Phenolic Acid Decarboxylase for Carbon Dioxide Fixation: Mining, Biochemical Characterization, and Regioselective Enzymatic β-carboxylation of para-hydroxystyrene Derivatives
Abstract
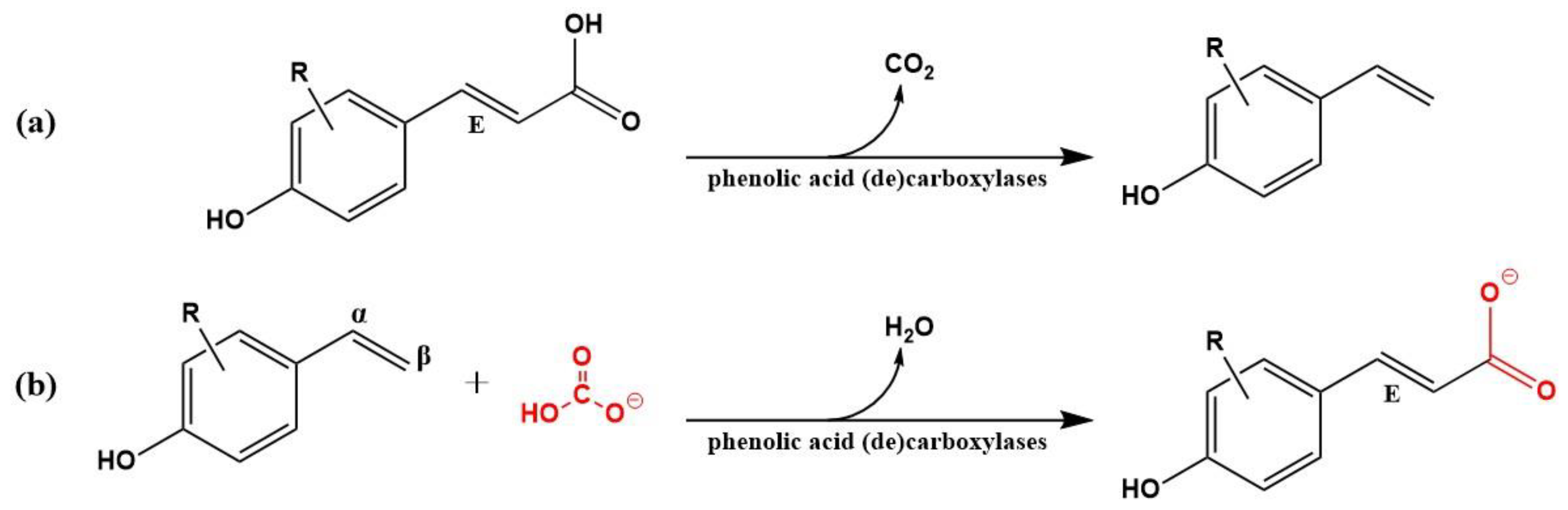
1. Introduction
2. Results and Discussion
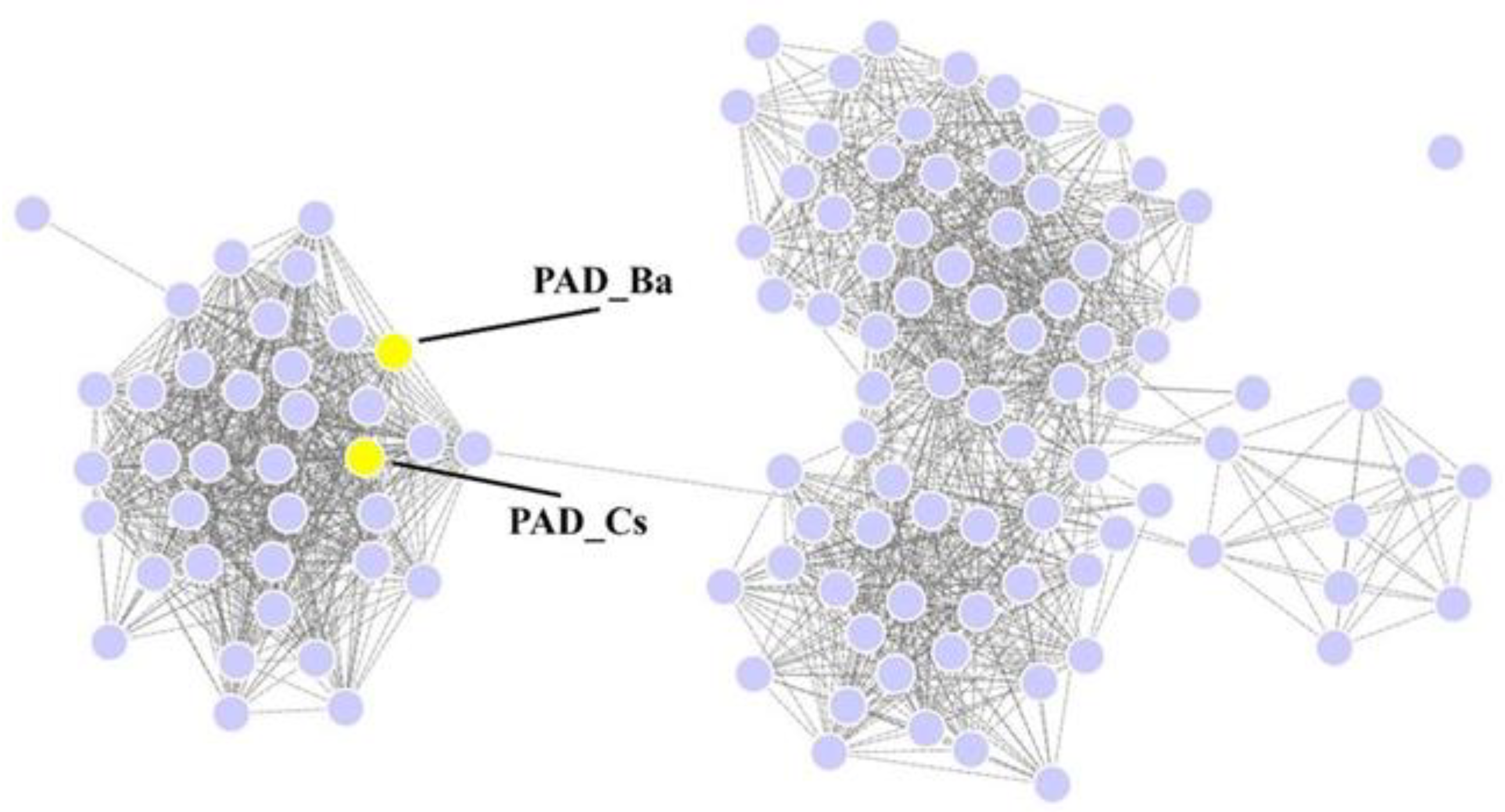
2.1. Mining of Amino Acid Sequences for (de)Carboxylase
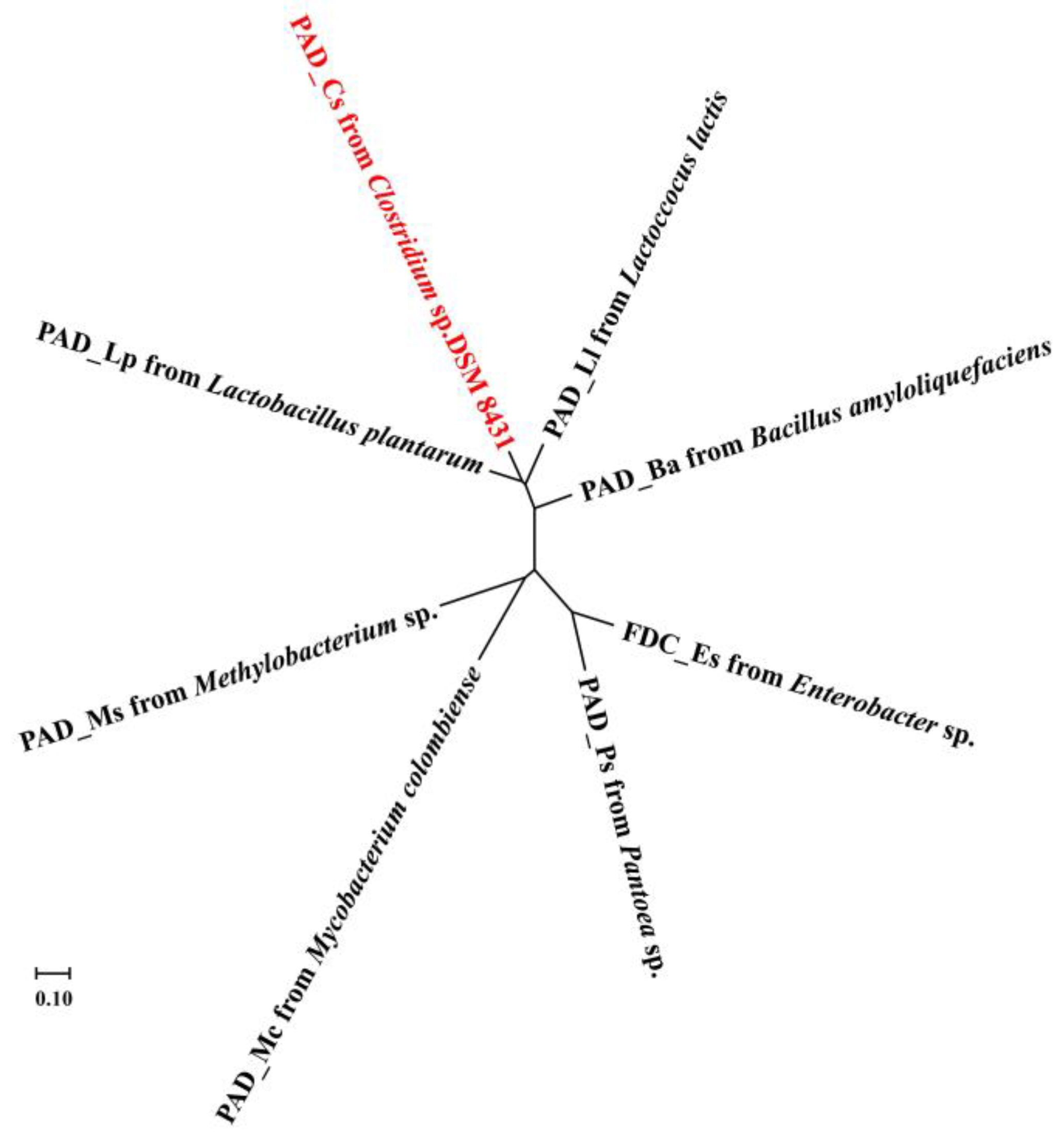
2.2. Sequence Analysis
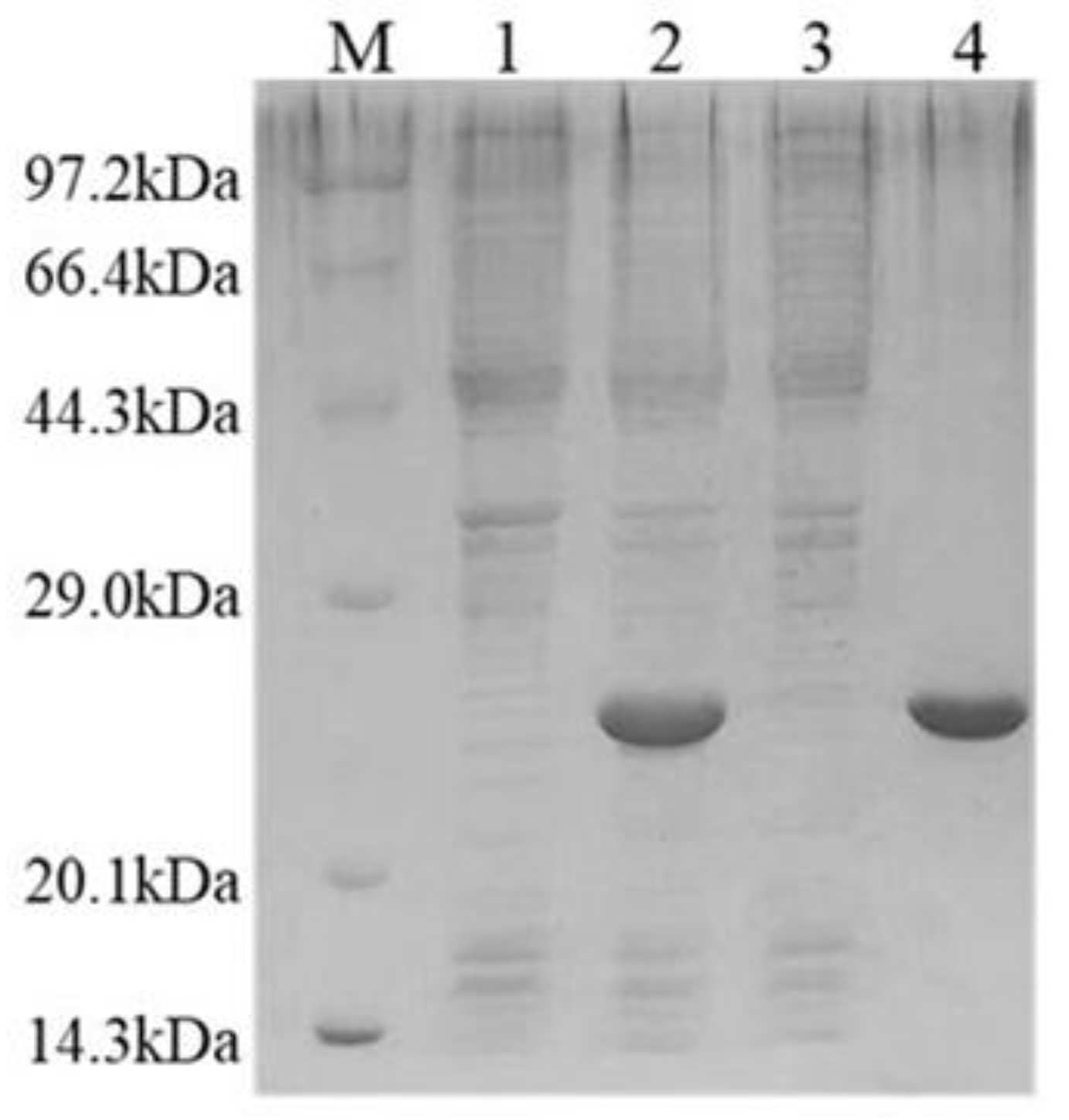
2.3. Overexpression and Purification of Recombinant PAD_Cs
2.4. Biochemical and Kinetic Properties of PAD_Cs
2.5. Investigating the β-carboxylation Synthesis Reaction System of p-coumaric Acid by E. coli Whole Cells
2.6. Substrate Scope of the β-carboxylation of para-hydroxystyrenes
3. Experimental Procedures
3.1. Materials
3.2. Mining and Sequence Analysis of Phenolic Acid (de)carboxylases
3.3. Cloning, Overexpression and Purification of Enzymes
3.4. Enzyme Assay
3.5. Biochemical and Kinetic Properties of PAD_Cs
3.6. Investigating the β-carboxylation Synthesis Reaction System of p-coumaric Acid by E. coli Whole Cells
3.7. Substrate Specificity in a β-carboxylation Reaction
3.8. Standard Reaction Conditions and Post-Treatment Procedures for a β-carboxylation Reaction
3.9. Analytical Procedures
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cai, X.; Xie, B. Direct Carboxylative Reactions for the Transformation of Carbon Dioxide into Carboxylic Acids and Derivatives. Synthesis 2013, 45, 3305–3324. [Google Scholar] [CrossRef]
- Wang, L.; Sun, W.; Liu, C. Recent Advances in Homogeneous Carbonylation Using CO2 as CO Surrogate. Chin. J. Chem. 2018, 36, 353–362. [Google Scholar] [CrossRef]
