Genomic Identification of the Levansucrase Operon in Novel Bacillus velezensis HL25 in Sucrose Utilizing Pathway and Functional Characterization of Its Levansucrase
Abstract
1. Introduction
2. Results and Discussion
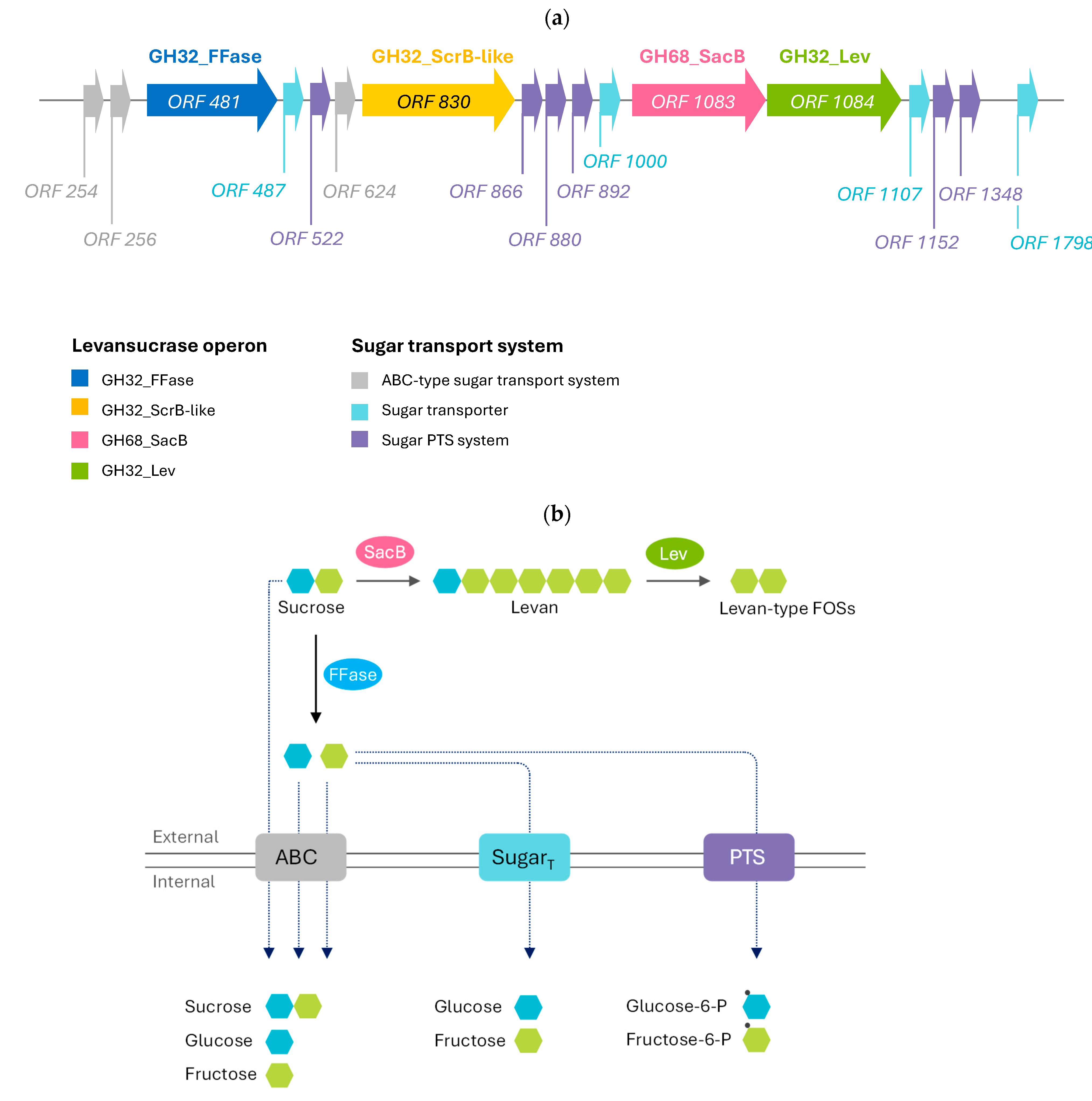
2.1. Identification of Sucrose-Utilizing Bacteria and Genomic Analysis of Related Enzymes in Levansucrase Operon
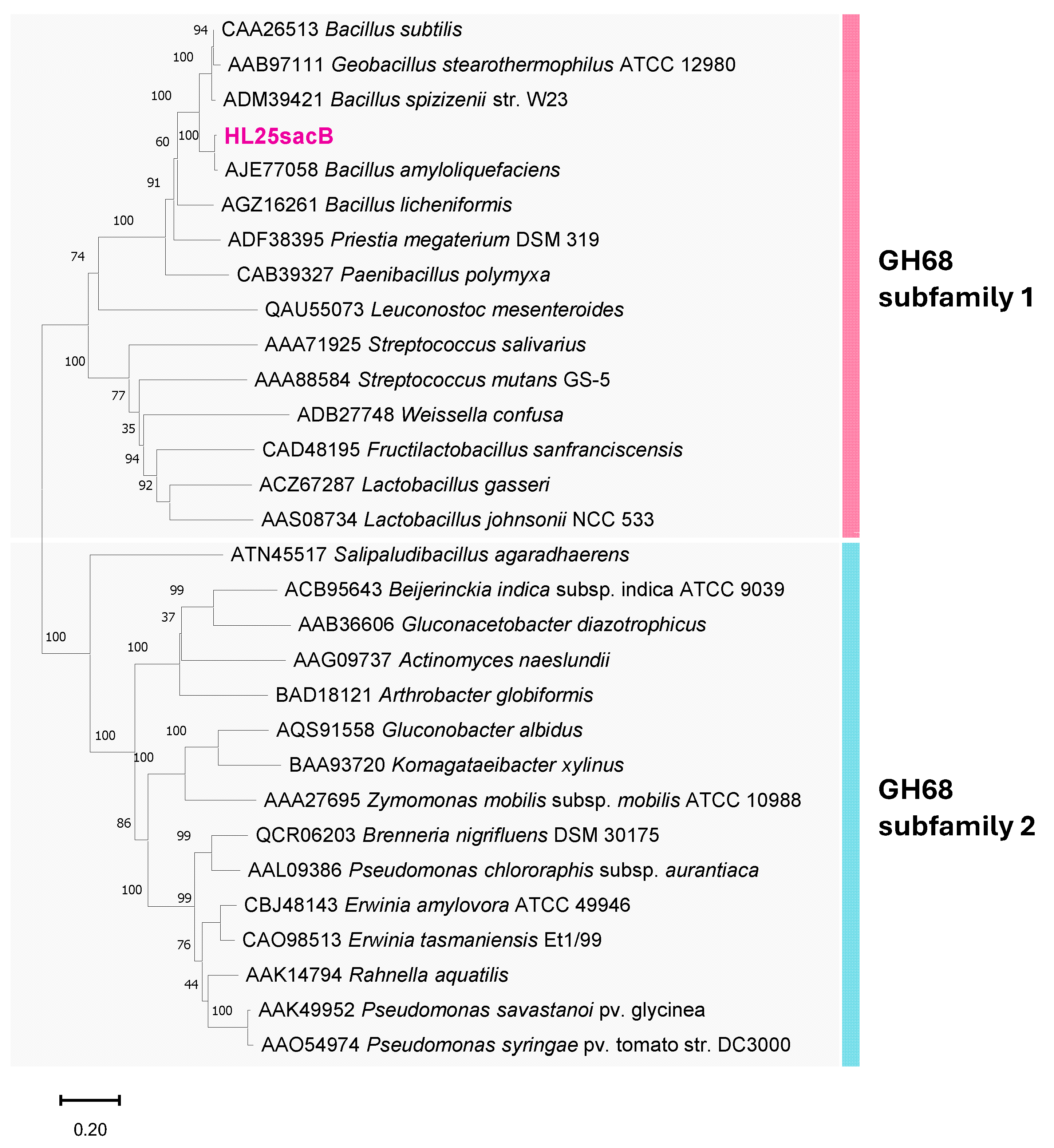
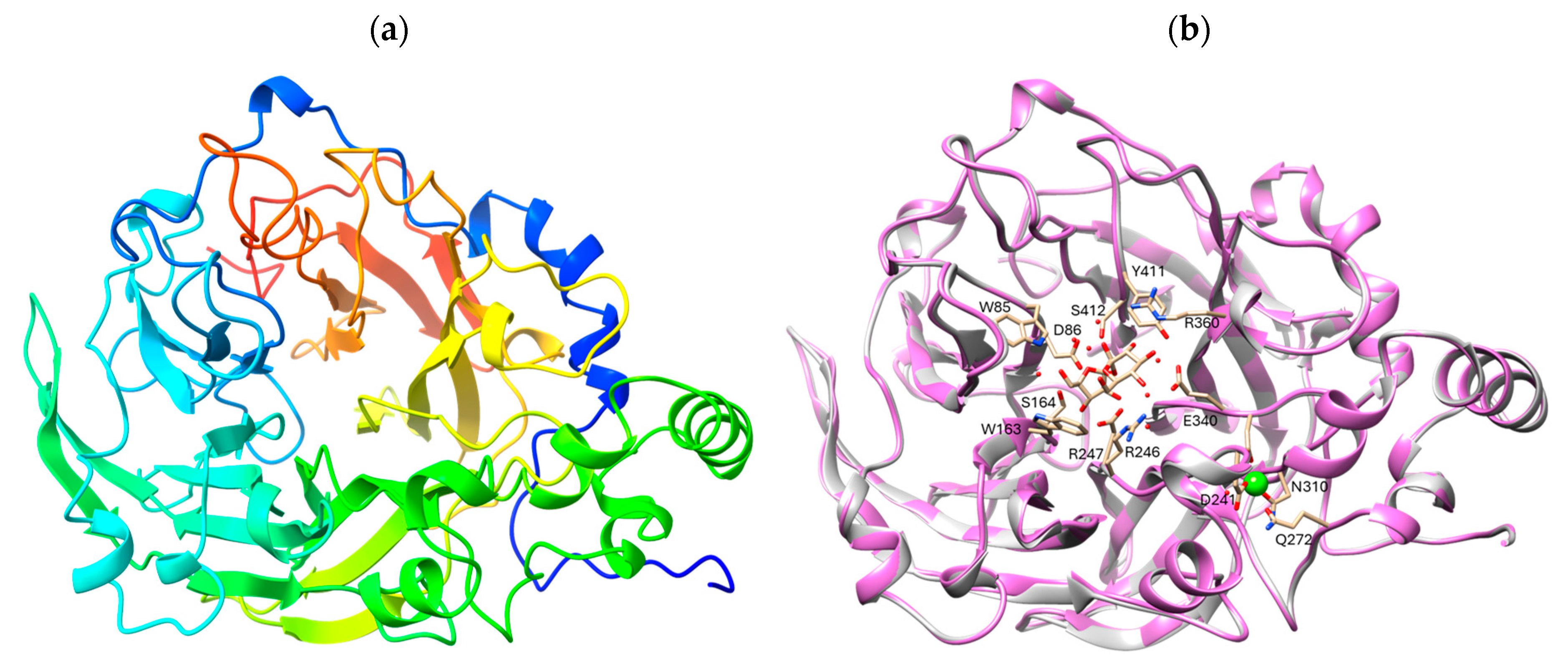
2.2. In Silico Analysis of Levansucrase from B. velezensis HL25
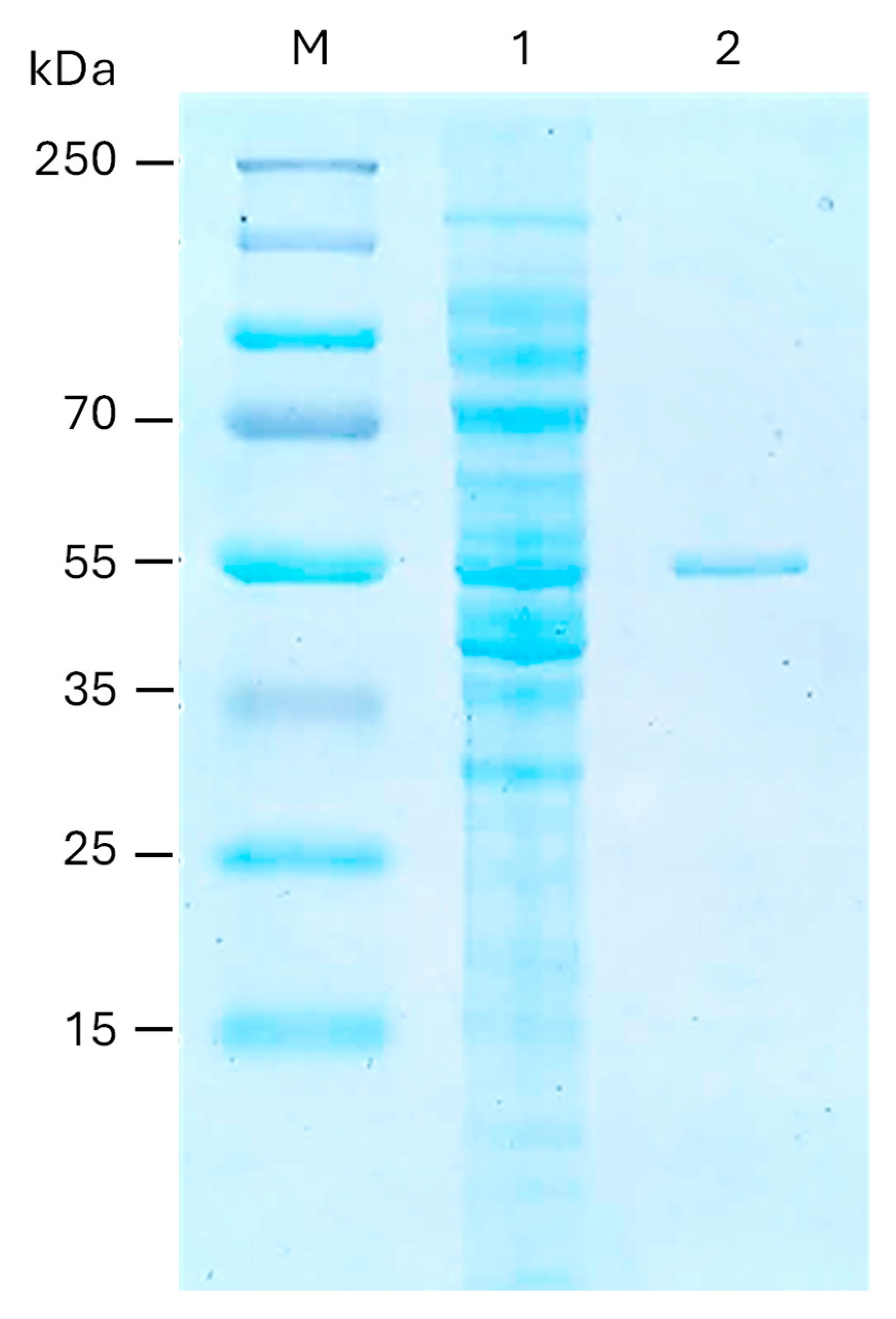
2.3. Heterologous Expression and Purification
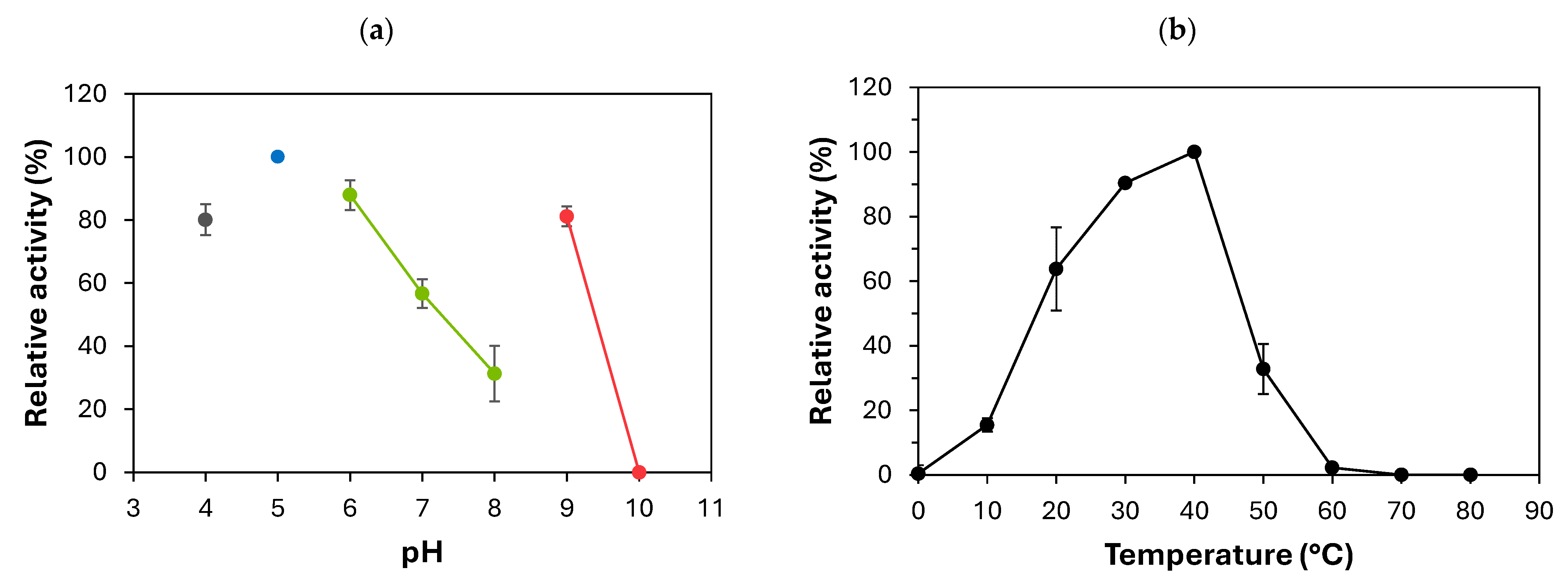
2.4. Enzymatic Activity and Biochemical Characteristics of Recombinant HL25SacB
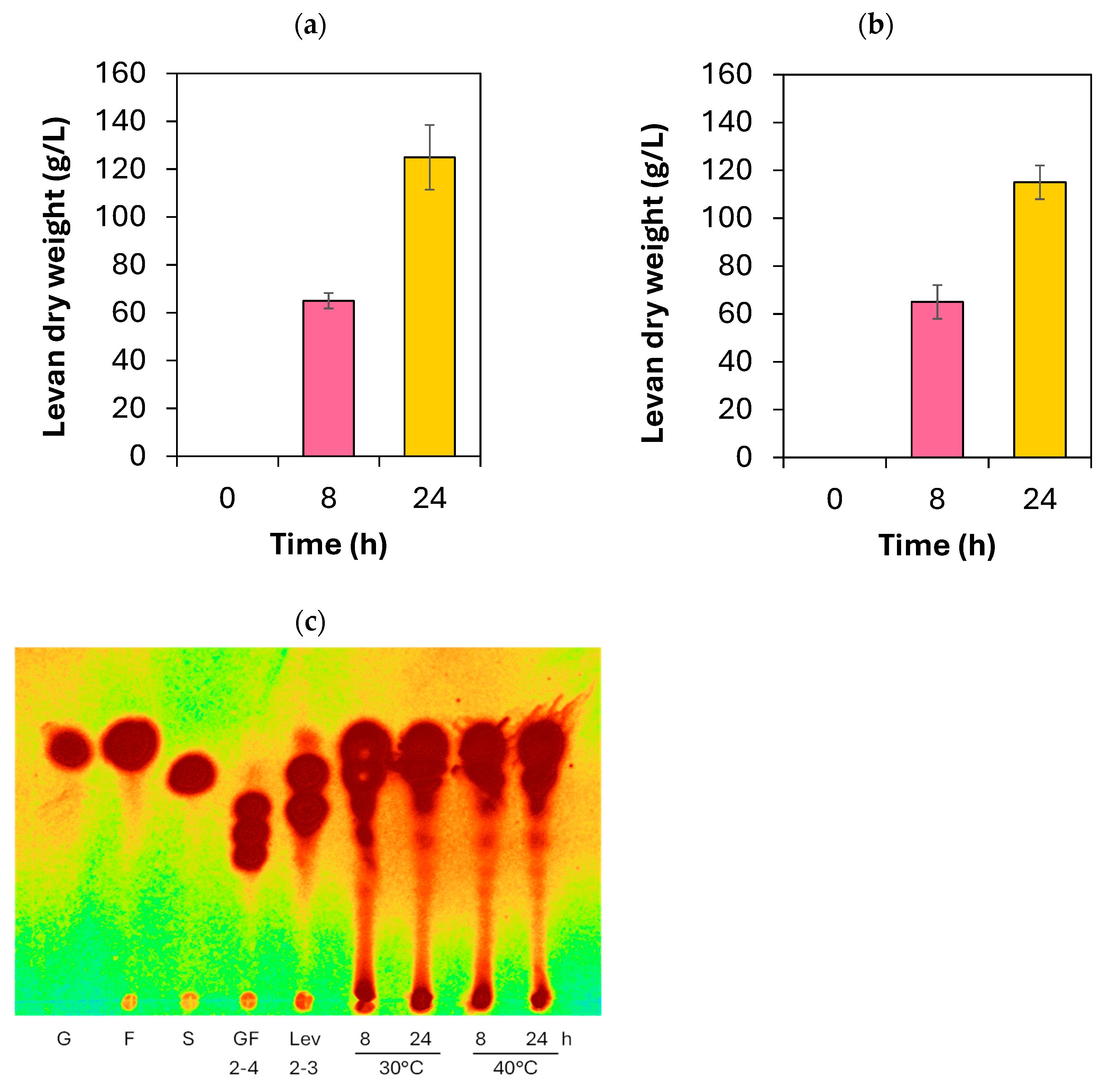
2.5. Biosynthesis of Fructooligosaccharides and Levan
3. Materials and Methods
3.1. Chemicals, Bacterial Strains and Plasmids
3.2. Bacterial Taxonomic Identification and Genome Sequence Analysis
3.3. Construction of pET32a-HL25SacB Recombinant Plasmid
3.4. Recombinant Protein Production and Purification
3.5. Enzyme Activity and Biochemical Property Characterization
3.6. Fructooligosaccharides and Levan Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Bourgot, C.; Rigaudier, F.; Juhel, C.; Herpin, F.; Meunier, C. Gastrointestinal tolerance of short-chain fructo-oligosaccharides from sugar beet: An observational, connected, dose-ranging study in healthy volunteers. Nutrients 2022, 14, 1461. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, A.L.; Rodrigues, L.R.; Lima, N.M.; Teixeira, J.A. An overview of the recent developments on fructooligosaccharide production and applications. Food Bioprocess Technol. 2014, 7, 324–337. [Google Scholar] [CrossRef]
- Domżał-Kędzia, M.; Ostrowska, M.; Lewińska, A.; Łukaszewicz, M. Recent developments and applications of microbial levan, a versatile polysaccharide-based biopolymer. Molecules 2023, 28, 5407. [Google Scholar] [CrossRef]
- Bruni, G.O.; Qi, Y.; Terrell, E.; Dupre, R.A.; Mattison, C.P. Characterization of levan fructan produced by a Gluconobacter japonicus strain isolated from a sugarcane processing facility. Microorganisms 2024, 12, 107. [Google Scholar] [CrossRef]
- Erdal Altıntaş, Ö.; Toksoy Öner, E.; Çabuk, A.; Aytar Çelik, P. Biosynthesis of levan by Halomonas elongata 153B: Optimization for enhanced production and potential biological activities for pharmaceutical field. J. Polym. Environ. 2022, 31, 1440–1455. [Google Scholar] [CrossRef]
