A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases
Abstract
1. Introduction
2. Biochemical Characteristics
3. Structural Characteristics
3.1. Structural Determinants of Cutinase Stability
3.2. Structural Determinants of Cutinase Activity
4. Improving the Heat and Operational Stability of Cutinases
4.1. Host Selection and Protein Engineering Techniques
4.2. Immobilization
5. Environmental Biocatalysis
5.1. Plastics Degradation
5.1.1. Degradation of Succinate Polyesters
5.1.2. Degradation of PCL
5.1.3. Degradation of PET
5.1.4. Degradation of Polyethylene Furanoate
5.1.5. Degradation of Other Synthetic Polymers
5.2. Engineering Cutinase Depolymerization Activity
5.3. Effect of Medium Components and Synergistic Interaction in Cutinase Depolymerase Activity
5.4. Other Bioremediation Applications
6. Superficial Functionalization of Polymers
6.1. Surface Modification of PET
6.2. Surface Modification of PLA
7. Cutinases as a Tool for Synthetic Chemistry
Author Contributions
Funding
Conflicts of Interest
References
- Fich, E.A.; Segerson, N.A.; Rose, J.K.C. The plant polyester cutin: Biosynthesis, structure, and biological roles. Annu. Rev. Plant Biol. 2016, 67, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.A.L.; van’t Klooster, J.W.; Wagemakers, C.A.M.; Dees, D.C.T.; van der Vlugt-Bergmans, C.J.B. Cutinase A of Botrytis cinerea is expressed, but not essential, during penetration of gerbera and tomato. Mol. Plant-Microbe Interact. 1997, 10, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Rong, W.; Massart, S.; Zhang, Z. Genome-wide identification and expression analysis of cutinase gene family in Rhizoctonia cerealis and functional study of an active cutinase RcCUT1 in the fungal–wheat interaction. Front. Microbiol. 2018, 9, 1813. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Li, D.-W.; Zheng, L.; Huang, J. CglCUT1 gene required for cutinase activity and pathogenicity of Colletotrichum gloeosporioides causing anthracnose of Camellia oleifera. Eur. J. Plant Pathol. 2017, 147, 103–114. [Google Scholar] [CrossRef]
- Li, D.; Ashby, A.M.; Johnstone, K. Molecular evidence that the extracellular cutinase Pbc1 is required for pathogenicity of Pyrenopeziza brassicae on oilseed rape. Mol. Plant-Microbe Interact. 2003, 16, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.M.; Flaishman, M.A.; Kolattukudy, P.E. Cutinase gene disruption in Fusarium solani f sp. pisi decreases its virulence on pea. Plant Cell 1994, 6, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.B.; Patil, S.S. Cutinase deficient mutants of Colletotrichum gloeosporioides are nonpathogenic to papaya fruit. Physiol. Mol. Plant Pathol. 1986, 28, 235–242. [Google Scholar] [CrossRef]
- Dickman, M.B.; Podila, G.K.; Kolattukudy, P.E. Insertion of cutinase gene into a wound pathogen enables it to infect intact host. Nature 1989, 342, 446–448. [Google Scholar] [CrossRef]
- Heinen, W. On the enzymatic degradation of cutin. IV. Separation of cutinase from associated oxidative enzymes. Enzymologia 1963, 25, 281–291. [Google Scholar]
- Shishiyama, J.; Araki, F.; Akai, S. Studies on cutin-esterase II. Characteristics of cutin-esterase from Botrytis cinerea and its activity on tomato-cutin. Plant Cell Physiol. 1970, 11, 937–945. [Google Scholar] [CrossRef]
- Kolattukudy, P.E. Biosynthesis of a lipid polymer, cutin: The structural component of plant cuticle. Biochem. Biophys. Res. Commun. 1970, 41, 299–305. [Google Scholar] [CrossRef]
- Purdy, R.E.; Kolattukudy, P.E. Depolymerization of a hydroxy fatty acid biopolymer, cutin, by an extracellular enzyme from Fusarium solani f. pisi: Isolation and some properties of the enzyme. Arch. Biochem. Biophys. 1973, 159, 61–69. [Google Scholar] [CrossRef]
- Purdy, R.E.; Kolattukudy, P.E. Hydrolysis of plant cuticle by plant pathogens. Properties of cutinase I, cutinase II, and a nonspecific esterase isolated from Fusarium solani pisi. Biochemistry 1975, 14, 2832–2840. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase structure, function and biocatalytic applications. Electron. J. Biotechnol. 1998, 1, 160–173. [Google Scholar] [CrossRef]
- Dutta, K.; Sen, S.; Veeranki, V.D. Production, characterization and applications of microbial cutinases. Process Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Morita, T.; Cao, X.; Yoshida, S.; Koitabashi, M.; Watanabe, T.; Suzuki, K.; Sameshima-Yamashita, Y.; Nakajima-Kambe, T.; Fujii, T.; et al. Biodegradable plastic-degrading enzyme from Pseudozyma antarctica: Cloning, sequencing, and characterization. Appl. Microbiol. Biotechnol. 2013, 97, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Roussel, A.; Amara, S.; Nyyssölä, A.; Mateos-Diaz, E.; Blangy, S.; Kontkanen, H.; Westerholm-Parvinen, A.; Carrière, F.; Cambillau, C. A cutinase from Trichoderma reesei with a lid-covered active site and kinetic properties of true lipases. J. Mol. Biol. 2014, 426, 3757–3772. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.; Litwińska, K.; Cordes, A.; Baronian, K.; Bode, R.; Schauer, F.; Kunze, G. Three new cutinases from the yeast Arxula adeninivorans that are suitable for biotechnological applications. Appl. Environ. Microbiol. 2015, 81, 5497–5510. [Google Scholar] [CrossRef]
- Sebastião, M.J.; Cabral, J.M.S.; Aires-Barros, M.R. Improved purification protocol of a Fusarium solani pisi recombinant cutinase by phase partitioning in aqueous two-phase systems of polyethylene glycol and phosphate. Enzyme Microb. Technol. 1996, 18, 251–260. [Google Scholar] [CrossRef]
- Dimarogona, M.; Nikolaivits, E.; Kanelli, M.; Christakopoulos, P.; Sandgren, M.; Topakas, E. Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential. Biochim. Biophys. Acta—Gen. Subj. 2015, 1850, 2308–2317. [Google Scholar] [CrossRef] [PubMed]
- Nikolaivits, E.; Kokkinou, A.; Karpusas, M.; Topakas, E. Microbial host selection and periplasmic folding in Escherichia coli affect the biochemical characteristics of a cutinase from Fusarium oxysporum. Protein Expr. Purif. 2016, 127, 1–7. [Google Scholar] [CrossRef]