- Zhang, Z.; Muschiol, J.; Huang, Y.; Sigurdardóttir, S.B.; von Solms, N.; Daugaard, A.E.; Wei, J.; Luo, J.; Xu, B.-H.; Zhang, S.; et al. Efficient ionic liquid-based platform for multi-enzymatic conversion of carbon dioxide to methanol. Green Chem. 2018, 20, 4339–4348. [Google Scholar] [CrossRef]
- Wang, S.; Xu, M.; Peng, T.; Zhang, C.; Li, T.; Hussain, I.; Wang, J.; Tan, B. Porous hypercrosslinked polymer-TiO2-graphene composite photocatalysts for visible-light-driven CO2 conversion. Nat. Commun. 2019, 10, 676. [Google Scholar] [CrossRef] [PubMed]
- Ishida, N.; Shimamoto, Y.; Murakami, M. Solar-Driven Incorporation of Carbon Dioxide into α-Amino Ketones. Angew. Chem. Int. Ed. 2012, 51, 11750–11752. [Google Scholar] [CrossRef]
- Song, J.; Liu, Q.; Liu, H.; Jiang, X. Recent Advances in Palladium-Catalyzed Carboxylation with CO2. Eur. J. Org. Chem. 2018, 2018, 696–713. [Google Scholar] [CrossRef]
- Senboku, H. Electrochemical Fixation of Carbon Dioxide: Synthesis of Carboxylic Acids. Chem. Rec. 2021, 21, 2354–2374. [Google Scholar] [CrossRef] [PubMed]
- Bierbaumer, S.; Nattermann, M.; Schulz, L.; Zschoche, R.; Erb, T.J.; Winkler, C.K.; Tinzl, M.; Glueck, S.M. Enzymatic Conversion of CO2: From Natural to Artificial Utilization. Chem. Rev. 2023, 123, 5702–5754. [Google Scholar] [CrossRef] [PubMed]
- Roger, M.; Reed, T.C.P.; Sargent, F.; Semrau, J.D. Harnessing Escherichia coli for Bio-Based Production of Formate under Pressurized H2 and CO2 Gases. Appl. Environ. Microbiol. 2021, 87, e00299-21. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Mokrushina, L.; Arlt, W. Thermodynamic Constraints for the Utilization of CO2. Chem. Ing. Tech. 2014, 86, 497–503. [Google Scholar] [CrossRef]
