Oxidoreductase Activity: Towards an Electron-Transfer Nanodevice. Macromol. Chem. Phys. 2010, 211, 229–238. [Google Scholar] [CrossRef]
- Nardin, C.; Widmer, J.; Winterhalter, M.; Meier, W. Amphiphilic block copolymer nanocontainers as bioreactors. Eur. Phys. J. E 2001, 4, 403–410. [Google Scholar] [CrossRef]
- Dean, H.; Benjamin, C.; Hyeseung, L.; Carlo, D.M. Protein-driven energy transduction across polymeric biomembranes. Nanotechnology 2004, 15, 1084. [Google Scholar]
- Denise, W.; Tae-Joon, J.; Jacob, S. Single molecule measurements of channel proteins incorporated into biomimetic polymer membranes. Nanotechnology 2006, 17, 3710. [Google Scholar]
- Lee, H.; Ho, D.; Kuo, K.; Montemagno, C.D. Vectorial insertion of bacteriorhodopsin for directed orientation assays in various polymeric biomembranes. Polymer 2006, 47, 2935–2941. [Google Scholar] [CrossRef]
- Choi, H.; Montemagno, C.D. Light-Driven Hybrid Bioreactor Based on Protein-Incorporated Polymer Vesicles. IEEE Trans. Nanotechnol. 2007, 6, 171–176. [Google Scholar] [CrossRef]
- Hua, D.; Kuang, L.; Liang, H. Self-Directed Reconstitution of Proteorhodopsin with Amphiphilic Block Copolymers Induces the Formation of Hierarchically Ordered Proteopolymer Membrane Arrays. J. Am. Chem. Soc. 2011, 133, 2354–2357. [Google Scholar] [CrossRef] [PubMed]
- Ihle, S.; Onaca, O.; Rigler, P.; Hauer, B.; Rodríguez-Ropero, F.; Fioroni, M.; Schwaneberg, U. Nanocompartments with a pH release system based on an engineered OmpF channel protein. Soft Matter 2011, 7, 532–539. [Google Scholar] [CrossRef]
- Einfalt, T.; Goers, R.; Dinu, I.A.; Najer, A.; Spulber, M.; Onaca-Fischer, O.; Palivan, C.G. Stimuli-Triggered Activity of Nanoreactors by Biomimetic Engineering Polymer Membranes. Nano Lett. 2015, 15, 7596–7603. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, G.; Discher, D.E.; Klein, M.L. Key roles for chain flexibility in block copolymer membranes that contain pores or make tubes. Nano Lett. 2005, 5, 2343–2349. [Google Scholar] [CrossRef] [PubMed]
- Itel, F.; Chami, M.; Najer, A.; Lorcher, S.; Wu, D.L.; Dinu, I.A.; Meier, W. Molecular Organization and Dynamics in Polymersome Membranes: A Lateral Diffusion Study. Macromolecules 2014, 47, 7588–7596. [Google Scholar] [CrossRef]
- Dan, N.; Safran, S.A. Self-Assembly in Mixtures of Diblock Copolymers. Macromolecules 1994, 27, 5766–5772. [Google Scholar] [CrossRef]
- Pata, V.; Dan, N. The effect of chain length on protein solubilization in polymer-based vesicles (polymersomes). Biophys. J. 2003, 85, 2111–2118. [Google Scholar] [CrossRef]