- Hövels, M.; Kosciow, K.; Kniewel, J.; Jakob, F.; Deppenmeier, U. High yield production of levan-type fructans by Gluconobacter japonicus LMG 1417. Int. J. Biol. Macromol. 2020, 164, 295–303. [Google Scholar] [CrossRef]
- Kim, K.H.; Chung, C.B.; Kim, Y.H.; Kim, K.S.; Han, C.S.; Kim, C.H. Cosmeceutical properties of levan produced by Zymomonas mobilis. J. Cosmet. Sci. 2005, 56, 395–406. [Google Scholar] [CrossRef]
- Ni, D.; Xu, W.; Bai, Y.; Zhang, W.; Zhang, T.; Mu, W. Biosynthesis of levan from sucrose using a thermostable levansucrase from Lactobacillus reuteri LTH5448. Int. J. Biol. Macromol. 2018, 113, 29–37. [Google Scholar] [CrossRef]
- Sarilmiser, H.K.; Ates, O.; Ozdemir, G.; Arga, K.Y.; Oner, E.T. Effective stimulating factors for microbial levan production by Halomonas smyrnensis AAD6T. J. Biosci. Bioeng. 2015, 119, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Lekakarn, H.; Prongjit, D.; Mhuantong, W.; Trakarnpaiboon, S.; Bunterngsook, B. Exploring levansucrase operon regulating levan-type fructooligosaccharides (L-FOSs) production in Priestia koreensis HL12. J. Microbiol. Biotechnol. 2024, 34, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Zhou, Y.; Wu, X.; Montalban-Lopez, M.; Wang, L.; Li, X.; Zheng, Z. Secretion of Bacillus amyloliquefaciens levansucrase from Bacillus subtilis and its application in the enzymatic synthesis of levan. ACS Food Sci. Technol. 2021, 1, 249–259. [Google Scholar] [CrossRef]
- Xu, W.; Ni, D.; Zhang, W.; Guang, C.; Zhang, T.; Mu, W. Recent advances in levansucrase and inulosucrase: Evolution, characteristics, and application. Crit. Rev. Food. Sci. Nutr. 2019, 59, 3630–3647. [Google Scholar] [CrossRef]
- Tezgel, N.; Kırtel, O.; Van den Ende, W.; Toksoy Oner, E. Fructosyltransferase enzymes for microbial fructan production. In Microbial Enzymes: Roles and Applications in Industries; Arora, N.K., Mishra, J., Mishra, V., Eds.; Springer: Singapore, 2020; pp. 1–39. [Google Scholar]
- Seibel, J.; Moraru, R.; Götze, S.; Buchholz, K.; Na’amnieh, S.; Pawlowski, A.; Hecht, H.-J. Synthesis of sucrose analogues and the mechanism of action of Bacillus subtilis fructosyltransferase (levansucrase). Carbohydr. Res. 2006, 341, 2335–2349. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Ni, D.; Dai, Q.; Guang, C.; Zhang, T.; Mu, W. An overview of levan-degrading enzyme from microbes. Appl. Microbiol. Biotechnol. 2019, 103, 7891–7902. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, X.; Ni, D.; Zhang, W.; Guang, C.; Mu, W. A review of fructosyl-transferases from catalytic characteristics and structural features to reaction mechanisms and product specificity. Food Chem. 2024, 440, 138250. [Google Scholar] [CrossRef] [PubMed]