- Kino, K.; Hirokawa, Y.; Gawasawa, R.; Murase, R.; Tsuchihashi, R.; Hara, R. Screening, gene cloning, and characterization of orsellinic acid decarboxylase from Arthrobacter sp. K8 for regio-selective carboxylation of resorcinol derivatives. J. Biotechnol. 2020, 323, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Payer, S.E.; Faber, K.; Glueck, S.M. Non-Oxidative Enzymatic (De)Carboxylation of (Hetero)Aromatics and Acrylic Acid Derivatives. Adv. Synth. Catal. 2019, 361, 2402–2420. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.M.; Raushel, F.M. Structural and Catalytic Diversity within the Amidohydrolase Superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef] [PubMed]
- Kirimura, K.; Gunji, H.; Wakayama, R.; Hattori, T.; Ishii, Y. Enzymatic Kolbe–Schmitt reaction to form salicylic acid from phenol: Enzymatic characterization and gene identification of a novel enzyme, Trichosporon moniliiforme salicylic acid decarboxylase. Biochem. Biophys. Res. Commun. 2010, 394, 279–284. [Google Scholar] [CrossRef]
- Tinikul, R.; Chenprakhon, P.; Maenpuen, S.; Chaiyen, P. Biotransformation of Plant-Derived Phenolic Acids. Biotechnol. J. 2018, 13, 1700632. [Google Scholar] [CrossRef] [PubMed]
- Wuensch, C.; Glueck, S.M.; Gross, J.; Koszelewski, D.; Schober, M.; Faber, K. Regioselective Enzymatic Carboxylation of Phenols and Hydroxystyrene Derivatives. Org. Lett. 2012, 14, 1974–1977. [Google Scholar] [CrossRef] [PubMed]
- Wuensch, C.; Pavkov-Keller, T.; Steinkellner, G.; Gross, J.; Fuchs, M.; Hromic, A.; Lyskowski, A.; Fauland, K.; Gruber, K.; Glueck, S.M.; et al. Regioselective Enzymatic β-Carboxylation of para-Hydroxy- styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases. Adv. Synth. Catal. 2015, 357, 1909–1918. [Google Scholar] [CrossRef]