- Muhammad, N.; Dworeck, T.; Fioroni, M.; Schwaneberg, U. Engineering of the E. coli outer membrane protein FhuA to overcome the hydrophobic mismatch in thick polymeric membranes. J. Nanobiotechnol. 2011, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Vriezema, D.M.; Garcia, P.M.; Sancho Oltra, N.; Hatzakis, N.S.; Kuiper, S.M.; Nolte, R.J.; Rowan, A.E.; van Hest, J.C. Positional assembly of enzymes in polymersome nanoreactors for cascade reactions. Angew. Chem. Int. Ed. 2007, 46, 7378–7382. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, S.M.; Nallani, M.; Vriezema, D.M.; Cornelissen, J.J.; van Hest, J.C.; Nolte, R.J.; Rowan, A.E. Enzymes containing porous polymersomes as nano reaction vessels for cascade reactions. Org. Biomol. Chem. 2008, 6, 4315–4318. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, S.F.; Nallani, M.; Cornelissen, J.J.; Nolte, R.J.; van Hest, J.C. A three-enzyme cascade reaction through positional assembly of enzymes in a polymersome nanoreactor. Chemistry 2009, 15, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Gräfe, D.; Gaitzsch, J.; Appelhans, D.; Voit, B. Cross-linked polymersomes as nanoreactors for controlled and stabilized single and cascade enzymatic reactions. Nanoscale 2014, 6, 10752–10761. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.J.R.W.; Marguet, M.; Marais, S.; Fraaije, M.W.; van Hest, J.C.M.; Lecommandoux, S. Cascade Reactions in Multicompartmentalized Polymersomes. Angew. Chem. Int. Ed. 2014, 53, 146–150. [Google Scholar] [CrossRef]
- Messager, L.; Burns, J.R.; Kim, J.; Cecchin, D.; Hindley, J.; Pyne, A.L.; Gaitzsch, J.; Battaglia, G.; Howorka, S. Biomimetic Hybrid Nanocontainers with Selective Permeability. Angew. Chem. Int. Ed. 2016, 55, 11106–11109. [Google Scholar] [CrossRef]
- Schmidt, S.; Castiglione, K.; Kourist, R. Overcoming the Incompatibility Challenge in Chemoenzymatic and Multi-Catalytic Cascade Reactions. Chemistry 2018, 24, 1755–1768. [Google Scholar] [CrossRef]
- Choi, H.J.; Montemagno, C.D. Artificial organelle: ATP synthesis from cellular mimetic polymersomes. Nano Lett. 2005, 5, 2538–2542. [Google Scholar] [CrossRef]
- Wendell, D.; Todd, J.; Montemagno, C. Artificial Photosynthesis in Ranaspumin-2 Based Foam. Nano Lett. 2010, 10, 3231–3236. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Olson, T.L.; Lin, S.; Flores, M.; Jiang, Y.; Zheng, W.; Williams, J.C.; Allen, J.P.; Liang, H. Interface for Light-Driven Electron Transfer by Photosynthetic Complexes Across Block Copolymer Membranes. J. Phys. Chem. Lett. 2014, 5, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Altamura, E.; Milano, F.; Tangorra, R.R.; Trotta, M.; Omar, O.H.; Stano, P.; Mavelli, F. Highly oriented photosynthetic reaction centers generate a proton gradient in synthetic protocells. Proc. Natl. Acad. Sci. USA 2017, 114, 3837–3842. [Google Scholar] [CrossRef] [PubMed]