- Thumarat, U.; Nakamura, R.; Kawabata, T.; Suzuki, H.; Kawai, F. Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119. Appl. Microbiol. Biotechnol. 2012, 95, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Oeser, T.; Then, J.; Kühn, N.; Barth, M.; Schmidt, J.; Zimmermann, W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express 2014, 4, 44. [Google Scholar] [CrossRef]
- Xu, H.; Yan, Q.; Duan, X.; Yang, S.; Jiang, Z. Characterization of an acidic cold-adapted cutinase from Thielavia terrestris and its application in flavor ester synthesis. Food Chem. 2015, 188, 439–445. [Google Scholar] [CrossRef]
- Yang, S.; Xu, H.; Yan, Q.; Liu, Y.; Zhou, P.; Jiang, Z. A low molecular mass cutinase of Thielavia terrestris efficiently hydrolyzes poly(esters). J. Ind. Microbiol. Biotechnol. 2013, 40, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sakamoto, H.; Shinozaki, Y.; Tabata, J.; Watanabe, T.; Mochizuki, A.; Koitabashi, M.; Fujii, T.; Tsushima, S.; Kitamoto, H.K. Affinity purification and characterization of a biodegradable plastic-degrading enzyme from a yeast isolated from the larval midgut of a stag beetle, Aegus laevicollis. Appl. Microbiol. Biotechnol. 2013, 97, 7679–7688. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Noguchi, M.T.; Shinozaki, Y.; Koitabashi, M.; Sameshima-Yamashita, Y.; Yoshida, S.; Fujii, T.; Kitamoto, H.K. Purification, characterization, and cloning of the gene for a biodegradable plastic-degrading enzyme from Paraphoma-related fungal strain B47-9. Appl. Microbiol. Biotechnol. 2014, 98, 4457–4465. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Yamagata, Y.; Abe, K.; Hasegawa, F.; Machida, M.; Ishioka, R.; Gomi, K.; Nakajima, T. Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2005, 67, 778–788. [Google Scholar] [CrossRef]
- Bermúdez-García, E.; Peña-Montes, C.; Castro-Rodríguez, J.A.; González-Canto, A.; Navarro-Ocaña, A.; Farrés, A. ANCUT2, a thermo-alkaline cutinase from Aspergillus nidulans and its potential applications. Appl. Biochem. Biotechnol. 2017, 182, 1014–1036. [Google Scholar] [CrossRef]
- Wang, G.Y.; Michailides, T.J.; Hammock, B.D.; Lee, Y.-M.; Bostock, R.M. Molecular cloning, characterization, and expression of a redox-responsive cutinase from Monilinia fructicola (wint.) honey. Fungal Genet. Biol. 2002, 35, 261–276. [Google Scholar] [CrossRef]
- Thumarat, U.; Kawabata, T.; Nakajima, M.; Nakajima, H.; Sugiyama, A.; Yazaki, K.; Tada, T.; Waku, T.; Tanaka, N.; Kawai, F. Comparison of genetic structures and biochemical properties of tandem cutinase-type polyesterases from Thermobifida alba AHK119. J. Biosci. Bioeng. 2015, 120, 491–497. [Google Scholar] [CrossRef]
- Nyyssölä, A.; Pihlajaniemi, V.; Järvinen, R.; Mikander, S.; Kontkanen, H.; Kruus, K.; Kallio, H.; Buchert, J. Screening of microbes for novel acidic cutinases and cloning and expression of an acidic cutinase from Aspergillus niger CBS 513.88. Enzyme Microb. Technol. 2013, 52, 272–278. [Google Scholar] [CrossRef]
- Nyyssölä, A.; Pihlajaniemi, V.; Häkkinen, M.; Kontkanen, H.; Saloheimo, M.; Nakari-Setälä, T. Cloning and characterization of a novel acidic cutinase from Sirococcus conigenus. Appl. Microbiol. Biotechnol. 2014, 98, 3639–3650. [Google Scholar] [CrossRef]
- Martinez, C.; De Geus, P.; Lauwereys, M.; Matthyssens, G.; Cambillau, C. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature 1992, 356, 615–618. [Google Scholar] [CrossRef]
- Longhi, S.; Cambillau, C. Structure-activity of cutinase, a small lipolytic enzyme. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 1999, 1441, 185–196. [Google Scholar] [CrossRef]
- Longhi, S.; Czjzek, M.; Lamzin, V.; Nicolas, A.; Cambillau, C. Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: Stereochemical analysis. J. Mol. Biol. 1997, 268, 779–799. [Google Scholar] [CrossRef]
- Roth, C.; Wei, R.; Oeser, T.; Then, J.; Föllner, C.; Zimmermann, W.; Sträter, N. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 2014, 98, 7815–7823. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.-W.W.; Ma, J.; Zheng, Y.; Ko, T.-P.P.; Xu, L.; Cheng, Y.-S.S.; Chen, C.-C.C.; Guo, R.-T.T. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Chen, S.; Tong, X.; Woodard, R.W.; Du, G.; Wu, J.; Chen, J. Identification and characterization of bacterial cutinase. J. Biol. Chem. 2008, 283, 25854–25862. [Google Scholar] [CrossRef]
- Herrero Acero, E.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Wei, R.; Zimmermann, W.; Zinn, M.; et al. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 2011, 44, 4632–4640. [Google Scholar] [CrossRef]
- Ribitsch, D.; Acero, E.H.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Freddi, G.; Schwab, H.; Guebitz, G.M. Characterization of a new cutinase from Thermobifida alba for PET-surface hydrolysis. Biocatal. Biotransform. 2012, 30, 2–9. [Google Scholar] [CrossRef]
- Hegde, K.; Veeranki, V.D. Production optimization and characterization of recombinant cutinases from Thermobifida fusca sp. NRRL B-8184. Appl. Biochem. Biotechnol. 2013, 170, 654–675. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.-J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef]
- Kwon, M.-A.; Kim, H.S.; Yang, T.H.; Song, B.K.; Song, J.K. High-level expression and characterization of Fusarium solani cutinase in Pichia pastoris. Protein Expr. Purif. 2009, 68, 104–109. [Google Scholar] [CrossRef]
- Rubio, M.B.; Cardoza, R.E.; Hermosa, R.; Gutiérrez, S.; Monte, E. Cloning and characterization of the Thcut1 gene encoding a cutinase of Trichoderma harzianum T34. Curr. Genet. 2008, 54, 301–312. [Google Scholar] [CrossRef]
- Liu, Z.; Gosser, Y.; Baker, P.J.; Ravee, Y.; Lu, Z.; Alemu, G.; Li, H.; Butterfoss, G.L.; Kong, X.-P.; Gross, R.; et al. Structural and functional studies of Aspergillus oryzae cutinase: Enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation. J. Am. Chem. Soc. 2009, 131, 15711–15716. [Google Scholar] [CrossRef]