- Payne, K.A.P.; White, M.D.; Fisher, K.; Khara, B.; Bailey, S.S.; Parker, D.; Rattray, N.J.W.; Trivedi, D.K.; Goodacre, R.; Beveridge, R.; et al. New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 2015, 522, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Bloor, S.; Michurin, I.; Titchiner, G.R.; Leys, D. Prenylated flavins: Structures and mechanisms. FEBS J. 2022, 290, 2232–2245. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.A.; Payne, K.A.P.; Leys, D. The UbiX-UbiD system: The biosynthesis and use of prenylated flavin (prFMN). Arch. Biochem. Biophys. 2017, 632, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Heker, I.; Haberhauer, G.; Meckenstock, R.U. Naphthalene Carboxylation in the Sulfate-Reducing Enrichment Culture N47 Is Proposed to Proceed via 1,3-Dipolar Cycloaddition to the Cofactor Prenylated Flavin Mononucleotide. Appl. Environ. Microbiol. 2023, 89, e0192722. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Shibue, M.; Ogino, K.; Nakamura, H.; Maeda, H. Enzymatic synthesis of pyruvic acid from acetaldehyde and carbon dioxide. Chem. Commun. 2001, 18, 1800–1801. [Google Scholar] [CrossRef]
- Sasano, K.; Takaya, J.; Iwasawa, N. Palladium(II)-Catalyzed Direct Carboxylation of Alkenyl C-H Bonds with CO2. J. Am. Chem. Soc. 2013, 135, 10954–10957. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Kwok, K.C. Ferulic acid: Pharmaceutical functions, preparation and applications in foods. J. Sci. Food Agric. 2004, 84, 1261–1269. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Sinha, B.; Choudhury, B.P.; Jha, N.K.; Palit, P.; Kundu, S.; Mandal, S.C.; Kolesarova, A.; Yousef, M.I.; Ruokolainen, J.; et al. Scavenging Properties of Plant-Derived Natural Biomolecule Para-Coumaric Acid in the Prevention of Oxidative Stress-Induced Diseases. Antioxidants 2021, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Himo, F. Theoretical Study of Enzyme Promiscuity: Mechanisms of Hydration and Carboxylation Activities of Phenolic Acid Decarboxylase. ACS Catal. 2017, 7, 1733–1741. [Google Scholar] [CrossRef]