- Tunuguntla, R.; Bangar, M.; Kim, K.; Stroeve, P.; Ajo-Franklin, C.M.; Noy, A. Lipid Bilayer Composition Can Influence the Orientation of Proteorhodopsin in Artificial Membranes. Biophys. J. 2013, 105, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Harder, D.; Hirschi, S.; Ucurum, Z.; Goers, R.; Meier, W.; Muller, D.J.; Fotiadis, D. Engineering a Chemical Switch into the Light-driven Proton Pump Proteorhodopsin by Cysteine Mutagenesis and Thiol Modification. Angew. Chem. Int. Ed. 2016, 55, 8846–8849. [Google Scholar] [CrossRef] [PubMed]
- Ritzmann, N.; Thoma, J.; Hirschi, S.; Kalbermatter, D.; Fotiadis, D.; Muller, D.J. Fusion Domains Guide the Oriented Insertion of Light-Driven Proton Pumps into Liposomes. Biophys. J. 2017, 113, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Hazard, A.; Montemagno, C. Improved purification for thermophilic F1F0 ATP synthase using n-dodecyl β-d-maltoside. Arch. Biochem. Biophys. 2002, 407, 117–124. [Google Scholar] [CrossRef]
- Goers, R.; Thoma, J.; Ritzmann, N.; Di Silvestro, A.; Alter, C.; Gunkel-Grabole, G.; Fotiadis, D.; Müller, D.J.; Meier, W. Optimized reconstitution of membrane proteins into synthetic membranes. Commun. Chem. 2018, 1, 35. [Google Scholar] [CrossRef]
- Oesterhelt, D.; Stoeckenius, W. Rhodopsin-like Protein from the Purple Membrane of Halobacterium halobium. Nat. New Biol. 1971, 233, 149–152. [Google Scholar] [CrossRef]
- Haupts, U.; Tittor, J.; Oesterhelt, D. Closing in on bacteriorhodopsin: Progress in Understanding the Molecule. Ann. Rev. Biophys. Biomol. Struct. 1999, 28, 367–399. [Google Scholar] [CrossRef]
- Lang-Hinrichs, C.; Queck, I.; Büldt, G.; Stahl, U.; Hildebrandt, V. The archaebacterial membrane protein bacterio-opsin is expressed and N-terminally processed in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1994, 244, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Hildebrandt, V.; Heberle, J.; Büldt, G. Photoactive mitochondria: in vivo transfer of a light-driven proton pump into the inner mitochondrial membrane of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 1994, 91, 9367–9371. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, V.; Fendler, K.; Heberle, J.; Hoffmann, A.; Bamberg, E.; Büldt, G. Bacteriorhodopsin expressed in Schizosaccharomyces pombe pumps protons through the plasma membrane. Proc. Natl. Acad. Sci. USA 1993, 90, 3578–3582. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Möckel, B.; Büldt, G.; Bamberg, E. Functional expression of bacteriorhodopsin in oocytes allows direct measurement of voltage dependence of light induced H+ pumping. FEBS Lett. 1995, 377, 263–266. [Google Scholar] [CrossRef]