- Koschorreck, K.; Liu, D.; Kazenwadel, C.; Schmid, R.D.; Hauer, B. Heterologous expression, characterization and site-directed mutagenesis of cutinase CUTAB1 from Alternaria brassicicola. Appl. Microbiol. Biotechnol. 2010, 87, 991–997. [Google Scholar] [CrossRef]
- Kazenwadel, C.; Eiben, S.; Maurer, S.; Beuttler, H.; Wetzl, D.; Hauer, B.; Koschorreck, K. Thiol-functionalization of acrylic ester monomers catalyzed by immobilized Humicola insolens cutinase. Enzyme Microb. Technol. 2012, 51, 9–15. [Google Scholar] [CrossRef]
- Seman, W.M.K.W.; Bakar, S.A.; Bukhari, N.A.; Gaspar, S.M.; Othman, R.; Nathan, S.; Mahadi, N.M.; Jahim, J.; Murad, A.M.A.; Bakar, F.D.A. High level expression of Glomerella cingulata cutinase in dense cultures of Pichia pastoris grown under fed-batch conditions. J. Biotechnol. 2014, 184, 219–228. [Google Scholar] [CrossRef]
- Duan, X.; Liu, Y.; You, X.; Jiang, Z.; Yang, S.; Yang, S. High-level expression and characterization of a novel cutinase from Malbranchea cinnamomea suitable for butyl butyrate production. Biotechnol. Biofuels 2017, 10, 223. [Google Scholar] [CrossRef]
- Yang, S.; Liu, M.; Long, L.; Zhang, R.; Ding, S. Characterization of a cutinase from Myceliophthora thermophila and its application in polyester hydrolysis and deinking process. Process Biochem. 2018, 66, 106–112. [Google Scholar] [CrossRef]
- Kodama, Y.; Masaki, K.; Kondo, H.; Suzuki, M.; Tsuda, S.; Nagura, T.; Shimba, N.; Suzuki, E.; Iefuji, H. Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2. Proteins Struct. Funct. Bioinform. 2009, 77, 710–717. [Google Scholar] [CrossRef]
- Nyon, M.P.; Rice, D.W.; Berrisford, J.M.; Huang, H.; Moir, A.J.G.; Craven, C.J.; Nathan, S.; Mahadi, N.M.; Abu Bakar, F.D. Crystallization and preliminary X-ray analysis of recombinant Glomerella cingulata cutinase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 504–508. [Google Scholar] [CrossRef]
- Kold, D.; Dauter, Z.; Laustsen, A.K.; Brzozowski, A.M.; Turkenburg, J.P.; Nielsen, A.D.; Koldsø, H.; Petersen, E.; Schiøtt, B.; De Maria, L.; et al. Thermodynamic and structural investigation of the specific SDS binding of Humicola insolens cutinase. Protein Sci. 2014, 23, 1023–1035. [Google Scholar] [CrossRef]
- Kitadokoro, K.; Thumarat, U.; Nakamura, R.; Nishimura, K.; Karatani, H.; Suzuki, H.; Kawai, F. Crystal structure of cutinase Est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76 Å resolution. Polym. Degrad. Stab. 2012, 97, 771–775. [Google Scholar] [CrossRef]
- Sulaiman, S.; You, D.-J.; Kanaya, E.; Koga, Y.; Kanaya, S. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase. Biochemistry 2014, 53, 1858–1869. [Google Scholar] [CrossRef]
- Miyakawa, T.; Mizushima, H.; Ohtsuka, J.; Oda, M.; Kawai, F.; Tanokura, M. Structural basis for the Ca2+-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2015, 99, 4297–4307. [Google Scholar] [CrossRef]
- Ribitsch, D.; Hromic, A.; Zitzenbacher, S.; Zartl, B.; Gamerith, C.; Pellis, A.; Jungbauer, A.; Łyskowski, A.; Steinkellner, G.; Gruber, K.; et al. Small cause, large effect: Structural characterization of cutinases from Thermobifida cellulosilytica. Biotechnol. Bioeng. 2017, 114, 2481–2488. [Google Scholar] [CrossRef]
- Hegde, K.; Dasu, V.V. Structural stability and unfolding properties of cutinases from Thermobifida fusca. Appl. Biochem. Biotechnol. 2014, 174, 803–819. [Google Scholar] [CrossRef]
- Shirke, A.N.; Basore, D.; Butterfoss, G.L.; Bonneau, R.; Bystroff, C.; Gross, R.A. Toward rational thermostabilization of Aspergillus oryzae cutinase: Insights into catalytic and structural stability. Proteins Struct. Funct. Bioinform. 2016, 84, 60–72. [Google Scholar] [CrossRef]
- Numoto, N.; Kamiya, N.; Bekker, G.-J.; Yamagami, Y.; Inaba, S.; Ishii, K.; Uchiyama, S.; Kawai, F.; Ito, N.; Oda, M. Structural dynamics of the PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190 in substrate-bound states elucidates the Ca2+-driven catalytic cycle. Biochemistry 2018. [Google Scholar] [CrossRef]
- Herrero Acero, E.; Ribitsch, D.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Steinkellner, G.; Gruber, K.; Schwab, H.; Guebitz, G.M. Surface engineering of a cutinase from Thermobifida cellulosilytica for improved polyester hydrolysis. Biotechnol. Bioeng. 2013, 110, 2581–2590. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.-Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Fecker, T.; Galaz-Davison, P.; Engelberger, F.; Narui, Y.; Sotomayor, M.; Parra, L.P.; Ramírez-Sarmiento, C.A. Active site flexibility as a hallmark for efficient PET degradation by I. sakaiensis PETase. Biophys. J. 2018, 114, 1302–1312. [Google Scholar] [CrossRef]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef]
- Chen, C.-C.; Han, X.; Ko, T.-P.; Liu, W.; Guo, R.-T. Structural studies reveal the molecular mechanism of PETase. FEBS J. 2018, 285, 3717–3723. [Google Scholar] [CrossRef]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Chin, I.-S.; Murad, A.M.A.; Mahadi, N.M.; Nathan, S.; Bakar, F.D.A. Thermal stability engineering of Glomerella cingulata cutinase. Protein Eng. Des. Sel. 2013, 26, 369–375. [Google Scholar] [CrossRef]
- Shirke, A.N.; Basore, D.; Holton, S.; Su, A.; Baugh, E.; Butterfoss, G.L.; Makhatadze, G.; Bystroff, C.; Gross, R.A. Influence of surface charge, binding site residues and glycosylation on Thielavia terrestris cutinase biochemical characteristics. Appl. Microbiol. Biotechnol. 2016, 100, 4435–4446. [Google Scholar] [CrossRef] [PubMed]
- Shirke, A.N.; White, C.; Englaender, J.A.; Zwarycz, A.; Butterfoss, G.L.; Linhardt, R.J.; Gross, R.A. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: Mechanism and effect on PET hydrolysis. Biochemistry 2018, 57, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Shirke, A.N.; Su, A.; Jones, J.A.; Butterfoss, G.L.; Koffas, M.A.G.; Kim, J.R.; Gross, R.A. Comparative thermal inactivation analysis of Aspergillus oryzae and Thiellavia terrestris cutinase: Role of glycosylation. Biotechnol. Bioeng. 2017, 114, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Hanapi, W.N.W.; Iuan-Sheau, C.; Mahadi, N.M.; Murad, A.M.A.; Bakar, F.D.A. Site-saturation mutagenesis of Glomerella cingulata cutinase gene for enhanced enzyme thermostability. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2015; Volume 1678, p. 030021. [Google Scholar]