- Cui, L.; Cui, A.; Li, Q.; Yang, L.; Liu, H.; Shao, W.; Feng, Y. Molecular Evolution of an Aminotransferase Based on Substrate–Enzyme Binding Energy Analysis for Efficient Valienamine Synthesis. ACS Catal. 2022, 12, 13703–13714. [Google Scholar] [CrossRef]
- Kelly, S.A.; Mix, S.; Moody, T.S.; Gilmore, B.F. Transaminases for industrial biocatalysis: Novel enzyme discovery. Appl. Microbiol. Biotechnol. 2020, 104, 4781–4794. [Google Scholar] [CrossRef]
- Wang, J.; Cao, X.; Chen, W.; Xu, J.; Wu, B. Identification and Characterization of a Thermostable GH36 α-Galactosidase from Anoxybacillus vitaminiphilus WMF1 and Its Application in Synthesizing Isofloridoside by Reverse Hydrolysis. Int. J. Mol. Sci. 2021, 22, 10778. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhou, J.; Wang, J.; Xu, S.; Cheng, C.; Ma, J.; Gao, Z. Complementary Distant and Active Site Mutations Simultaneously Enhance Catalytic Activity and Thermostability of α-Galactosidase. J. Agric. Food Chem. 2025, 73, 3635–3644. [Google Scholar] [CrossRef]
- Huang, H.-K.; Tokashiki, M.; Maeno, S.; Onaga, S.; Taira, T.; Ito, S. Purification and properties of phenolic acid decarboxylase from Candida guilliermondii. J. Ind. Microbiol. Biotechnol. 2012, 39, 55–62. [Google Scholar] [CrossRef]
- Wang, X.; Tang, X.; Chen, H.; Zhang, H.; Chen, Y.Q.; Zhao, J.; Chen, W. Purification and characterization of isocitrate dehydrogenase from Mortierella alpina. Process Biochem. 2022, 121, 575–583. [Google Scholar] [CrossRef]