- Castrejón-Carrillo, S.; Morales-Moreno, L.A.; Rodríguez-Alegría, M.E.; Zavala-Padilla, G.T.; Bello-Pérez, L.A.; Moreno-Zaragoza, J.; López Munguía, A. Insights into the heterogeneity of levan polymers synthesized by levansucrase Bs-SacB from Bacillus subtilis 168. Carbohydr. Polym. 2024, 323, 121439. [Google Scholar] [CrossRef]
- Chandravanshi, M.; Sharma, A.; Dasgupta, P.; Mandal, S.K.; Kanaujia, S.P. Identification and characterization of ABC transporters for carbohydrate uptake in Thermus thermophilus HB8. Gene 2019, 696, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Cheng, L.; Zhao, Y.; Wang, L.; Wang, S.; Zhang, J. Biosynthesis and prebiotic activity of a linear levan from a new Paenibacillus isolate. Appl. Microbiol. Biotechnol. 2021, 105, 769–787. [Google Scholar] [CrossRef]
- Wang, J.; Xu, X.; Zhao, F.; Yin, N.; Zhou, Z.; Han, Y. Biosynthesis and structural characterization of levan by a recombinant levansucrase from Bacillus subtilis ZW019. Waste Biomass Valorization 2022, 13, 4599–4609. [Google Scholar] [CrossRef]
- Jadaun, J.S.; Narnoliya, L.K.; Agarwal, N.; Singh, S.P. Catalytic biosynthesis of levan and short-chain fructooligosaccharides from sucrose-containing feedstocks by employing the levansucrase from Leuconostoc mesenteroides MTCC10508. Int. J. Biol. Macromol. 2019, 127, 486–495. [Google Scholar] [CrossRef]
- Polsinelli, I.; Caliandro, R.; Salomone-Stagni, M.; Demitri, N.; Rejzek, M.; Field, R.A.; Benini, S. Comparison of the Levansucrase from the epiphyte Erwinia tasmaniensis vs its homologue from the phytopathogen Erwinia amylovora. Int. J. Biol. Macromol. 2019, 127, 496–501. [Google Scholar] [CrossRef]
- Khandekar, S.; Srivastava, A.; Pletzer, D.; Stahl, A.; Ullrich, M.S. The conserved upstream region of lscB/C determines expression of different levansucrase genes in plant pathogen Pseudomonas syringae. BMC Microbiol. 2014, 14, 79. [Google Scholar] [CrossRef]
- Martinez-Fleites, C.; Ortiz-Lombardia, M.; Pons, T.; Tarbouriech, N.; Taylor, E.J.; Arrieta, J.G.; Hernandez, L.; Davies, G.J. Crystal structure of levansucrase from the Gram-negative bacterium Gluconacetobacter diazotrophicus. Biochem. J. 2005, 390, 19–27. [Google Scholar] [CrossRef]
- Gao, S.; Qi, X.; Hart, D.J.; Gao, H.; An, Y. Expression and characterization of levansucrase from Clostridium acetobutylicum. J. Agric. Food Chem. 2017, 65, 867–871. [Google Scholar] [CrossRef]