- Then, J.; Wei, R.; Oeser, T.; Barth, M.; Belisário-Ferrari, M.R.; Schmidt, J.; Zimmermann, W. Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolases from Thermobifida fusca. Biotechnol. J. 2015, 10, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Kiyota, Y.; Miyazaki, M. Techniques for peparation of cross-linked enzyme aggregates and their applications in bioconversions. Catalysts 2018, 8, 174. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Silva, C.; Martins, M.; Jing, S.; Fu, J.; Cavaco-Paulo, A. Practical insights on enzyme stabilization. Crit. Rev. Biotechnol. 2018, 38, 335–350. [Google Scholar] [CrossRef]
- Sousa, I.T.; Lourenço, N.M.T.; Afonso, C.A.M.; Taipa, M.A. Protein stabilization with a dipeptide-mimic triazine-scaffolded synthetic affinity ligand. J. Mol. Recognit. 2013, 26, 104–112. [Google Scholar] [CrossRef]
- Zhang, C.; Zeng, G.; Huang, D.; Lai, C.; Huang, C.; Li, N.; Xu, P.; Cheng, M.; Zhou, Y.; Tang, W.; et al. Combined removal of di(2-ethylhexyl)phthalate (DEHP) and Pb( ii ) by using a cutinase loaded nanoporous gold-polyethyleneimine adsorbent. RSC Adv. 2014, 4, 55511–55518. [Google Scholar] [CrossRef]
- Su, A.; Shirke, A.; Baik, J.; Zou, Y.; Gross, R. Immobilized cutinases: Preparation, solvent tolerance and thermal stability. Enzyme Microb. Technol. 2018, 116, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Kumari, V.; Kumar, S.; Kaur, I.; Bhalla, T.C. Graft copolymerization of acrylamide on chitosan-co-chitin and its application for immobilization of Aspergillus sp. RL2Ct cutinase. Bioorg. Chem. 2017, 70, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Vastano, M.; Quartinello, F.; Herrero Acero, E.; Guebitz, G.M. His-tag immobilization of cutinase 1 from Thermobifida cellulosilytica for solvent-free synthesis of polyesters. Biotechnol. J. 2017, 12, 1700322. [Google Scholar] [CrossRef] [PubMed]
- Muley, A.B.; Chaudhari, S.A.; Singhal, R.S. Non-covalent conjugation of cutinase from Fusarium sp. ICT SAC1 with pectin for enhanced stability: Process minutiae, kinetics, thermodynamics and structural study. Int. J. Biol. Macromol. 2017, 102, 729–740. [Google Scholar] [CrossRef]
- Muley, A.B.; Chaudhari, S.A.; Mulchandani, K.H.; Singhal, R.S. Extraction and characterization of chitosan from prawn shell waste and its conjugation with cutinase for enhanced thermo-stability. Int. J. Biol. Macromol. 2018, 111, 1047–1058. [Google Scholar] [CrossRef]
- Hegde, K.; Veeranki, V. Studies on immobilization of cutinases from Thermobifida fusca on glutaraldehyde activated chitosan beads. Br. Biotechnol. J. 2014, 4, 1049–1063. [Google Scholar] [CrossRef]
- Sipponen, M.H.; Farooq, M.; Koivisto, J.; Pellis, A.; Seitsonen, J.; Österberg, M. Spatially confined lignin nanospheres for biocatalytic ester synthesis in aqueous media. Nat. Commun. 2018, 9, 2300. [Google Scholar] [CrossRef]
- Chaudhari, S.A.; Singhal, R.S. A strategic approach for direct recovery and stabilization of Fusarium sp. ICT SAC1 cutinase from solid state fermented broth by carrier free cross-linked enzyme aggregates. Int. J. Biol. Macromol. 2017, 98, 610–621. [Google Scholar] [CrossRef]
- Nikolaivits, E.; Makris, G.; Topakas, E. Immobilization of a cutinase from Fusarium oxysporum and application in pineapple flavor synthesis. J. Agric. Food Chem. 2017, 65, 3505–3511. [Google Scholar] [CrossRef]
- Koshti, R.; Mehta, L.; Samarth, N. Biological recycling of polyethylene terephthalate: A mini-review. J. Polym. Environ. 2018, 26, 3520–3529. [Google Scholar] [CrossRef]
- Katiyar, V.; Gaur, S.S.; Pal, A.K.; Kumar, A. Properties of plastics for packaging applications. In Polymers for Packaging Applications; Alavi, S., Thomas, S., Sandeep, K.P., Kalarikkal, N., Varghese, J., Yaragalla, S., Eds.; Apple Academic Press, Inc.: Palm Bay, FL, USA, 2015; pp. 3–38. [Google Scholar]
- Lyu, M.-Y.; Choi, T.G. Research trends in polymer materials for use in lightweight vehicles. Int. J. Precis. Eng. Manuf. 2015, 16, 213–220. [Google Scholar] [CrossRef]
- Hirschler, M.M. Flame retardants and heat release: Review of data on individual polymers. Fire Mater. 2015, 39, 232–258. [Google Scholar] [CrossRef]
- Vecchiato, S.; Ahrens, J.; Pellis, A.; Scaini, D.; Mueller, B.; Herrero Acero, E.; Guebitz, G.M. Enzymatic functionalization of HMLS-polyethylene terephthalate fabrics improves the adhesion to rubber. ACS Sustain. Chem. Eng. 2017, 5, 6456–6465. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed]
- Awaja, F.; Pavel, D. Recycling of PET. Eur. Polym. J. 2005, 41, 1453–1477. [Google Scholar] [CrossRef]
- Ferrario, V.; Pellis, A.; Cespugli, M.; Guebitz, G.; Gardossi, L.; Ferrario, V.; Pellis, A.; Cespugli, M.; Guebitz, G.M.; Gardossi, L. Nature inspired solutions for polymers: Will cutinase enzymes make polyesters and polyamides greener? Catalysts 2016, 6, 205. [Google Scholar] [CrossRef]
- Pan, W.; Bai, Z.; Su, T.; Wang, Z. Enzymatic degradation of poly(butylene succinate) with different molecular weights by cutinase. Int. J. Biol. Macromol. 2018, 111, 1040–1046. [Google Scholar] [CrossRef]
- Hu, X.; Su, T.; Pan, W.; Li, P.; Wang, Z. Difference in solid-state properties and enzymatic degradation of three kinds of poly(butylene succinate)/cellulose blends. RSC Adv. 2017, 7, 35496–35503. [Google Scholar] [CrossRef]
- Bai, Z.; Liu, Y.; Su, T.; Wang, Z.; Bai, Z.; Liu, Y.; Su, T.; Wang, Z. Effect of hydroxyl monomers on the enzymatic degradation of poly(ethylene succinate), poly(butylene succinate), and poly(hexylene succinate). Polymers (Basel) 2018, 10, 90. [Google Scholar] [CrossRef]