- Karnik, S.; Doi, T.; Molday, R.; Khorana, H.G. Expression of the archaebacterial bacterio-opsin gene with and without signal sequences in Escherichia coli: the expressed proteins are located in the membrane but bind retinal poorly. Proc. Natl. Acad. Sci. USA 1990, 87, 8955–8959. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.M.; Greenfield, D.; Bustamante, C.; Liphardt, J. Light-powering Escherichia coli with proteorhodopsin. Proc. Natl. Acad. Sci. USA 2007, 104, 2408–2412. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Bradley, A.S.; Waldbauer, J.R.; Summons, R.E.; DeLong, E.F. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. USA 2007, 104, 5590–5595. [Google Scholar] [CrossRef]
- Béjà, O.; Aravind, L.; Koonin, E.V.; Suzuki, M.T.; Hadd, A.; Nguyen, L.P.; Jovanovich, S.B.; Gates, C.M.; Feldman, R.A.; Spudich, J.L.; et al. Bacterial Rhodopsin: Evidence for a New Type of Phototrophy in the Sea. Science 2000, 289, 1902. [Google Scholar] [CrossRef]
- Johnson, E.T.; Schmidt-Dannert, C. Light-energy conversion in engineered microorganisms. Trends Biotechnol. 2008, 26, 682–689. [Google Scholar] [CrossRef]
- Caffarri, S.; Tibiletti, T.; Jennings, R.C.; Santabarbara, S. A comparison between plant photosystem I and photosystem II architecture and functioning. Curr. Protein Pept. Sci. 2014, 15, 296–331. [Google Scholar] [CrossRef]
- Nelson, N.; Yocum, C.F. Structure and function of photosystems I and II. Ann. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef] [PubMed]
- Feher, G.; Allen, J.P.; Okamura, M.Y.; Rees, D.C. Structure and function of bacterial photosynthetic reaction centres. Nature 1989, 339, 111. [Google Scholar] [CrossRef]
- Ferguson, S.J.; Jackson, J.B.; McEwan, A.G. Anaerobic respiration in the Rhodospirillaceae: Characterisation of pathways and evaluation of roles in redox balancing during photosynthesis. FEMS Microbiol. Lett. 1987, 46, 117–143. [Google Scholar] [CrossRef]
- Stowell, M.H.B.; McPhillips, T.M.; Rees, D.C.; Soltis, S.M.; Abresch, E.; Feher, G. Light-Induced Structural Changes in Photosynthetic Reaction Center: Implications for Mechanism of Electron-Proton Transfer. Science 1997, 276, 812. [Google Scholar] [CrossRef] [PubMed]
- Klamt, S.; Grammel, H.; Straube, R.; Ghosh, R.; Gilles, E.D. Modeling the electron transport chain of purple non-sulfur bacteria. Mol. Syst. Biol. 2008, 4, 156. [Google Scholar] [CrossRef] [PubMed]