- Shaheen, S.; Aman, A.; Qader, S.A.; Siddiqui, N.N. Hyper-production of levansucrase from Zymomonas mobilis KIBGEIB14 using submerged fermentation technique. Pak. J. Pharm. Sci. 2017, 30, 2053–2059. [Google Scholar] [PubMed]
- Kirtel, O.; Menéndez, C.; Versluys, M.; Van den Ende, W.; Hernández, L.; Toksoy Öner, E. Levansucrase from Halomonas smyrnensis AAD6(T): First halophilic GH-J clan enzyme recombinantly expressed, purified, and characterized. Appl. Microbiol. Biotechnol. 2018, 102, 9207–9220. [Google Scholar] [CrossRef] [PubMed]
- Lekakarn, H.; Bunterngsook, B.; Jaikaew, P.; Kuantum, T.; Wansuksri, R.; Champreda, V. Functional characterization of recombinant endo-levanase (LevBk) from Bacillus koreensis HL12 on short-chain levan-type fructooligosaccharides production. Protein J. 2022, 41, 477–488. [Google Scholar] [CrossRef]
- Han, S.; Liu, H.; Li, S.; Zheng, Z.; Yan, Q.; Jiang, Z. High level production of a β-fructofuranosidase in Aspergillus niger for the preparation of prebiotic bread using in situ enzymatic conversion. Food Res. Int. 2025, 208, 116225. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Izquierdo, Y.; Salomé-Abarca, L.F.; González-Hernández, J.C.; López, M.G. Fructooligosaccharides (FOS) production by microorganisms with fructosyltransferase activity. Fermentation 2023, 9, 968. [Google Scholar] [CrossRef]
- de Lima, M.Z.T.; de Almeida, L.R.; Mera, A.M.; Bernardes, A.; Garcia, W.; Muniz, J.R.C. Crystal structure of a sucrose-6-phosphate hydrolase from Lactobacillus gasseri with potential applications in fructan production and the food industry. J. Agric. Food Chem. 2021, 69, 10223–10234. [Google Scholar] [CrossRef]
- Daguer, J.P.; Geissmann, T.; Petit-Glatron, M.F.; Chambert, R. Autogenous modulation of the Bacillus subtilis sacB-levB-yveA levansucrase operon by the levB transcript. Microbiology 2004, 150, 3669–3679. [Google Scholar] [CrossRef]
- Liyaskina, E.V.; Rakova, N.A.; Kitykina, A.A.; Rusyaeva, V.V.; Toukach, P.V.; Fomenkov, A.; Vainauskas, S.; Roberts, R.J.; Revin, V.V. Production and characterization of the exopolysaccharide from strain Paenibacillus polymyxa 2020. PLoS ONE 2021, 16, e0253482. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumar, V.; Rajendhran, J.; Busby, S.J.; Gunasekaran, P. Characterization of multiple promoters and transcript stability in the sacB-sacC gene cluster in Zymomonas mobilis. Arch. Microbiol. 2009, 191, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Sun, B.; Shi, H.; Gao, T.; He, Y.; Li, Y.; Liu, Y.; Li, X.; Zhang, L.; Li, S.; et al. Sucrose triggers a novel signaling cascade promoting Bacillus subtilis rhizosphere colonization. ISME J. 2021, 15, 2723–2737. [Google Scholar] [CrossRef]