- Zhang, A.; Mo, X.; Zhou, N.; Wang, Y.; Wei, G.; Chen, J.; Chen, K.; Ouyang, P. A novel bacterial β-N-acetyl glucosaminidase from Chitinolyticbacter meiyuanensis possessing transglycosylation and reverse hydrolysis activities. Biotechnol. Biofuels 2020, 13, 115. [Google Scholar] [CrossRef]
- Hu, H.; Li, L.; Ding, S. An organic solvent-tolerant phenolic acid decarboxylase from Bacillus licheniformis for the efficient bioconversion of hydroxycinnamic acids to vinyl phenol derivatives. Appl. Microbiol. Biotechnol. 2014, 99, 5071–5081. [Google Scholar] [CrossRef]
- Landete, J.M.; Rodríguez, H.; Curiel, J.A.; de las Rivas, B.; Mancheño, J.M.; Muñoz, R. Gene cloning, expression, and characterization of phenolic acid decarboxylase from Lactobacillus brevis RM84. J. Ind. Microbiol. Biotechnol. 2010, 37, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Degrassi, G.; Polverino De Laureto, P.; Bruschi, C.V. Purification and Characterization of Ferulate and p-Coumarate Decarboxylase from Bacillus pumilus. Appl. Environ. Microbiol. 1995, 61, 326–332. [Google Scholar] [CrossRef]
- RODRiguez, H.; Landete, J.M.; Curiel, J.A.; de Las Rivas, B.; Mancheño, J.M.; Muñoz, R. Characterization of the p-coumaric Acid Decarboxylase from Lactobacillus plantarum CECT 748. J. Agric. Food Chem. 2008, 56, 3068–3072. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yu, H.-N.; Wu, Y.-F.; Liu, X.-Y.; Cheng, A.-X.; Lou, H.-X. Cloning and functional characterization of a phenolic acid decarboxylase from the liverwort Conocephalum japonicum. Biochem. Biophys. Res. Commun. 2016, 481, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Tokashiki, M.; Tokashiki, M.; Uechi, K.; Ito, S.; Taira, T. Characterization and induction of phenolic acid decarboxylase from Aspergillus luchuensis. J. Biosci. Bioeng. 2018, 126, 162–168. [Google Scholar] [CrossRef]