- Watanabe, T.; Shinozaki, Y.; Yoshida, S.; Koitabashi, M.; Sameshima-Yamashita, Y.; Fujii, T.; Fukuoka, T.; Kitamoto, H.K. Xylose induces the phyllosphere yeast Pseudozyma antarctica to produce a cutinase-like enzyme which efficiently degrades biodegradable plastics. J. Biosci. Bioeng. 2014, 117, 325–329. [Google Scholar] [CrossRef]
- Abdel-Motaal, F.F.; El-Sayed, M.A.; El-Zayat, S.A.; Ito, S. Biodegradation of poly (ε-caprolactone) (PCL) film and foam plastic by Pseudozyma japonica sp. nov., a novel cutinolytic ustilaginomycetous yeast species. 3 Biotech 2014, 4, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Ping, L.-F.; Chen, X.-Y.; Yuan, X.-L.; Zhang, M.; Chai, Y.-J.; Shan, S.-D. Application and comparison in biosynthesis and biodegradation by Fusarium solani and Aspergillus fumigatus cutinases. Int. J. Biol. Macromol. 2017, 104, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Adıgüzel, A.O.; Tunçer, M. Purification and characterization of cutinase from Bacillus sp. KY0701 isolated from plastic wastes. Prep. Biochem. Biotechnol. 2017, 47, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Gamerith, C.; Zartl, B.; Pellis, A.; Guillamot, F.; Marty, A.; Acero, E.H.; Guebitz, G.M. Enzymatic recovery of polyester building blocks from polymer blends. Process Biochem. 2017, 59, 58–64. [Google Scholar] [CrossRef]
- Pellis, A.; Gamerith, C.; Ghazaryan, G.; Ortner, A.; Herrero Acero, E.; Guebitz, G.M. Ultrasound-enhanced enzymatic hydrolysis of poly(ethylene terephthalate). Bioresour. Technol. 2016, 218, 1298–1302. [Google Scholar] [CrossRef] [PubMed]
- Alves, N.M.; Mano, J.F.; Balaguer, E.; Meseguer Dueñas, J.M.; Gómez Ribelles, J.L. Glass transition and structural relaxation in semi-crystalline poly(ethylene terephthalate): A DSC study. Polymer (Guildf) 2002, 43, 4111–4122. [Google Scholar] [CrossRef]
- Barth, M.; Oeser, T.; Wei, R.; Then, J.; Schmidt, J.; Zimmermann, W. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem. Eng. J. 2015, 93, 222–228. [Google Scholar] [CrossRef]
- Castro, A.M.D.; Carniel, A. A novel process for poly(ethylene terephthalate) depolymerization via enzyme-catalyzed glycolysis. Biochem. Eng. J. 2017, 124, 64–68. [Google Scholar] [CrossRef]
- Castro, A.M.D.; Carniel, A.; Sirelli, L.; Dias, M.L.; Menezes, S.M.C.D.; Chinelatto Junior, L.S.; Honorato, H.D.A. Enzyme-catalyzed simultaneous hydrolysis-glycolysis reactions reveals tunability on PET depolymerization products. Biochem. Eng. J. 2018, 137, 239–246. [Google Scholar] [CrossRef]
- Weinberger, S.; Canadell, J.; Quartinello, F.; Yeniad, B.; Arias, A.; Pellis, A.; Guebitz, G.; Weinberger, S.; Canadell, J.; Quartinello, F.; et al. Enzymatic degradation of poly(ethylene 2,5-furanoate) powders and amorphous films. Catalysts 2017, 7, 318. [Google Scholar] [CrossRef]
- Weinberger, S.; Haernvall, K.; Scaini, D.; Ghazaryan, G.; Zumstein, M.T.; Sander, M.; Pellis, A.; Guebitz, G.M. Enzymatic surface hydrolysis of poly(ethylene furanoate) thin films of various crystallinities. Green Chem. 2017, 19, 5381–5384. [Google Scholar] [CrossRef]
- Schmidt, J.; Wei, R.; Oeser, T.; Dedavid e Silva, L.; Breite, D.; Schulze, A.; Zimmermann, W.; Schmidt, J.; Wei, R.; Oeser, T.; et al. Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers (Basel) 2017, 9, 65. [Google Scholar] [CrossRef]
- Hou, S.; Hoyle, D.M.; Blackwell, C.J.; Haernvall, K.; Perz, V.; Guebitz, G.M.; Khosravi, E. Hydrolytic degradation of ROMP thermosetting materials catalysed by bio-derived acids and enzymes: From networks to linear materials. Green Chem. 2016, 18, 5190–5199. [Google Scholar] [CrossRef]
- Haernvall, K.; Zitzenbacher, S.; Biundo, A.; Yamamoto, M.; Schick, M.B.; Ribitsch, D.; Guebitz, G.M. Enzymes as enhancers for the biodegradation of synthetic polymers in wastewater. ChemBioChem 2018, 19, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Whiteford, J.R.; Spanu, P.D. Hydrophobins and the interactions between fungi and plants. Mol. Plant Pathol. 2002, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Ribitsch, D.; Herrero Acero, E.; Przylucka, A.; Zitzenbacher, S.; Marold, A.; Gamerith, C.; Tscheließnig, R.; Jungbauer, A.; Rennhofer, H.; Lichtenegger, H.; et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl. Environ. Microbiol. 2015, 81, 3586–3592. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, Y.; Kim, Y.-K.; Tanaka, T.; Nanatani, K.; Takahashi, T.; Abe, K. Asp30 of Aspergillus oryzae cutinase CutL1 is involved in the ionic interaction with fungal hydrophobin RolA. Biosci. Biotechnol. Biochem. 2017, 81, 1363–1368. [Google Scholar] [CrossRef]
- Takahashi, T.; Tanaka, T.; Tsushima, Y.; Muragaki, K.; Uehara, K.; Takeuchi, S.; Maeda, H.; Yamagata, Y.; Nakayama, M.; Yoshimi, A.; et al. Ionic interaction of positive amino acid residues of fungal hydrophobin RolA with acidic amino acid residues of cutinase CutL1. Mol. Microbiol. 2015, 96, 14–27. [Google Scholar] [CrossRef]
- Tanaka, T.; Nakayama, M.; Takahashi, T.; Nanatani, K.; Yamagata, Y.; Abe, K. Analysis of the ionic interaction between the hydrophobin RodA and two cutinases of Aspergillus nidulans obtained via an Aspergillus oryzae expression system. Appl. Microbiol. Biotechnol. 2017, 101, 2343–2356. [Google Scholar] [CrossRef]
- Ribitsch, D.; Yebra, A.O.; Zitzenbacher, S.; Wu, J.; Nowitsch, S.; Steinkellner, G.; Greimel, K.; Doliska, A.; Oberdorfer, G.; Gruber, C.C.; et al. Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis. Biomacromolecules 2013, 14, 1769–1776. [Google Scholar] [CrossRef]