- Steinberg-Yfrach, G.; Liddell, P.A.; Hung, S.-C.; Moore, A.L.; Gust, D.; Moore, T.A. Conversion of light energy to proton potential in liposomes by artificial photosynthetic reaction centres. Nature 1997, 385, 239. [Google Scholar] [CrossRef]
- Einfalt, T.; Witzigmann, D.; Edlinger, C.; Sieber, S.; Goers, R.; Najer, A.; Spulber, M.; Onaca-Fischer, O.; Huwyler, J.; Palivan, C.G. Biomimetic artificial organelles with in vitro and in vivo activity triggered by reduction in microenvironment. Nat. Commun. 2018, 9, 1127. [Google Scholar] [CrossRef]
- Dai, L.; Tan, L.-M.; Jiang, Y.-L.; Shi, Y.; Wang, P.; Zhang, J.-P.; Otomo, Z.-Y. Orientation assignment of LH2 and LH1-RC complexes from Thermochromatium tepidum reconstituted in PC liposome and their ultrafast excitation dynamics comparison between in artificial and in natural chromatophores. Chem. Phys. Lett. 2018, 705, 78–84. [Google Scholar] [CrossRef]
- Nagy, L.; Milano, F.; Dorogi, M.; Agostiano, A.; Laczkó, G.; Szebényi, K.; Váró, G.; Trotta, M.; Maróti, P. Protein/Lipid Interaction in the Bacterial Photosynthetic Reaction Center: Phosphatidylcholine and Phosphatidylglycerol Modify the Free Energy Levels of the Quinones. Biochemistry 2004, 43, 12913–12923. [Google Scholar] [CrossRef]
- Trotta, M.; Milano, F.; Nagy, L.; Agostiano, A. Response of membrane protein to the environment: the case of photosynthetic Reaction Centre. Mater. Sci. Eng. C 2002, 22, 263–267. [Google Scholar] [CrossRef]
- Palazzo, G.; Mallardi, A.; Giustini, M.; Berti, D.; Venturoli, G. Cumulant Analysis of Charge Recombination Kinetics in Bacterial Reaction Centers Reconstituted into Lipid Vesicles. Biophys. J. 2000, 79, 1171–1179. [Google Scholar] [CrossRef]
- Baciou, L.; Rivas, E.; Sebban, P. P+QA− and P+QB− charge recombinations in Rhodopseudomonas viridis chromatophores and in reaction centers reconstituted in phosphatidylcholine liposomes. Existence of two conformational states of the reaction centers and effects of pH and o-phenanthroline. Biochemistry 1990, 29, 2966–2976. [Google Scholar] [CrossRef] [PubMed]
- Overfield, R.E.; Wraight, C.A. Oxidation of cytochromes c and c2 by bacterial photosynthetic reaction centers in phospholipid vesicles. 1. Studies with neutral membranes. Biochemistry 1980, 19, 3322–3327. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.P.; Feher, G.; Yeates, T.O.; Komiya, H.; Rees, D.C. Structure of the reaction center from Rhodobacter sphaeroides R-26: protein-cofactor (quinones and Fe2+) interactions. Proc. Natl. Acad. Sci. USA 1988, 85, 8487–8491. [Google Scholar] [CrossRef] [PubMed]