- Vu, T.H.N.; Quach, N.T.; Xuan, D.; Le, H.; Chi, L.; Lam, N.; Ngo, C.; Hà, H.; Chu, H.; Phi, Q.-T. A genomic perspective on the potential of termite-associated Cellulosimicrobium cellulans MP1 as producer of plant biomass-acting enzymes and exopolysaccharides. PeerJ 2021, 9, e11839. [Google Scholar] [CrossRef]
- Zeng, L.; Burne, R.A. Sucrose- and fructose-specific effects on the transcriptome of Streptococcus mutans, as determined by RNA sequencing. Appl. Environ. Microbiol. 2016, 82, 146–156. [Google Scholar] [CrossRef]
- Carreón-Rodríguez, O.E.; Gosset, G.; Escalante, A.; Bolívar, F. Glucose transport in Escherichia coli: From basics to transport engineering. Microorganisms 2023, 11, 1588. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, M.; Le Coq, D.; Aymerich, S.; Gonzy-Tréboul, G.; Gay, P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol. Gen. Genet. 1985, 200, 220–228. [Google Scholar] [CrossRef]
- Waldherr, F.W.; Meissner, D.; Vogel, R.F. Genetic and functional characterization of Lactobacillus panis levansucrase. Arch. Microbiol. 2008, 190, 497–505. [Google Scholar] [CrossRef]
- Rathsam, C.; Jacques, N.A. Role of C-terminal domains in surface attachment of the fructosyltransferase of Streptococcus salivarius ATCC 25975. J. Bacteriol. 1998, 180, 6400–6403. [Google Scholar] [CrossRef]
- Borchert, T.V.; Nagarajan, V. Structure-function studies on the Bacillus amyloliquefaciens levansucrase signal peptide. In Genetics and Biotechnology of Bacilli; Zukowski, M.M., Ganesan, A.T., Hoch, J.A., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 171–177. [Google Scholar]
- Lekakarn, H.; Phusiri, N.; Komonmusik, T.; Jaikaew, P.; Trakarnpaiboon, S.; Bunterngsook, B. Novel cold-active levansucrase (SacBPk) from Priestia koreensis HL12 for short-chain fructooligosaccharides and levan synthesis. Catalysts 2025, 15, 216. [Google Scholar] [CrossRef]
- Homann, A.; Biedendieck, R.; Götze, S.; Jahn, D.; Seibel, J. Insights into polymer versus oligosaccharide synthesis: Mutagenesis and mechanistic studies of a novel levansucrase from Bacillus megaterium. Biochem. J. 2007, 407, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Strube, C.P.; Homann, A.; Gamer, M.; Jahn, D.; Seibel, J.; Heinz, D.W. Polysaccharide synthesis of the levansucrase SacB from Bacillus megaterium is controlled by distinct surface motifs. J. Biol. Chem. 2011, 286, 17593–17600. [Google Scholar] [CrossRef]