- Wei, R.; Oeser, T.; Schmidt, J.; Meier, R.; Barth, M.; Then, J.; Zimmermann, W. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol. Bioeng. 2016, 113, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Kawai, F.; Oda, M.; Tamashiro, T.; Waku, T.; Tanaka, N.; Yamamoto, M.; Mizushima, H.; Miyakawa, T.; Tanokura, M. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2014, 98, 10053–10064. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Chen, J.; Wu, J. Enhanced activity toward PET by site-directed mutagenesis of Thermobifida fusca cutinase–CBM fusion protein. Carbohydr. Polym. 2013, 97, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; He, L.; Wang, L.; Li, T.; Li, C.; Liu, H.; Luo, Y.; Bao, R. Protein crystallography and site-direct mutagenesis analysis of the poly(ethylene terephthalate) hydrolase PETase from Ideonella sakaiensis. ChemBioChem 2018, 19, 1471–1475. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Gao, Z.; Wang, Z.; Su, T.; Yang, L.; Li, P. Enzymatic degradation of poly(butylene succinate) by cutinase cloned from Fusarium solani. Polym. Degrad. Stab. 2016, 134, 211–219. [Google Scholar] [CrossRef]
- Groß, C.; Hamacher, K.; Schmitz, K.; Jager, S. Cleavage product accumulation decreases the activity of cutinase during PET hydrolysis. J. Chem. Inf. Model. 2017, 57, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Carniel, A.; Valoni, É.; Nicomedes, J.; Gomes, A.D.C.; Castro, A.M. de Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]
- De Castro, A.M.; Carniel, A.; Nicomedes Junior, J.; da Conceição Gomes, A.; Valoni, É. Screening of commercial enzymes for poly(ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J. Ind. Microbiol. Biotechnol. 2017, 44, 835–844. [Google Scholar] [CrossRef]
- Barth, M.; Honak, A.; Oeser, T.; Wei, R.; Belisário-Ferrari, M.R.; Then, J.; Schmidt, J.; Zimmermann, W. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnol. J. 2016, 11, 1082–1087. [Google Scholar] [CrossRef]
- Schmidt, J.; Wei, R.; Oeser, T.; Belisário-Ferrari, M.R.; Barth, M.; Then, J.; Zimmermann, W. Effect of Tris, MOPS, and phosphate buffers on the hydrolysis of polyethylene terephthalate films by polyester hydrolases. FEBS Open Bio 2016, 6, 919–927. [Google Scholar] [CrossRef]
- Saxena, A.; Singh Chauhan, P. Role of various enzymes for deinking paper: A review. Crit. Rev. Biotechnol. 2017, 37, 598–612. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, X.; Zhang, G.; Chen, J.; Sang, M.; Long, Z.; Wang, B. Studies on the environmentally friendly deinking process employing biological enzymes and composite surfactant. Cellulose 2018, 25, 3079–3089. [Google Scholar] [CrossRef]
- Hong, R.; Su, L.; Chen, S.; Long, Z.; Wu, J. Comparison of cutinases in enzymic deinking of old newsprint. Cellulose 2017, 24, 5089–5099. [Google Scholar] [CrossRef]
- Liu, M.; Yang, S.; Long, L.; Cao, Y.; Ding, S. Engineering a chimeric lipase-cutinase (Lip-Cut) for efficient enzymatic deinking of waste paper. BioResources 2017, 13, 981–996. [Google Scholar] [CrossRef]
- Hong, R.; Su, L.; Wu, J. Cutinases catalyze polyacrylate hydrolysis and prevent their aggregation. Polym. Degrad. Stab. 2019, 159, 23–30. [Google Scholar] [CrossRef]
- Gao, D.-W.; Wen, Z.-D. Phthalate esters in the environment: A critical review of their occurrence, biodegradation, and removal during wastewater treatment processes. Sci. Total Environ. 2016, 541, 986–1001. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Min, J.; Bae, K.-D.; Gu, M.B.; Lee, J. Biodegradation of dipropyl phthalate and toxicity of its degradation products: A comparison of Fusarium oxysporum f. sp. pisi cutinase and Candida cylindracea esterase. Arch. Microbiol. 2005, 184, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Lee, J. Enzymatic degradation of dibutyl phthalate and toxicity of its degradation products. Biotechnol. Lett. 2005, 27, 635–639. [Google Scholar] [CrossRef]
- Ahn, J.-Y.; Kim, Y.-H.; Min, J.; Lee, J. Accelerated degradation of dipentyl phthalate by Fusarium oxysporum f. sp. pisi cutinase and toxicity evaluation of its degradation products using bioluminescent bacteria. Curr. Microbiol. 2006, 52, 340–344. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Seo, H.-S.; Min, J.; Kim, Y.-C.; Ban, Y.-H.; Han, K.Y.; Park, J.-S.; Bae, K.-D.; Gu, M.B.; Lee, J. Enhanced degradation and toxicity reduction of dihexyl phthalate by Fusarium oxysporum f. sp. pisi cutinase. J. Appl. Microbiol. 2007, 102, 221–228. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Lee, J.; Ahn, J.-Y.; Gu, M.B.; Moon, S.-H. Enhanced degradation of an endocrine-disrupting chemical, butyl benzyl phthalate, by Fusarium oxysporum f. sp. pisi cutinase. Appl. Environ. Microbiol. 2002, 68, 4684–4688. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Lee, J.; Moon, S.-H. Degradation of an endocrine disrupting chemical, DEHP [di-(2-ethylhexyl)-phthalate], by Fusarium oxysporum f. sp. pisi cutinase. Appl. Microbiol. Biotechnol. 2003, 63, 75–80. [Google Scholar] [CrossRef]
- Viksoe-Nielsen, A.; Hauerbach Soerensen, B. Cutinase for Detoxification of Feed Products 2008. WO/2009/080701, 20 July 2009. [Google Scholar]
- Viskoe-Nielsen, A.; Hauerbach Soerensen, B. Detoxification of Feed Products 2009. WO/2009/109607, 11 September 2009. [Google Scholar]
- Kim, Y.-H.; Ahn, J.-Y.; Moon, S.-H.; Lee, J. Biodegradation and detoxification of organophosphate insecticide, malathion by Fusarium oxysporum f. sp. pisi cutinase. Chemosphere 2005, 60, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Kawai, F.; Kawase, T.; Shiono, T.; Urakawa, H.; Sukigara, S.; Tu, C.; Yamamoto, M. Enzymatic hydrophilization of polyester fabrics using a recombinant cutinase Cut 190 and their surface characterization. J. Fiber Sci. Technol. 2017, 73, 8–18. [Google Scholar] [CrossRef]
- Kanelli, M.; Vasilakos, S.; Nikolaivits, E.; Ladas, S.; Christakopoulos, P.; Topakas, E. Surface modification of poly(ethylene terephthalate) (PET) fibers by a cutinase from Fusarium oxysporum. Process Biochem. 2015, 50, 1885–1892. [Google Scholar] [CrossRef]
- Ruoming, Y.; Qiang, W.; Xuerong, F.; Yuanyuan, Y.; Jiugang, Y.; Ping, W. A new model substrate for cutinase hydrolyzing polyethylene terephthalate. Fibers Polym. 2013, 14, 1128–1133. [Google Scholar]
- Silva, C.; Da, S.; Silva, N.; Matamá, T.; Araújo, R.; Martins, M.; Chen, S.; Chen, J.; Wu, J.; Casal, M.; Cavaco-Paulo, A. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates. Biotechnol. J. 2011, 6, 1230–1239. [Google Scholar] [CrossRef]