- Hellingwerf, K.J. Reaction centers fromRhodopseudomonas sphaeroides in reconstituted phospholipid vesicles. I. Structural studies. J. Bioenerg. Biomembr. 1987, 19, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Ueno, T.; Fujii, T.; Yang, Q.; Asada, Y.; Miyake, J. Orientation of Photosynthetic Reaction Center Reconstituted in Neutral and Charged Liposomes. Biosci. Biotechnol. Biochem. 1997, 61, 1577–1579. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Iwamoto, M.; Kato, A.; Yoshikawa, K.; Oiki, S. Oriented Reconstitution of a Membrane Protein in a Giant Unilamellar Vesicle: Experimental Verification with the Potassium Channel KcsA. J. Am. Chem. Soc. 2011, 133, 11774–11779. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Whited, G.; Nguyen, C.; Stucky, G.D. The directed cooperative assembly of proteorhodopsin into 2D and 3D polarized arrays. Proc. Natl. Acad. Sci. USA 2007, 104, 8212–8217. [Google Scholar] [CrossRef]
- Luo, L.; Eisenberg, A. Thermodynamic Size Control of Block Copolymer Vesicles in Solution. Langmuir 2001, 17, 6804–6811. [Google Scholar] [CrossRef]
- Luo, L.; Eisenberg, A. Thermodynamic Stabilization Mechanism of Block Copolymer Vesicles. J. Am. Chem. Soc. 2001, 123, 1012–1013. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, F.; Yuan, W.; Jin, T. Polymersomes of asymmetric bilayer membrane formed by phase-guided assembly. J. Controlled Release 2010, 147, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.F.; Thordarson, P. Polymersomes as protocellular constructs. J. Polym. Sci., Part A: Polym. Chem. 2017, 55, 3817–3825. [Google Scholar] [CrossRef]
- Stoenescu, R.; Graff, A.; Meier, W. Asymmetric ABC-triblock copolymer membranes induce a directed insertion of membrane proteins. Macromol. Biosci. 2004, 4, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Romsicki, Y.; Sharom, F.J. Phospholipid flippase activity of the reconstituted P-glycoprotein multidrug transporter. Biochemistry 2001, 40, 6937–6947. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, N.; Wörner, A.C.; Yang, J.; Shastri, S.; Hellmich, U.A.; Aslimovska, L.; Maier, M.S.M.; Glaubitz, C. Solid-state NMR and functional studies on proteorhodopsin. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-Speed Atomic Force Microscopy Reveals Rotary Catalysis of Rotorless F1-ATPase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Stock, D.; Leslie, A.G.W.; Walker, J.E. Molecular Architecture of the Rotary Motor in ATP Synthase. Science 1999, 286, 1700–1705. [Google Scholar] [CrossRef]