- Harada, T.; Mino, Y. Some properties of p-coumarate decarboxylase from Cladosporium phlei. Can. J. Microbiol. 1976, 22, 1258–1262. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Benítez, A.; Narayan, A.R.H. Frontiers in Biocatalysis: Profiling Function across Sequence Space. ACS Cent. Sci. 2019, 5, 1747–1749. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J.; Valencia, A. Protein–Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
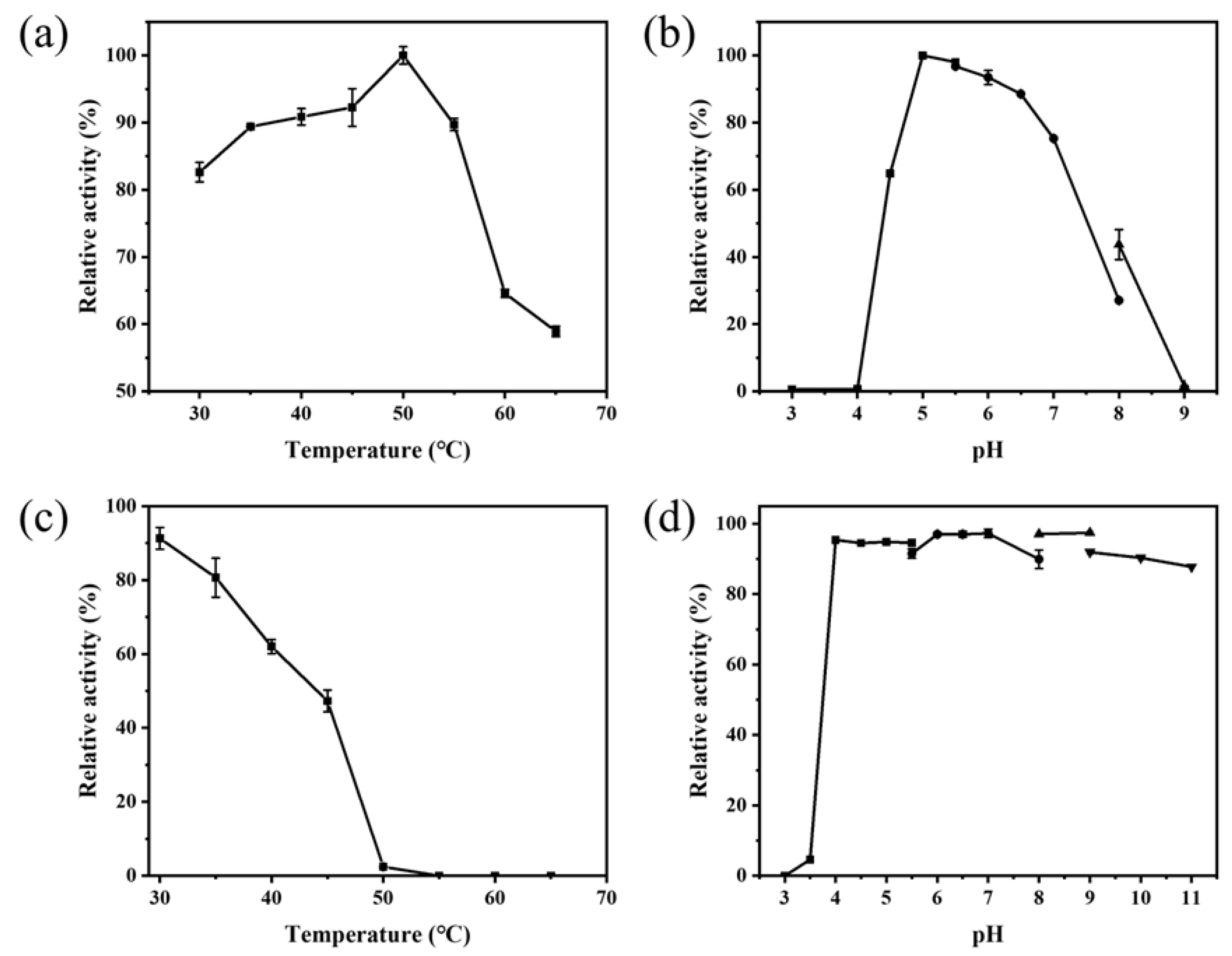

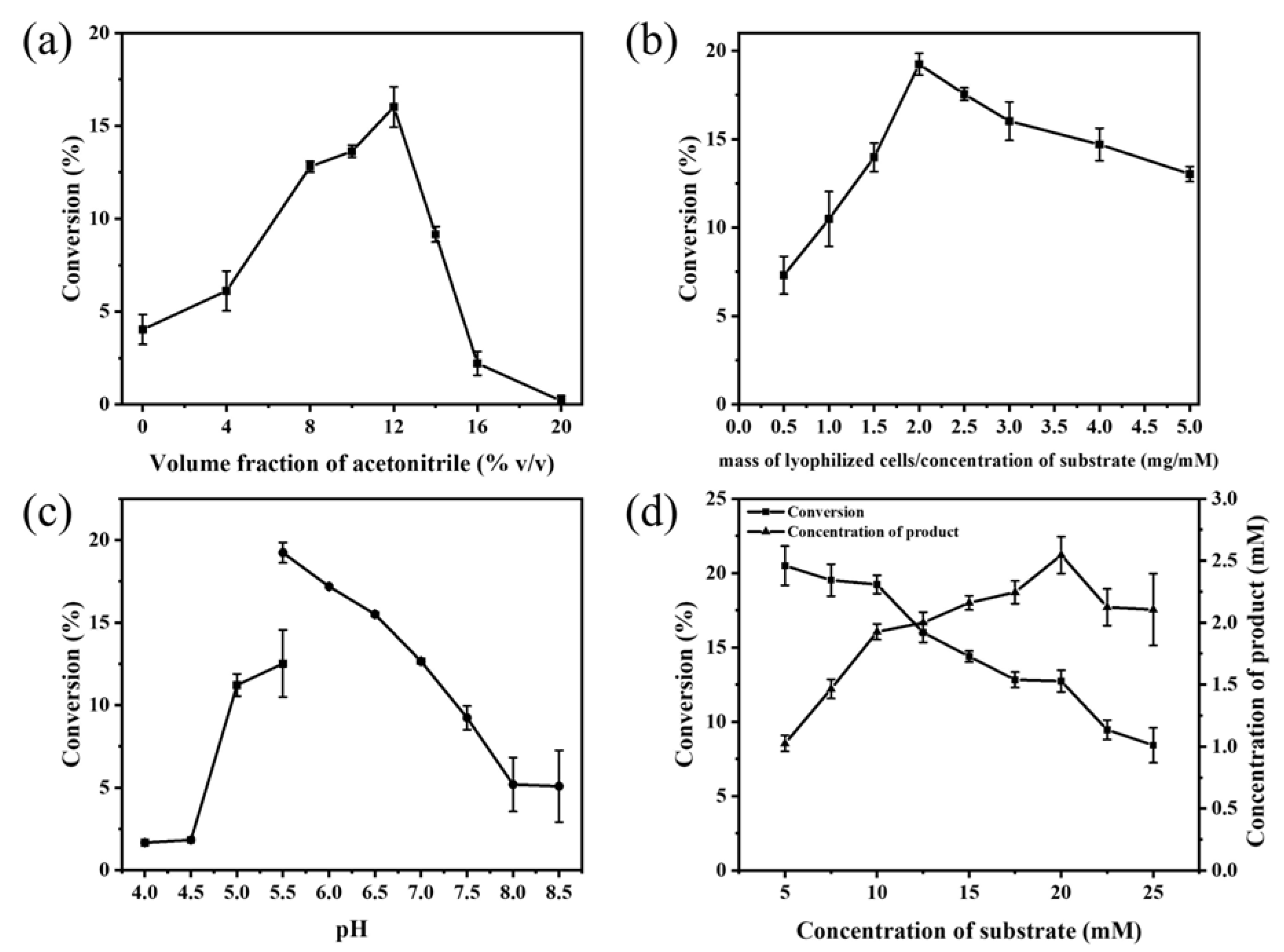
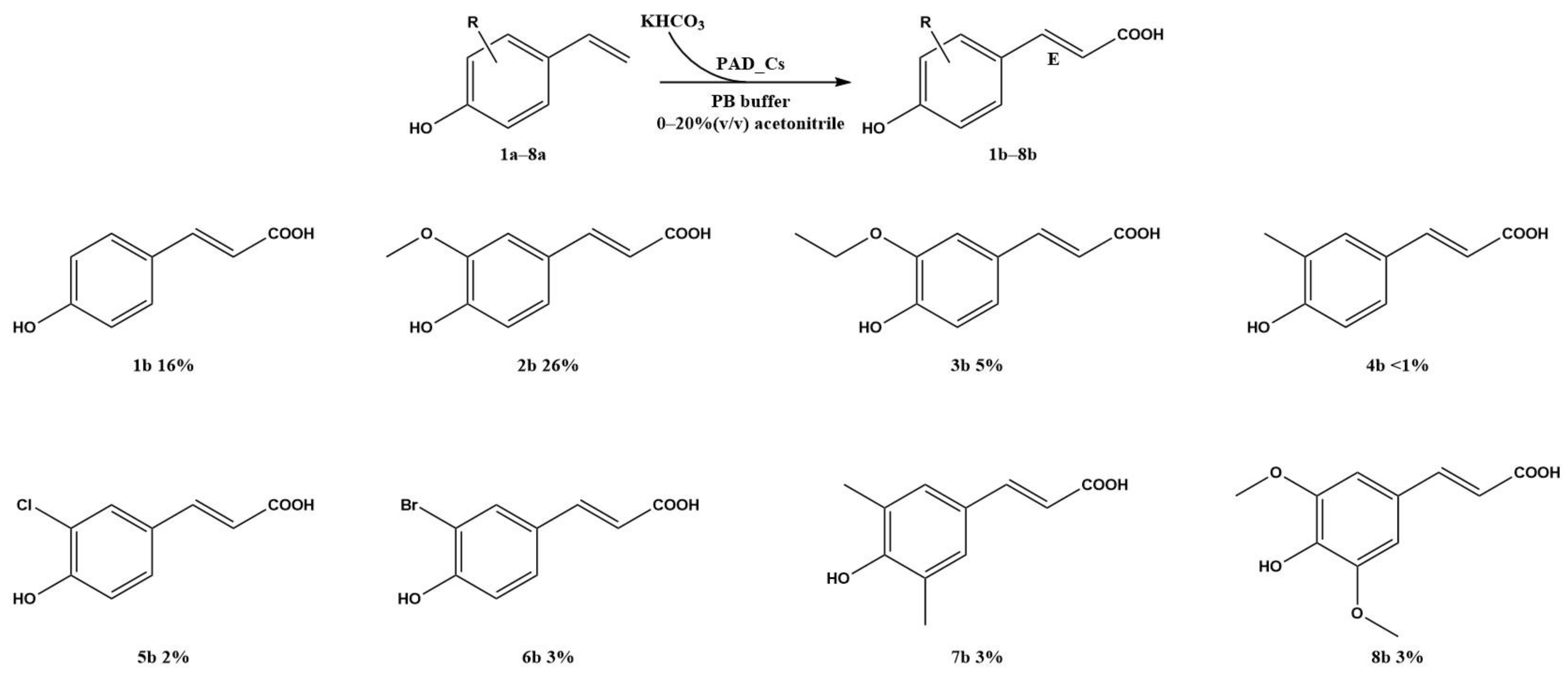
| Organism | Optimum Temperature (°C) | Optimum pH | Thermostability | pH Stability | Ref |
|---|---|---|---|---|---|
| Clostridium sp. DSM 8431 | 50 | 5.0 | 48%, 45 °C, 2 h | >85%, 4–11 | This study |
| Bacillus licheniformis | 37 | 6.0 | 45%, 45 °C, 1 h | >80%, 5–9 | [34] |
| Lactobacillus brevis RM84 | 30 | 6.5 | 65%, 37 °C, 2 h | NR | [35] |
| Bacillus pumilus | 37 | 5.5 | 0%, 42 °C, 30 min | >70%, 5–7 | [36] |
| Lactobacillus plantarum CECT 748T | 30 | 5.0–6.0 | 1%, 30 °C, 12 h | NR | [37] |
| Conocephalum japonicum | 25 | 5.5 | NR | NR | [38] |
| Aspergillus luchuensis | 40 | 5.7 | 100%, 50 °C, 1 h | >80%, 5–10 | [39] |
| Cladosporium phlei | 23 | 6.0 | 52%, 20 °C, 24 h | NR | [40] |
| Organism | Vmax (IU/mg) | Km (mM) | Kcat (s−1) | Kcat/Km (mM−1 s−1) | Ref |
|---|---|---|---|---|---|
| Clostridium sp. DSM 8431 | 315.53 ± 7.90 | 8.03 ± 0.50 | 50.95 ± 1.28 | 6.35 ± 0.55 | This study |
| Bacillus licheniformis | 268.43 ± 8.23 | 1.64 ± 0.12 | 93.14 ± 2.85 | 56.75 ± 2.45 | [34] |
| Lactobacillus brevis RM84 | 9.97 | 0.98 | NR | NR | [35] |
| Bacillus pumilus | 220 | 1.38 | NR | NR | [36] |