- Polsinelli, I.; Caliandro, R.; Demitri, N.; Benini, S. The structure of sucrose-soaked levansucrase crystals from Erwinia tasmaniensis reveals a binding pocket for levanbiose. Int. J. Mol. Sci. 2020, 21, 83. [Google Scholar] [CrossRef]
- Xu, W.; Ni, D.; Hou, X.; Pijning, T.; Guskov, A.; Rao, Y.; Mu, W. Crystal structure of levansucrase from the Gram-negative bacterium Brenneria provides insights into its product size specificity. J. Agric. Food Chem. 2022, 70, 5095–5105. [Google Scholar] [CrossRef]
- Raga-Carbajal, E.; Díaz-Vilchis, A.; Rojas-Trejo, S.P.; Rudiño-Piñera, E.; Olvera, C. The molecular basis of the nonprocessive elongation mechanism in levansucrases. J. Biol. Chem. 2021, 296, 100178. [Google Scholar] [CrossRef] [PubMed]
- Sahyoun, A.M.; Karboune, S. Unveiling four levansucrase enzymes: Insights into catalytic properties, kinetics, and end-product profiles. Process Biochem. 2024, 141, 39–49. [Google Scholar] [CrossRef]
- Phengnoi, P.; Thakham, N.; Rachphirom, T.; Teerakulkittipong, N.; Lirio, G.A.; Jangiam, W. Characterization of levansucrase produced by novel Bacillus siamensis and optimization of culture condition for levan biosynthesis. Heliyon 2022, 8, e12137. [Google Scholar] [CrossRef]
- Guang, C.; Zhang, X.; Ni, D.; Zhang, W.; Xu, W.; Mu, W. Identification of a thermostable levansucrase from Pseudomonas orientalis that allows unique product specificity at different temperatures. Polymers 2023, 15, 1435. [Google Scholar] [CrossRef]
- Ben Ammar, Y.; Matsubara, T.; Ito, K.; Iizuka, M.; Limpaseni, T.; Pongsawasdi, P.; Minamiura, N. Characterization of a thermostable levansucrase from Bacillus sp. TH4-2 capable of producing high molecular weight levan at high temperature. J. Biotechnol. 2002, 99, 111–119. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.; Qian, H.; Mu, W.; Miao, M.; Jiang, B. Biosynthesis of levan by levansucrase from Bacillus methylotrophicus SK 21.002. Carbohydr. Polym. 2014, 101, 975–981. [Google Scholar] [CrossRef]
- Hill, A.; Karboune, S.; Narwani, T.J.; de Brevern, A.G. Investigating the product profiles and structural relationships of new levansucrases with conventional and non-conventional substrates. Int. J. Mol. Sci. 2020, 21, 5402. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Huang, F.; Yang, H.; Cong, H.; Zhang, X.; Xie, X.; Yang, H.; Tong, Q.; Luo, N.; Zhu, P.; et al. Factors affecting the production and molecular weight of levan in enzymatic synthesis by recombinant Bacillus subtilis levansucrase SacB-T305A. Polym. Int. 2021, 70, 185–192. [Google Scholar] [CrossRef]