- Tkavc, T.; Vesel, A.; Acero, E.H.; Fras Zemljič, L. Comparison of oxygen plasma and cutinase effect on polyethylene terephthalate surface. J. Appl. Polym. Sci. 2013, 128, 3570–3575. [Google Scholar] [CrossRef]
- Panseri, S.; Martino, P.A.; Cagnardi, P.; Celano, G.; Tedesco, D.; Castrica, M.; Balzaretti, C.; Chiesa, L.M. Feasibility of biodegradable based packaging used for red meat storage during shelf-life: A pilot study. Food Chem. 2018, 249, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Cayla, A.; Rault, F.; Giraud, S.; Salaün, F.; Fierro, V.; Celzard, A.; Cayla, A.; Rault, F.; Giraud, S.; Salaün, F.; et al. PLA with intumescent system containing lignin and ammonium polyphosphate for flame retardant textile. Polymers (Basel) 2016, 8, 331. [Google Scholar] [CrossRef]
- Sabek, O.M.; Farina, M.; Fraga, D.W.; Afshar, S.; Ballerini, A.; Filgueira, C.S.; Thekkedath, U.R.; Grattoni, A.; Gaber, A.O. Three-dimensional printed polymeric system to encapsulate human mesenchymal stem cells differentiated into islet-like insulin-producing aggregates for diabetes treatment. J. Tissue Eng. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Cosme, J.G.L.; Xu, T.; Miszuk, J.M.; Picciani, P.H.S.; Fong, H.; Sun, H. Three dimensional electrospun PCL/PLA blend nanofibrous scaffolds with significantly improved stem cells osteogenic differentiation and cranial bone formation. Biomaterials 2017, 115, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Silvestrini, L.; Scaini, D.; Coburn, J.M.; Gardossi, L.; Kaplan, D.L.; Herrero Acero, E.; Guebitz, G.M. Enzyme-catalyzed functionalization of poly(L-lactic acid) for drug delivery applications. Process Biochem. 2017, 59, 77–83. [Google Scholar] [CrossRef]
- Ortner, A.; Pellis, A.; Gamerith, C.; Orcal Yebra, A.; Scaini, D.; Kaluzna, I.; Mink, D.; de Wildeman, S.; Herrero Acero, E.; Guebitz, G.M. Superhydrophobic functionalization of cutinase activated poly(lactic acid) surfaces. Green Chem. 2017, 19, 816–822. [Google Scholar] [CrossRef]
- Zaks, A.; Klibanov, A.M. Enzymatic catalysis in organic media at 100 degrees C. Science 1984, 224, 1249–1251. [Google Scholar] [CrossRef] [PubMed]
- De Barros, D.P.C.; Pinto, F.; Fonseca, L.P.; Cabral, J.M.S.; Lemos, F. Kinetic model for the esterification of ethyl caproate for reaction optimization. J. Mol. Catal. B Enzym. 2014, 101, 16–22. [Google Scholar] [CrossRef]
- Dutta, K.; Hegde, K.; Dasu, V.V. Synthesis of methyl esters by transesterification catalyzed by cutinase from Pseudomonas cepacia NRRL B 2320 and kinetic analysis. Curr. Trends Biotechnol. Pharm. 2014, 8, 1–10. [Google Scholar]
- Su, L.; Hong, R.; Guo, X.; Wu, J.; Xia, Y. Short-chain aliphatic ester synthesis using Thermobifida fusca cutinase. Food Chem. 2016, 206, 131–136. [Google Scholar] [CrossRef]
- De Barros, D.P.; Pinto, F.; Pfluck, A.C.; Dias, A.S.; Fernandes, P.; Fonseca, L.P. Improvement of enzyme stability for alkyl esters synthesis in miniemulsion systems by using media engineering. J. Chem. Technol. Biotechnol. 2018, 93, 1338–1346. [Google Scholar] [CrossRef]
- Nikolaivits, E.; Norra, G.-F.; Voutsas, E.; Topakas, E. Cutinase from Fusarium oxysporum catalyzes the acylation of tyrosol in an aqueous medium: Optimization and thermodynamic study of the reaction. J. Mol. Catal. B Enzym. 2016, 129, 29–36. [Google Scholar] [CrossRef]
- Douka, A.; Vouyiouka, S.; Papaspyridi, L.-M.; Papaspyrides, C.D. A review on enzymatic polymerization to produce polycondensation polymers: The case of aliphatic polyesters, polyamides and polyesteramides. Prog. Polym. Sci. 2018, 79, 1–25. [Google Scholar] [CrossRef]
- Feder, D.; Gross, R.A. Exploring chain length selectivity in HiC-catalyzed polycondensation reactions. Biomacromolecules 2010, 11, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Hunsen, M.; Azim, A.; Mang, H.; Wallner, S.R.; Ronkvist, A.; Wenchun Xie, A.; Gross, R.A. A cutinase with polyester synthesis activity. Macromolecules 2006, 40, 148–150. [Google Scholar] [CrossRef]
- Hunsen, M.; Abul, A.; Xie, W.; Gross, R. Humicola insolens cutinase-catalyzed lactone ring-opening polymerizations: Kinetic and mechanistic studies. Biomacromolecules 2008, 9, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Pellis, A.; Ferrario, V.; Zartl, B.; Brandauer, M.; Gamerith, C.; Herrero Acero, E.; Ebert, C.; Gardossi, L.; Guebitz, G.M. Enlarging the tools for efficient enzymatic polycondensation: Structural and catalytic features of cutinase 1 from Thermobifida cellulosilytica. Catal. Sci. Technol. 2016, 6, 3430–3442. [Google Scholar] [CrossRef]
- Stavila, E.; Arsyi, R.Z.; Petrovic, D.M.; Loos, K. Fusarium solani pisi cutinase-catalyzed synthesis of polyamides. Eur. Polym. J. 2013, 49, 834–842. [Google Scholar] [CrossRef]
- Stavila, E.; Alberda van Ekenstein, G.O.R.; Loos, K. Enzyme-catalyzed synthesis of aliphatic–aromatic oligoamides. Biomacromolecules 2013, 14, 1600–1606. [Google Scholar] [CrossRef] [PubMed]
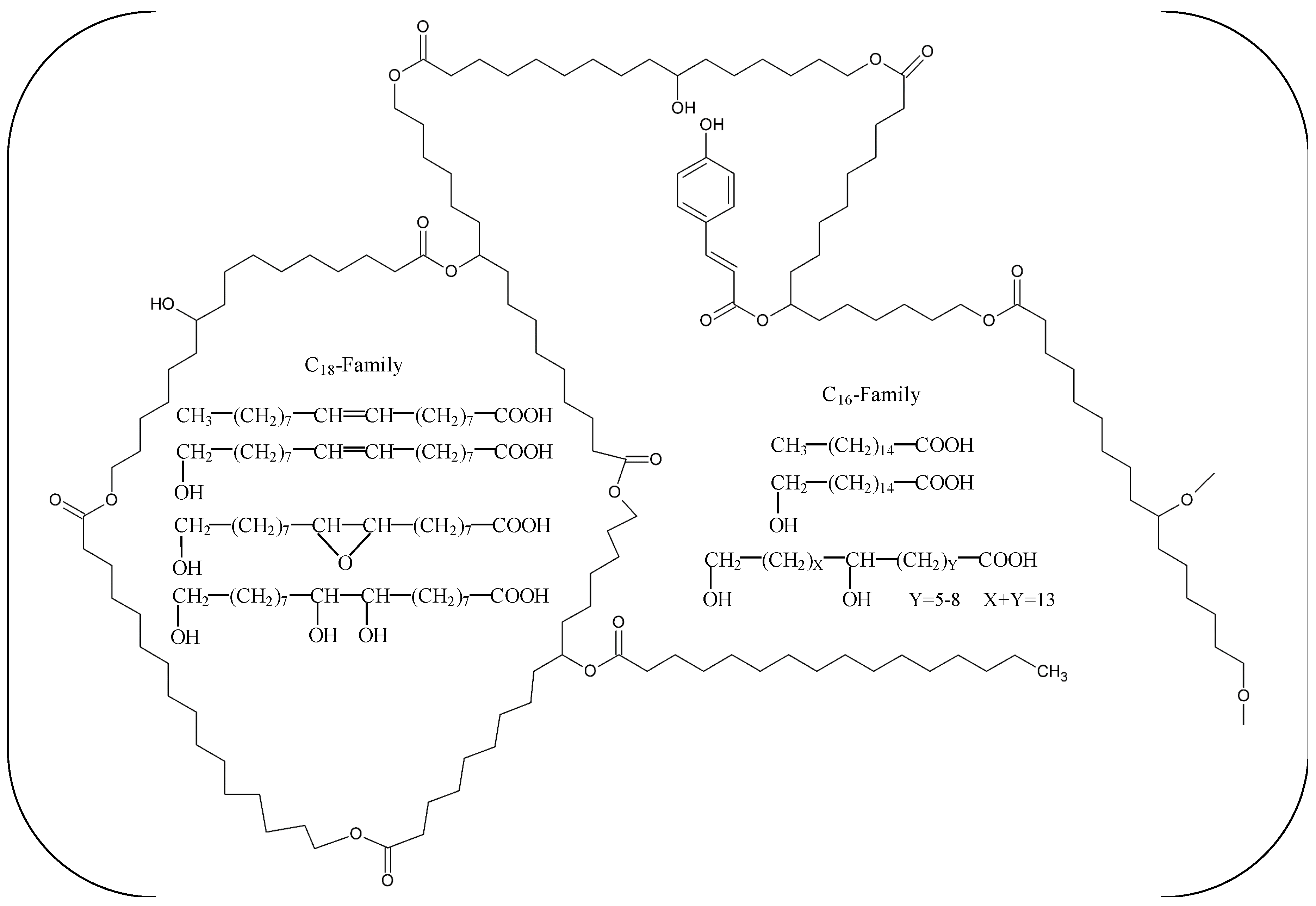
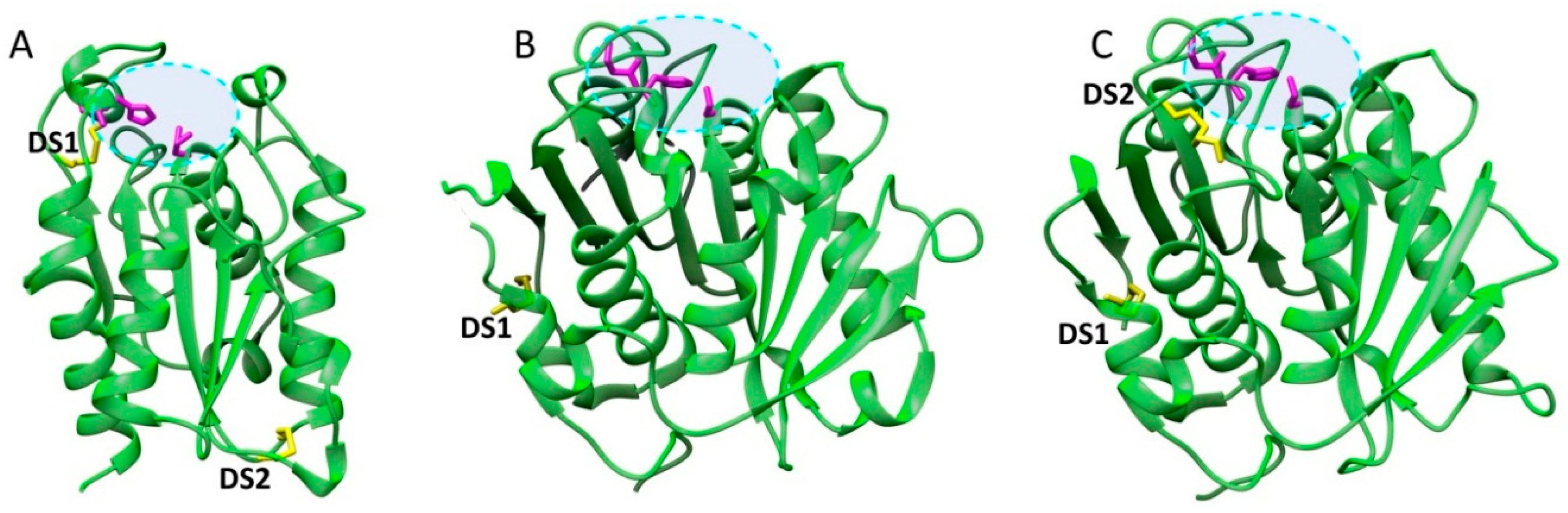
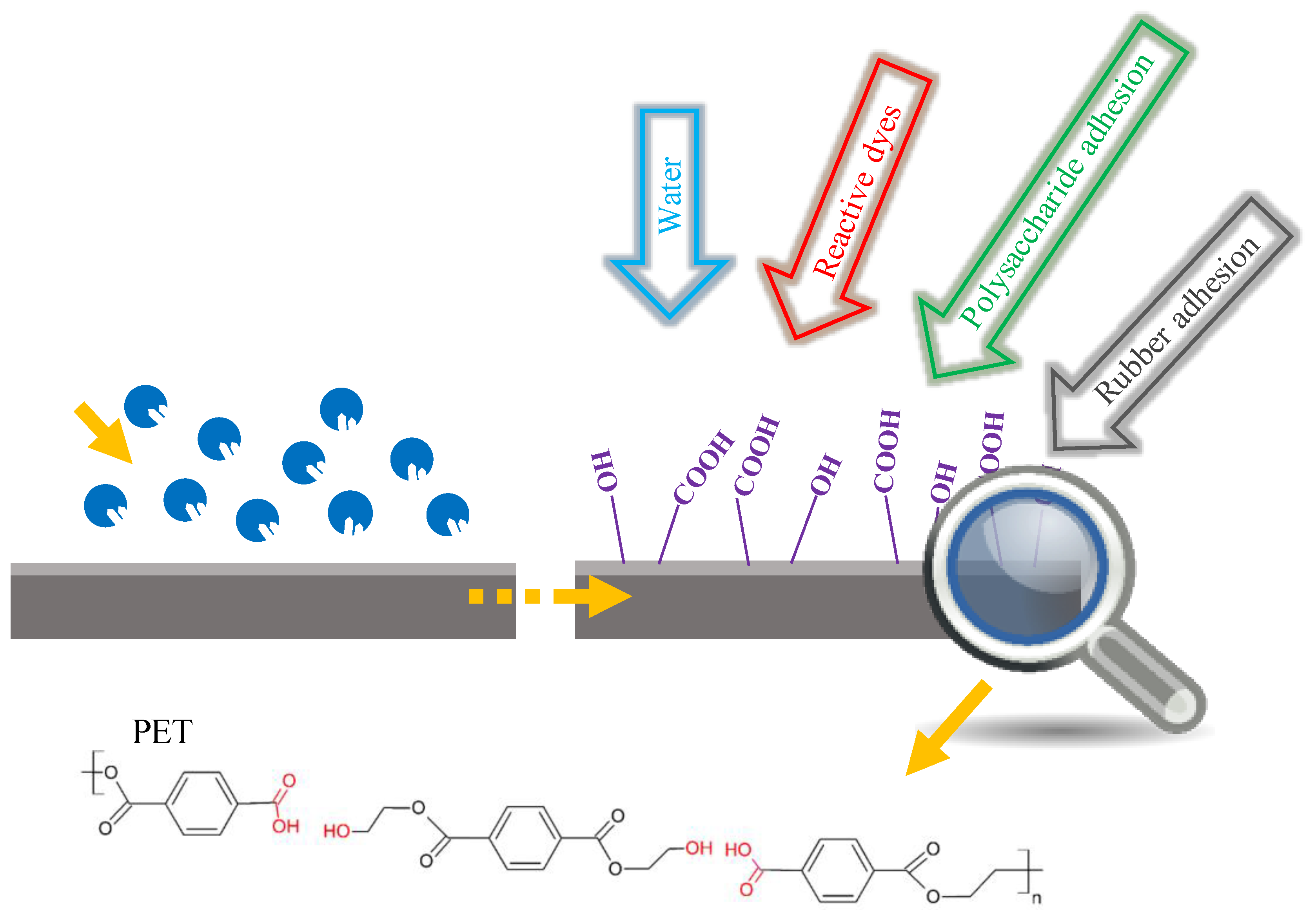
| Name | Origin | Gene Accession Number | Host | MW (kDa) | Topt (°C) | pHopt | pI | Kinetics/Activity on pNPB | pNP-Ester Specificity (kcat/KM) | Thermostability Half-Life (t1/2) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial Sources | |||||||||||
| Tfu_0883 | Thermobifida fusca | YP_288943 | E. coli BL21 Rosetta (DE3) PlysS cells | 29 | 60 | 8 | 458 U/mg | t1/2 (60 °C) = 40 h | [40] | ||
| Tfu_0882 | Thermobifida fusca | YP_288944 | E. coli BL21 Rosetta (DE3) PlysS cells | 29 | 60 | 8 | 223 U/mg | [40] | |||
| Thc_Cut1 | Thermobifida cellulosilytica | HQ147785 | E.coli BL21-Gold (DE3) | 29.4 | 1.48 mM 195.1 s−1 | C2 | [41] | ||||
| Thc_Cut2 | Thermobifida cellulosilytica | HQ147786 | E.coli BL21-Gold (DE3) | 29.7 | 2.13 mM 5.3 s−1 | C2 | [41] | ||||