| Lactobacillus plantarum CECT 748T | 711 | 1.12 | NR | NR | [37] |
| Aspergillus luchuensis | NR | 6.63 | 1122 | 169.3 | [39] |
| Cladosporium phlei | NR | 0.65 | NR | NR | [40] |
| Product | Identity (%) | p-coumaric Acid | Ferulic Acid | Sinapic Acid |
|---|---|---|---|---|
| Biocatalyst | PAD_Cs vs. | Conversion (%) a | ||
| PAD_Cs | —— | 20 b | 26 | 3 |
| PAD_Lp | 79.8 | 2 c | 30 | 3 |
| PAD_Ll | 76.8 | 3 c | 2 c | 4 |
| PAD_Ba | 74.1 | 18 c | 26 c | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Wang, S.; Zhou, J.; Xu, J.; Wu, B.; Gao, Z.; He, B. Phenolic Acid Decarboxylase for Carbon Dioxide Fixation: Mining, Biochemical Characterization, and Regioselective Enzymatic β-carboxylation of para-hydroxystyrene Derivatives. Catalysts 2025, 15, 210. https://doi.org/10.3390/catal15030210
Chen J, Wang S, Zhou J, Xu J, Wu B, Gao Z, He B. Phenolic Acid Decarboxylase for Carbon Dioxide Fixation: Mining, Biochemical Characterization, and Regioselective Enzymatic β-carboxylation of para-hydroxystyrene Derivatives. Catalysts. 2025; 15(3):210. https://doi.org/10.3390/catal15030210
Chicago/Turabian StyleChen, Jie, Shirong Wang, Junru Zhou, Jiaxing Xu, Bin Wu, Zhen Gao, and Bingfang He. 2025. "Phenolic Acid Decarboxylase for Carbon Dioxide Fixation: Mining, Biochemical Characterization, and Regioselective Enzymatic β-carboxylation of para-hydroxystyrene Derivatives" Catalysts 15, no. 3: 210. https://doi.org/10.3390/catal15030210
APA StyleChen, J., Wang, S., Zhou, J., Xu, J., Wu, B., Gao, Z., & He, B. (2025). Phenolic Acid Decarboxylase for Carbon Dioxide Fixation: Mining, Biochemical Characterization, and Regioselective Enzymatic β-carboxylation of para-hydroxystyrene Derivatives. Catalysts, 15(3), 210. https://doi.org/10.3390/catal15030210