- Peng, J.; Xu, W.; Ni, D.; Zhang, W.; Zhang, T.; Guang, C.; Mu, W. Preparation of a novel water-soluble gel from Erwinia amylovora levan. Int. J. Biol. Macromol. 2019, 122, 469–478. [Google Scholar] [CrossRef]
- Magri, A.; Oliveira, M.R.; Baldo, C.; Tischer, C.A.; Sartori, D.; Mantovani, M.S.; Celligoi, M. Production of fructooligosaccharides by Bacillus subtilis natto CCT7712 and their antiproliferative potential. J. Appl. Microbiol. 2020, 128, 1414–1426. [Google Scholar] [CrossRef]
- Lu, L.; Fu, F.; Zhao, R.; Jin, L.; He, C.; Xu, L.; Xiao, M. A recombinant levansucrase from Bacillus licheniformis 8-37-0-1 catalyzes versatile transfructosylation reactions. Process Biochem. 2014, 49, 1503–1510. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, S.; Zhang, T.; Jiang, B.; Mu, W. Efficient biosynthesis of levan from sucrose by a novel levansucrase from Brenneria goodwinii. Carbohydr. Polym. 2017, 157, 1732–1740. [Google Scholar] [CrossRef]
- Xu, W.; Ni, D.; Yu, S.; Zhang, T.; Mu, W. Insights into hydrolysis versus transfructosylation: Mutagenesis studies of a novel levansucrase from Brenneria sp. EniD312. Int. J. Biol. Macromol. 2018, 116, 335–345. [Google Scholar] [CrossRef]
- Lekakarn, H.; Bunterngsook, B.; Pajongpakdeekul, N.; Prongjit, D.; Champreda, V. A novel low temperature active maltooligosaccharides-forming amylase from Bacillus koreensis HL12 as biocatalyst for maltooligosaccharide production. 3 Biotech 2022, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2015, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.J. Confidence limits on phylogenies: The bootstrap revisited. Cladistics 1989, 5, 113–129. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2014, 43, D222–D226. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Huang, C.C.; Ferrin, T.E. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform. 2006, 7, 339. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Belghith, K.S.; Dahech, I.; Belghith, H.; Mejdoub, H. Microbial production of levansucrase for synthesis of fructooligosaccharides and levan. Int. J. Biol. Macromol. 2012, 50, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, J.; Veerapandian, B.; Sarwareddy, K.K.; Mani, K.P.; Shanmugam, S.R.; Venkatachalam, P. Studies on solvent precipitation of levan synthesized using Bacillus subtilis MTCC 441. Heliyon 2019, 5, e02414. [Google Scholar] [CrossRef] [PubMed]