- Boyer, P.D. The ATP Synthase—A Splendid Molecular Machine. Ann. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Luo, T.-J.M.; Soong, R.; Lan, E.; Dunn, B.; Montemagno, C. Photo-induced proton gradients and ATP biosynthesis produced by vesicles encapsulated in a silica matrix. Nat. Mater. 2005, 4, 220–224. [Google Scholar] [CrossRef]
- Duan, L.; He, Q.; Wang, K.; Yan, X.; Cui, Y.; Möhwald, H.; Li, J. Adenosine Triphosphate Biosynthesis Catalyzed by FoF1 ATP Synthase Assembled in Polymer Microcapsules. Angew. Chem. 2007, 119, 7126–7130. [Google Scholar] [CrossRef]
- Qi, W.; Duan, L.; Wang, K.W.; Yan, X.H.; Cui, Y.; He, Q.; Li, J. Motor Protein CF0F1 Reconstituted in Lipid-Coated Hemoglobin Microcapsules for ATP Synthesis. Adv. Mater. 2008, 20, 601–605. [Google Scholar] [CrossRef]
- Li, J.-H.; Wang, Y.-F.; Ha, W.; Liu, Y.; Ding, L.-S.; Li, B.-J.; Zhang, S. Cyclodextrin-Based Microcapsules as Bioreactors for ATP Biosynthesis. Biomacromolecules 2013, 14, 2984–2988. [Google Scholar] [CrossRef] [PubMed]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55. [Google Scholar] [CrossRef] [PubMed]
- Andreiadis, E.S.; Chavarot-Kerlidou, M.; Fontecave, M.; Artero, V. Artificial Photosynthesis: From Molecular Catalysts for Light-driven Water Splitting to Photoelectrochemical Cells. Photochem. Photobiol. 2011, 87, 946–964. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.I.; Volodkin, D.V.; Sukhorukov, G.B. Protein—Calcium Carbonate Coprecipitation: A Tool for Protein Encapsulation. Biotechnol. Progr. 2008, 21, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Klermund, L.; Castiglione, K. Polymersomes as nanoreactors for preparative biocatalytic applications: current challenges and future perspectives. Bioprocess. Biosyst. Eng. 2018, 41, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Michel, H. Editorial: The Nonsense of Biofules. Angew. Chem. Int. Ed. 2012, 51, 2516–2518. [Google Scholar] [CrossRef]
- Dimroth, F.; Roesener, T.; Essig, S.; Weuffen, C.; Wekkeli, A.; Oliva, E.; Siefer, G.; Volz, K.; Hannappel, T.; Häussler, D.; et al. Comparison of Direct Growth and Wafer Bonding for the Fabrication of GaInP/GaAs Dual-Junction Solar Cells on Silicon. IEEE J. Photovoltaics 2014, 4, 620–625. [Google Scholar] [CrossRef]
- Hall, D.; Rao, K. Photosynthesis, 6th ed.; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
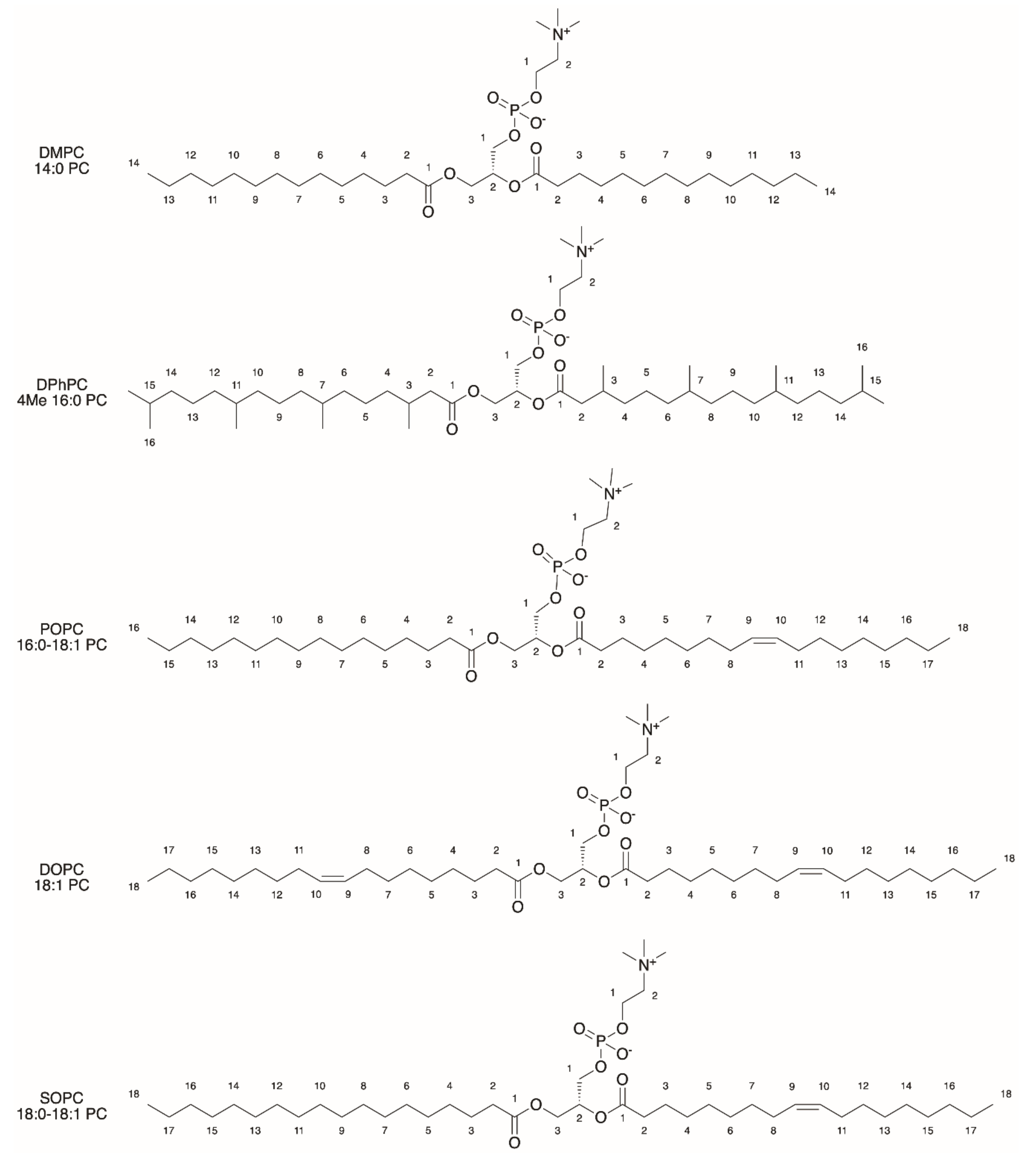
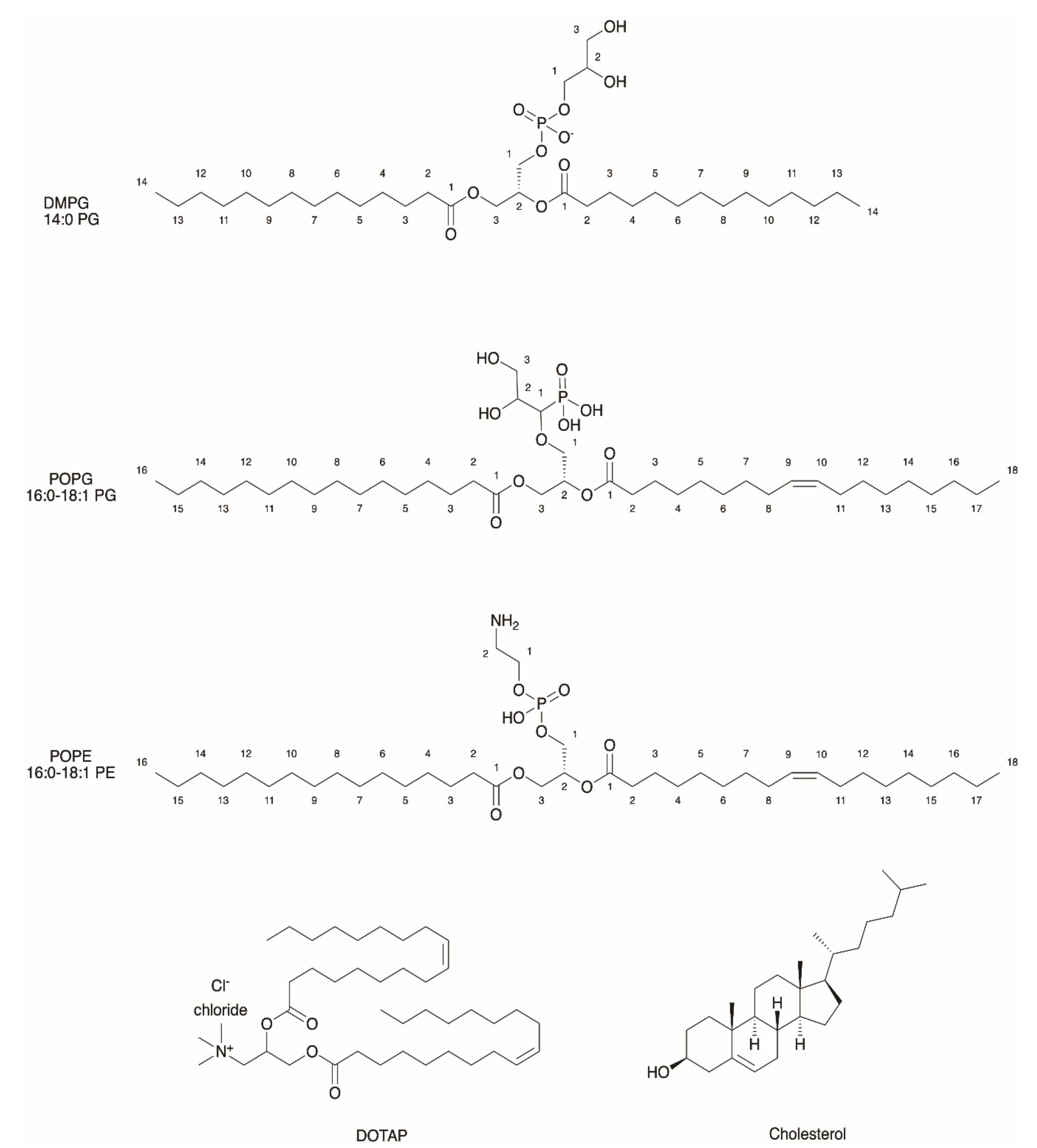
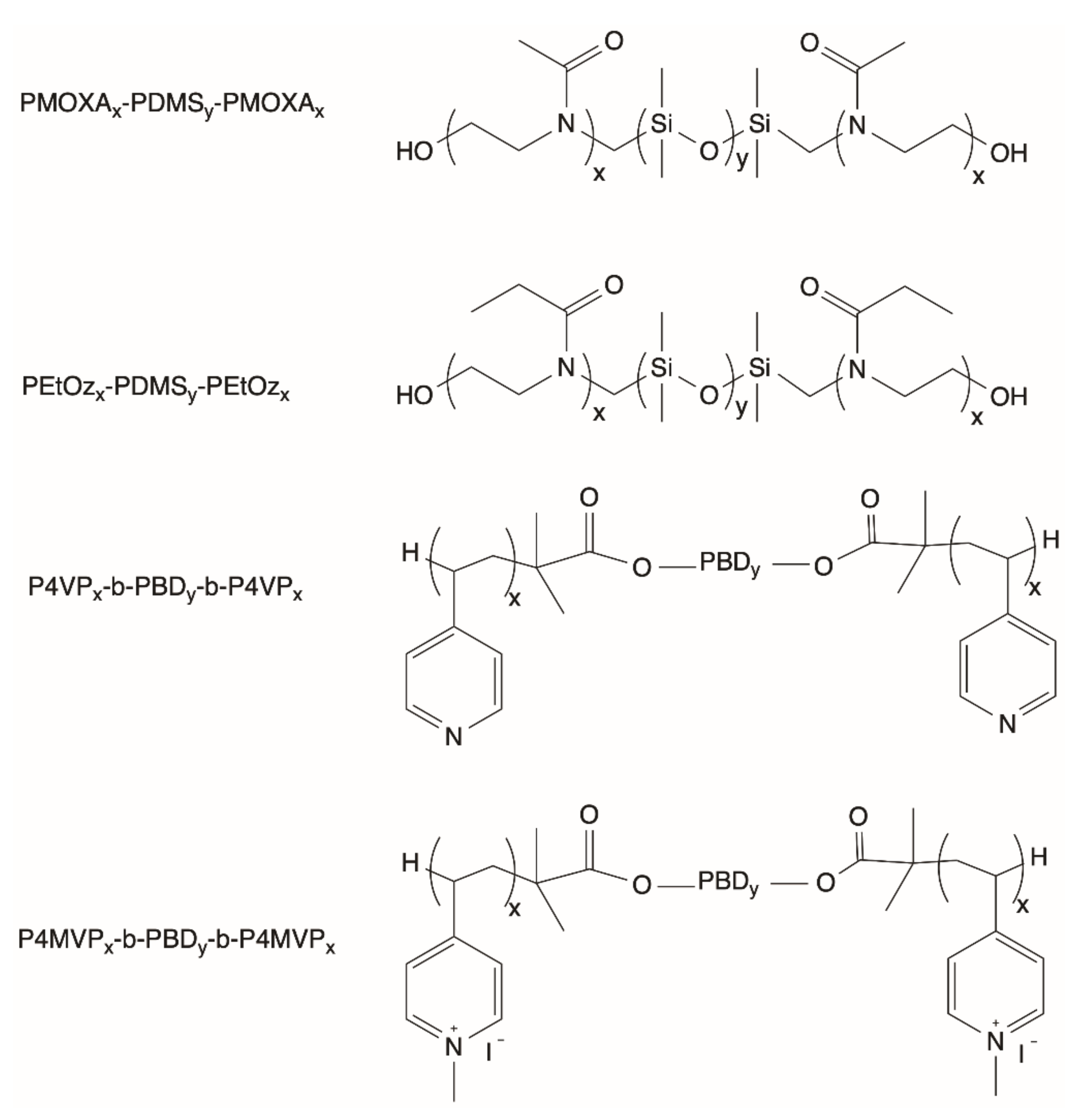

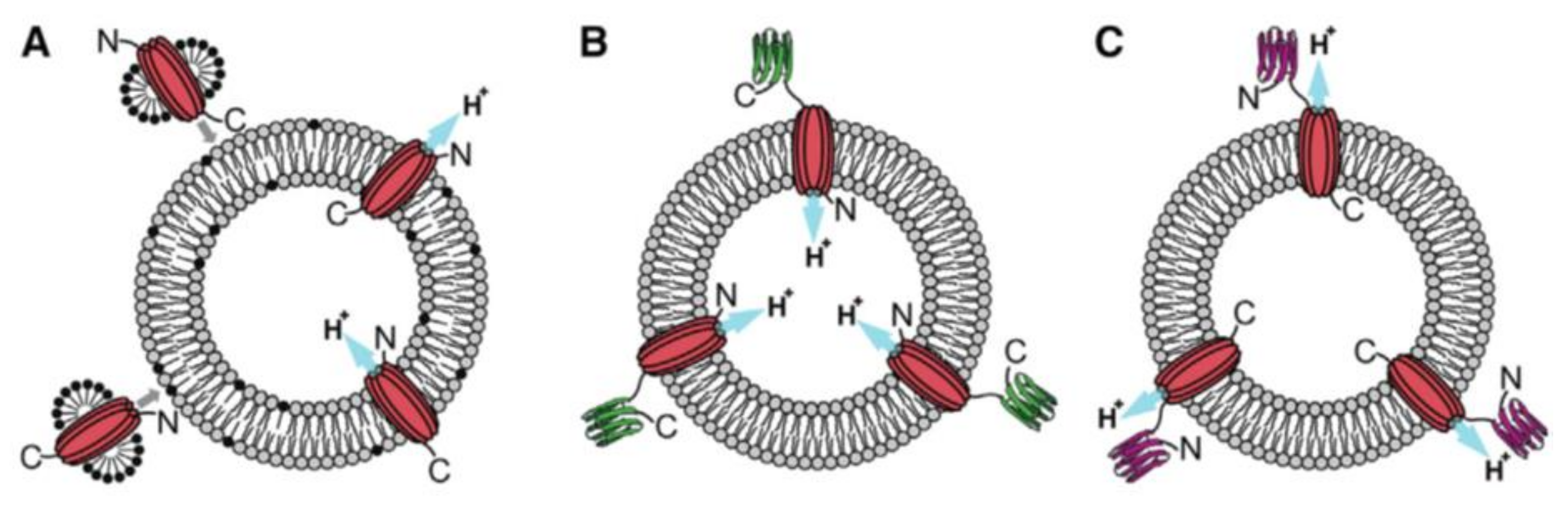
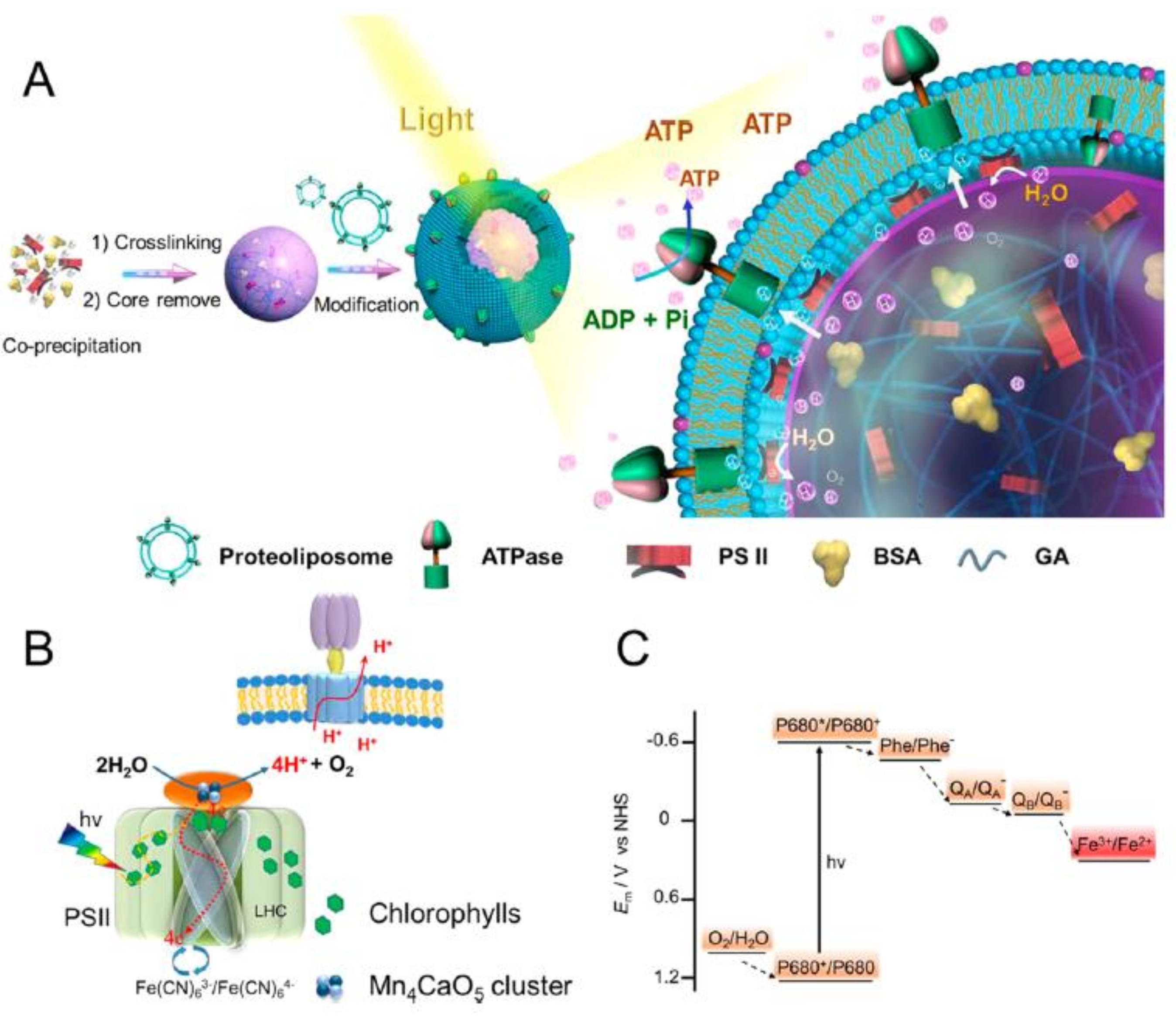
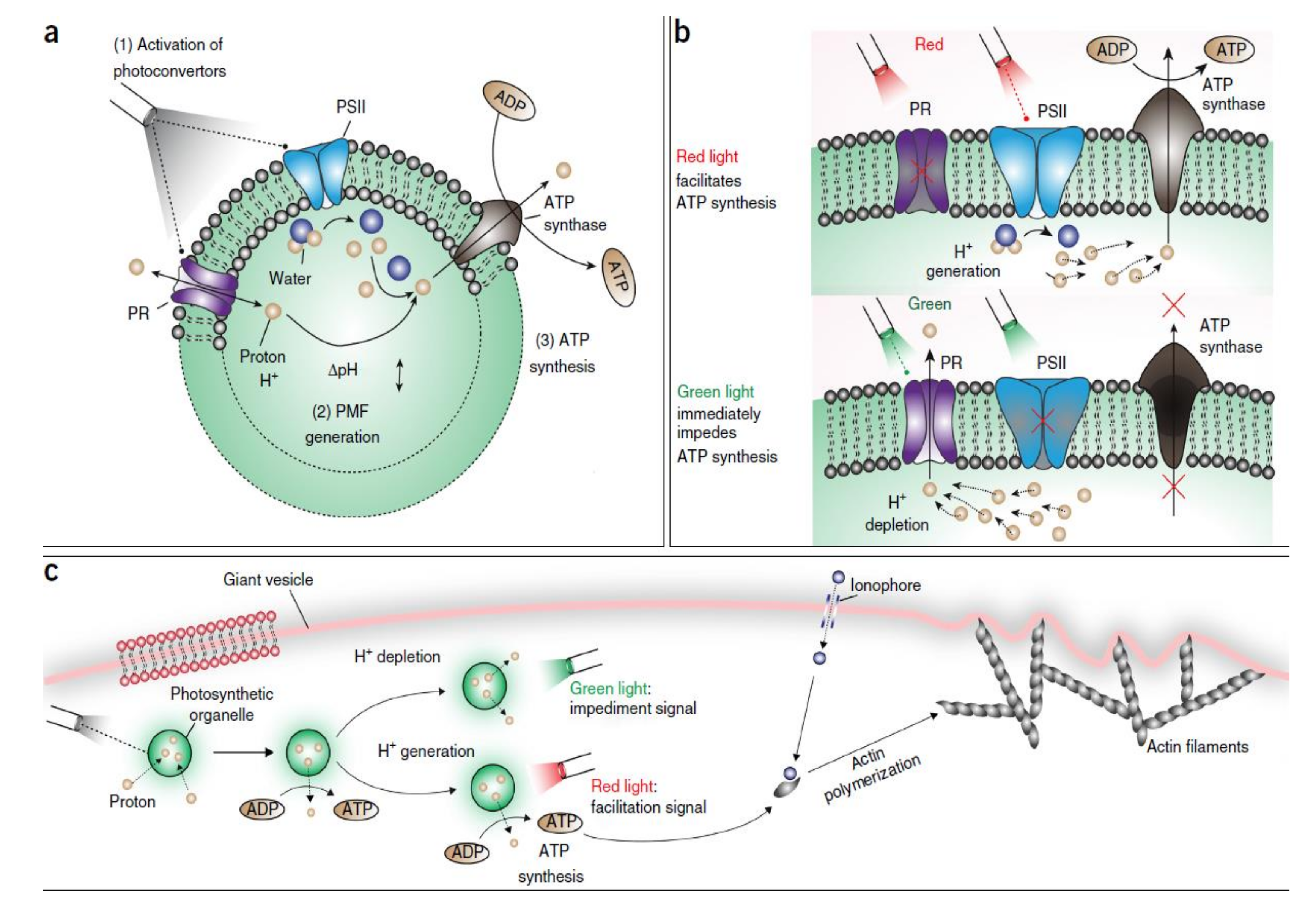
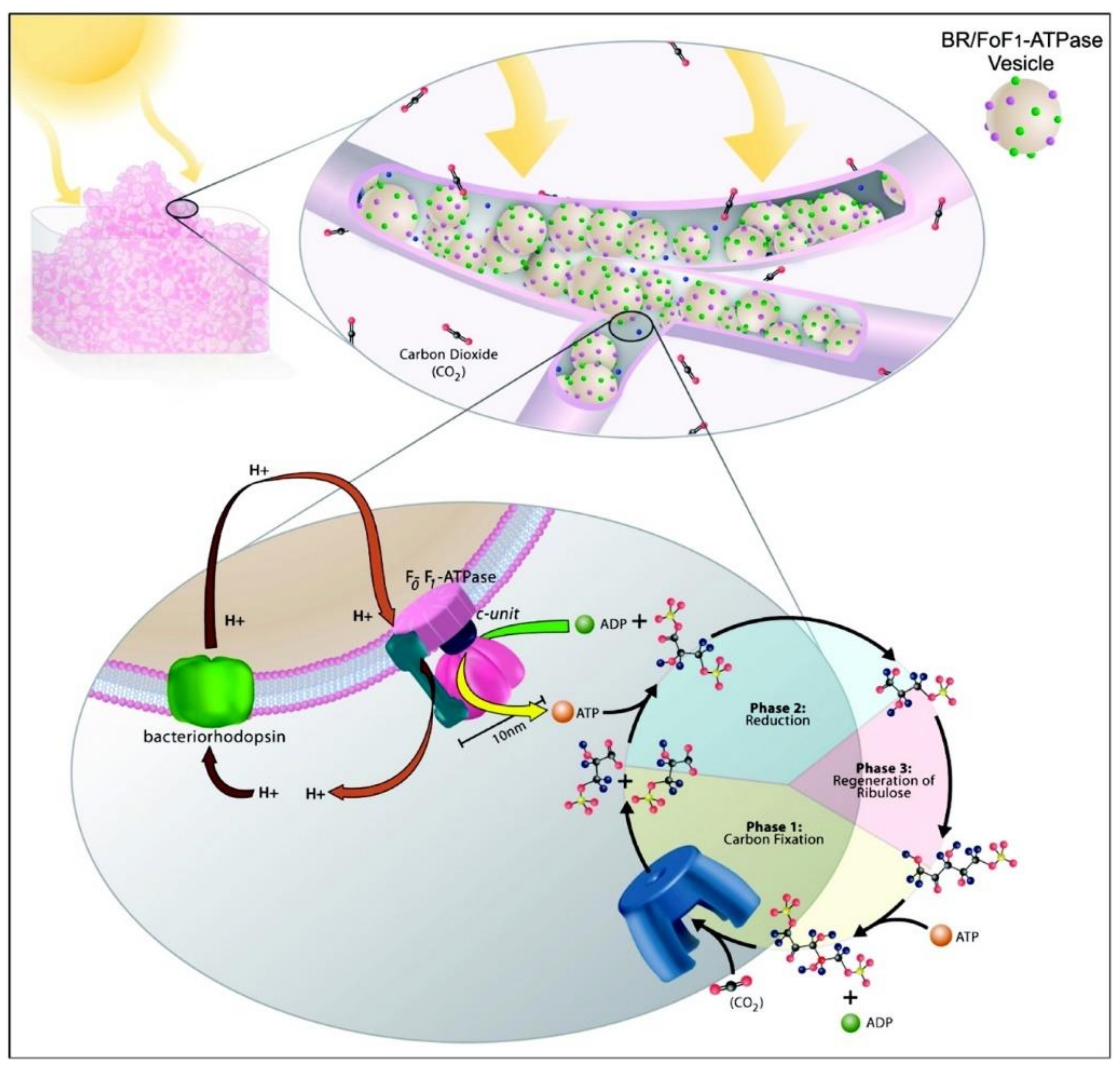