| Thf42_cut1 | Thermobifida fusca | HQ147787 | E.coli BL21-Gold (DE3) | 29.6 | 2.10 mM 30.9 s−1 | C2 | [41] | ||||
| Tha_Cut1 | Thermobifida alba | HQ147784 | E.coli BL21-Gold (DE3) | 28.1 | 1.93 mΜ 6.03 s−1 | C2 | [42] | ||||
| Est119 | Thermobifida alba AHK119 | est119 | E. coli Rosetta-gami B (DE3) | 30 | 50 | 6 | 2.3 U/mg 3.41 mM 4.48 s−1 1.31 s−1 mM−1 | C6 | [23] | ||
| Cut1 | Thermobifida fusca | JN129499.1 | E. coli BL21 (DE3) | 30.1 | 55 | 8 | 542.5 U/mg 0.13 mM 178 s−1 13.69 s−1 mM−1 | C4 * | Stable at 37 °C | [43] | |
| Cut2 | Thermobifida fusca | JN129500.1 | E. coli BL21 (DE3) | 29.6 | 55 | 8 | 643.4 U/mg 0.09 mM 253 s−1 2840 s−1 mM−1 | C4 * | Stable at 37 °C t1/2 (55 °C) = 30 h | [43] | |
| Tcur1278 | Thermomonospora curvata | HG939554 | E. coli TOP10 | 35 | 60 | 8.5 | 3 U/mg | 80% act at 50 °C and 55 °C for 60 min | [24] | ||
| Tcur0390 | Thermomonospora curvata | HG939555 | E. coli TOP10 | 35 | 55 | 8.5 | 17.9 U/mg | 40% act at 50 °C for 60 min 15% action at 55 °C and 60 °C for 10 min | [24] | ||
| LC-cutinase | Leaf-branch compost metagenome | HQ704839 | E. coli BL21-CodonPlus(DE3)-RP | 29 | 50 | 8.5 | 9.3 | C4 * | t1/2 (60 °C) = 80 min t1/2 (70 °C) = 40 min | [44] | |
| Fungal Sources | |||||||||||
| F. solani cutinase | Fusarium solani | K02640 | Pichia pastoris X-33 | 20 | 40 | 8 | [45] | ||||
| MFCUT1 | Monilinia fructicola | AF305598 | Pichia pastoris | 22 | 8.4 | 40.8 min−1 1.23 μM 5.5·10−4 s−1 mM−1 | [31] | ||||
| CutL1 | Aspergillus oryzae | P52956 | WT | 21.6 | 50 | 9 | 0.22 mM 18 s−1 81.82 s−1 mM−1 | C5 | Stable at 40 °C for 30 min | [29] | |
| THCUT1 | Trichoderma harzianum | AJ896891 | Pichia pastoris GS115 | 29 | 7.5–8 | 4.2 | 0.74 U/mg 0.57 mM | C2 | [46] | ||
| Aspergillus oryzae | Pichia pastoris | 0.21 μM 5.8·10−5 s−1 mM−1 | C4 | [47] | |||||||
| CUTAB1 | Alternaria brassicicola | U03393.1 | Pichia pastoris X-33 | 24 | 40 | 7–9 | 5.2–7 | 1057 U/mg | C4 * | Stable at 40 °C | [48] |
| HiC | Humicola insolens | hic | Pichia pastoris X-33 | 50 | 0.45 mM 191 s−1 424.4 s−1 mM−1 | C8 | Stable at 50 °C for 48 h t1/2 (85 °C) = 1 h | [49] | |||
| TtcutA | Thielavia terrestris | WT | 25.3 | 50 | 4 | 1200 U/mg 1 mM 0.62 s−1 0.62 s−1 mM−1 | C4 | Stable at 65 °C for 30 min | [26] | ||
| AnCut5 | Aspergillus niger | anig5 | Pichia pastoris X-33 | 22.8 | 6 | [33] | |||||
| CmCut1 | Cryptococcus magnus | WT | 21 | 40 | 7.5 | C12 * | [27] | ||||
| PaE | Pseudozyma antarctica | DM067526 | Saccharomyces cerevisiae | 20.4 | 40 | 9.5 | ~220 U/mg | t1/2 (40 °C) = ~1 h | [17] | ||
| G. cingulata cutinase | Glomerella cingulata | AF444194 | Pichia pastoris X-33 | 25 | 25 | 8 | C8 | [50] | |||
| Tr cutinase | Trichoderma reesei | tre60489 v2.0 | Trichoderma reesei | 23.8 | 6 | 57 U/mg | 80% act after 20 h at 50 °C or 1 h at 60 °C | [18] | |||
| ScCut1 | Sirococcus conigenus | KF193402 | Pichia pastoris X-33 | 20 | 4.7–5.2 | 1.7 mM | C2 * | t1/2 (55 °C) = 9.5 h t1/2 (85 °C) = 25 min | [34] | ||
| PCLE | Paraphoma-related strain | AB823702 | WT | 19.7 | 45 | 7.2 | ~13 U/mg | C5 * | [28] | ||
| TtcutB | Thielavia terrestris | WT | 27.3 | 55 | 4 | 983 U/mg 1.1 mM 0.85 s−1 0.77 s−1 mM−1 | C4 | t1/2 (70 °C) = 51 min t1/2 (80 °C) = 49 min | [25] | ||
| FoCut5a | Fusarium oxysporum | foqg_13916.1 | E. coli BL21 (DE3) | 23 | 40 | 8 | 7.9 | 0.7 mM 111.9 s−1 152.5 s−1 mM−1 | C4 | Deactivation after 2 h at 35 °C | [21] |
| Acut1-6hp | Arxula adeninivorans | LN828946 | A. adeninivorans G1212 | 21.6 | 20 | 5 | 66.1 U/mg 1.6 mM | C6 * | Stable at 25 °C for 2 h | [19] | |
| Acut2-6hp | Arxula adeninivorans | LN828947 | A. adeninivorans G1212 | 21.6 | 30 | 5 | 1747 U/mg 1.5 mM | C6 * | Stable at 40 °C for 2 h | [19] | |
| Acut3-6hp | Arxula adeninivorans | LN828948 | A. adeninivorans G1212 | 29.2 | 30 | 5.5 | 1251 U/mg 1.9 mM | C4 * | Stable at 50 °C for 4 h | [19] | |
| ANCUT2 | Aspergillus nidulans | ABF50887 (protein) | WT | 29 | 60 | 9 | 5.05 | ~350 U/mg | C2 * | 60% act after 1 h at 60 °C | [30] |
| McCut | Malbranchea cinnamomea | KY568910.1 | Pichia pastoris GS115 | 21.9 | 45 | 8 | 1147.9 U/mg 0.66 mM 0.46 s−1 0.70 s−1 mM−1 | C6 | t1/2 (80 °C) = 92 min t1/2 (85 °C) = 67 min | [51] | |
| MtCUT | Myceliophthora thermophila | XP_003663956.1 | Pichia pastoris KM71H | 23.4 | 30 | 8.5 | 2.34 mM | C4 | Stable at 25 °C for 20 min | [52] | |
| No | PDB ID | Microorganism | Reference | |
|---|---|---|---|---|
| Fungal cutinases | 1 | 1CUS | Fusarium solani pisi | [35] |
| 2 | 2CZQ | Cryptococcus sp. | [53] | |
| 3 | 3DCN | Glomerella cingulata | [54] | |
| 4 | 3GBS | Aspergillus oryzae | [47] | |
| 5 | 4OYY | Humicola insolens | [55] | |
| 6 | 4PSC | Trichoderma reesei | [18] | |
| 7 | 5AJH | Fusarium oxysporum | [21] | |
| 8 | 5X88 | Malbranchea cinnamomea | [51] | |
| Bacterial cutinases | 1 | 3VIS | Thermobifida alba | [56] |
| 2 | 4EBO | Leaf branch compost bacterial cutinase | [57] | |
| 3 | 4CG1 | Thermobifida fusca | [38] | |
| 4 | 4WFI | Saccharomonospora viridis | [58] | |
| 5 | 5LUI | Thermobifida cellulosilytica (Thc_Cut1) | [59] | |
| 6 | 5LUJ | T. cellulosilytica (Thc_Cut2) | [59] | |
| 7 | 5XG0 | Ideonella sakaiensis | [39] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolaivits, E.; Kanelli, M.; Dimarogona, M.; Topakas, E. A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases. Catalysts 2018, 8, 612. https://doi.org/10.3390/catal8120612
Nikolaivits E, Kanelli M, Dimarogona M, Topakas E. A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases. Catalysts. 2018; 8(12):612. https://doi.org/10.3390/catal8120612
Chicago/Turabian StyleNikolaivits, Efstratios, Maria Kanelli, Maria Dimarogona, and Evangelos Topakas. 2018. "A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases" Catalysts 8, no. 12: 612. https://doi.org/10.3390/catal8120612
APA StyleNikolaivits, E., Kanelli, M., Dimarogona, M., & Topakas, E. (2018). A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases. Catalysts, 8(12), 612. https://doi.org/10.3390/catal8120612