| Organism | Optimal pH | Optimal Temp (°C) | Enzyme Activity (U/mg Protein) | Km (mM) | Vmax | Sucrose Concentration (g/L) | Yield of Levan (g/L) | Reference |
|---|---|---|---|---|---|---|---|---|
| Bacillus velezensis HL25 | 5 | 40 | 28.68 | 22.97 | 156 µmol/mg/min | 200 | 125 | This study |
| Bacillus siamensis | 5 | 37 | 15.95 | N.D. | N.D. | 200 | 145.9 | [50] |
| Bacillus amyloliquefaciens | 7.0 | 35 | 93.4 | 0.71 | 0.49 μmol/mL/min | 200 | 78.34 | [11] |
| Bacillus amyloliquefaciens | 6 | 37 | 82 | N.D. | N.D. | 500 | 200 | [50] |
| Bacillus subtilis | 5.2 | 40 | N.D. | 25.63 | N.D. | 100 | 30.6 | [20] |
| Bacillus licheniformis | 6.5 | 45 | N.D. | 152.79 | 0.03 mM/min | 274 | 7.1 | [58] |
| Brenneria goodwinii | 6 | 35 | N.D. | N.D. | N.D. | 500 | 185 | [59] |
| Priestia koreensis HL12 | 6 | 35–40 | 167.46 | 46.15 | 28.54 mole/mg/min | 500 | 129 | [43] |
| Brenneria sp. EniD312 | 6.5 | 45 | N.D. | N.D. | N.D. | 250 | 85 | [60] |
| Erwinia amylovora | 6.5 | 35 | N.D. | N.D. | N.D. | 500 | 295 | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lekakarn, H.; Chaisuriyaphun, J.; Junsuk, R.; Kornpitak, P.; Komonmusik, T.; Mhuantong, W.; Bunterngsook, B. Genomic Identification of the Levansucrase Operon in Novel Bacillus velezensis HL25 in Sucrose Utilizing Pathway and Functional Characterization of Its Levansucrase. Catalysts 2025, 15, 1059. https://doi.org/10.3390/catal15111059
Lekakarn H, Chaisuriyaphun J, Junsuk R, Kornpitak P, Komonmusik T, Mhuantong W, Bunterngsook B. Genomic Identification of the Levansucrase Operon in Novel Bacillus velezensis HL25 in Sucrose Utilizing Pathway and Functional Characterization of Its Levansucrase. Catalysts. 2025; 15(11):1059. https://doi.org/10.3390/catal15111059
Chicago/Turabian StyleLekakarn, Hataikarn, Jiruchaya Chaisuriyaphun, Ruethaikan Junsuk, Putanat Kornpitak, Teeranart Komonmusik, Wuttichai Mhuantong, and Benjarat Bunterngsook. 2025. "Genomic Identification of the Levansucrase Operon in Novel Bacillus velezensis HL25 in Sucrose Utilizing Pathway and Functional Characterization of Its Levansucrase" Catalysts 15, no. 11: 1059. https://doi.org/10.3390/catal15111059
APA StyleLekakarn, H., Chaisuriyaphun, J., Junsuk, R., Kornpitak, P., Komonmusik, T., Mhuantong, W., & Bunterngsook, B. (2025). Genomic Identification of the Levansucrase Operon in Novel Bacillus velezensis HL25 in Sucrose Utilizing Pathway and Functional Characterization of Its Levansucrase. Catalysts, 15(11), 1059. https://doi.org/10.3390/catal15111059