| Liposomes/Polymersomes | Embedded Membrane Proteins/(Artificial) Photosynthetic Systems | References |
|---|---|---|
| Crude soybean phospholipids or pure PC/PE/(cardiolipin) | Purple membrane from Halobacterium halobium/mitochondrial ATPase | [12] |
| PC/PA/cholesterol (Molar ratio 6.4:0.6:0.3) (optimized composition) | BR from Halobacterium salinarium/chloroplastic F0F1-ATPase (mass ratio 1:10) | [14] |
| PC/PA/cholesterol (20 mol%) (Mass ratio 10:1) | Artificial RC/chloroplastic F0F1-ATPase | [15] |
| SOPC | BR from Halobacterium salinarium | [16] |
| POPC/PG/cholesterol (Molar ratio: 5:1:1) | RC from Rhodobacter sphaeroides R-26 | [17] |
| PC, PC/PA, DPhPC | BR from Halobacterium salinarium | [18] |
| DMPC/DMPG (Mass ratio 9:1) | PSII from spinach (microsphere) /chloroplastic F0F1-ATPase (proteoliposomes) | [19] |
| POPC/POPE/POPG/Cholesterol (Molar ratio 2:1:1:1) | PSII from spinach/heterologously expressed PR/ATP synthase from Bacillus pseudofirmus | [20] |
| POPC/POPG (Molar ratio 9:1) | RC from Rhodobacter sphaeroides R-26 | [73] |
| POPC/POPG (Molar ratio 4:1) DOPC:DOTAP (Molar ratio 4:1) | Heterologously expressed PR | [74] |
| DOPC | Heterologously expressed PR | [75] |
| DOPC | Heterologously expressed PR | [76] |
| PC/PA/cholesterol (Molar ratio 9:1:3) | BR from Halobacterium halobium/F0F1-ATP synthase from Bacillus PS3 | [77] |
| DOPC PMOXA17-PDMS65-PMOXA17 | Heterologously expressed PR | [78] |
| PMOXA-PDMS-PMOXA | BR from Halobacterium salinarium | [53] |
| PEtOz-PDMS-PEtOz | BR from Halobacterium salinarium/F0F1-ATP synthase from Bacillus PS3 | [54,70] |
| P4VP28-b-PBD22-b-P4VP28 P4MVP18-b-PBD93-b-P4MVP18 | Heterologously expressed PR | [55] |
| PMOXA12-PDMS33-PMOXA12 | BR from Halobacterium/F0F1-ATP synthase from Bacillus PS3 | [71] |
| P4MVP28-b-PBD22-b-P4MVP28 P4MVP29-b-PBD56-b-P4MVP29 | RC from Rhodobacter sphaeroides | [72] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Castiglione, K. Light-Driven Biocatalysis in Liposomes and Polymersomes: Where Are We Now? Catalysts 2019, 9, 12. https://doi.org/10.3390/catal9010012
Wang G, Castiglione K. Light-Driven Biocatalysis in Liposomes and Polymersomes: Where Are We Now? Catalysts. 2019; 9(1):12. https://doi.org/10.3390/catal9010012
Chicago/Turabian StyleWang, Guoshu, and Kathrin Castiglione. 2019. "Light-Driven Biocatalysis in Liposomes and Polymersomes: Where Are We Now?" Catalysts 9, no. 1: 12. https://doi.org/10.3390/catal9010012
APA StyleWang, G., & Castiglione, K. (2019). Light-Driven Biocatalysis in Liposomes and Polymersomes: Where Are We Now? Catalysts, 9(1), 12. https://doi.org/10.3390/catal9010012





