Improving the Catalytic Efficiency of an AA9 Lytic Polysaccharide Monooxygenase MtLPMO9G by Consensus Mutagenesis
Abstract
1. Introduction
2. Results and Discussion
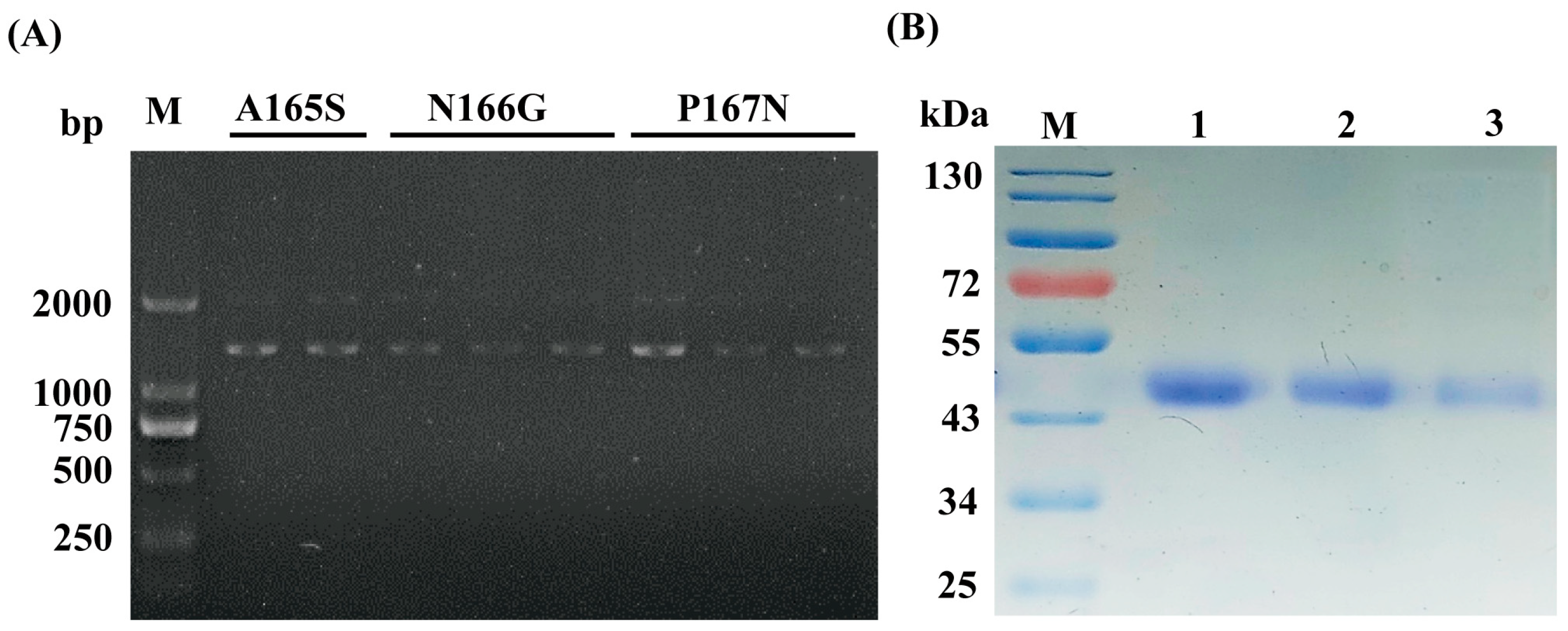
2.1. Selection of Target Residues to Improve MtLPMO9G Activity and Site-Directed Mutagenesis
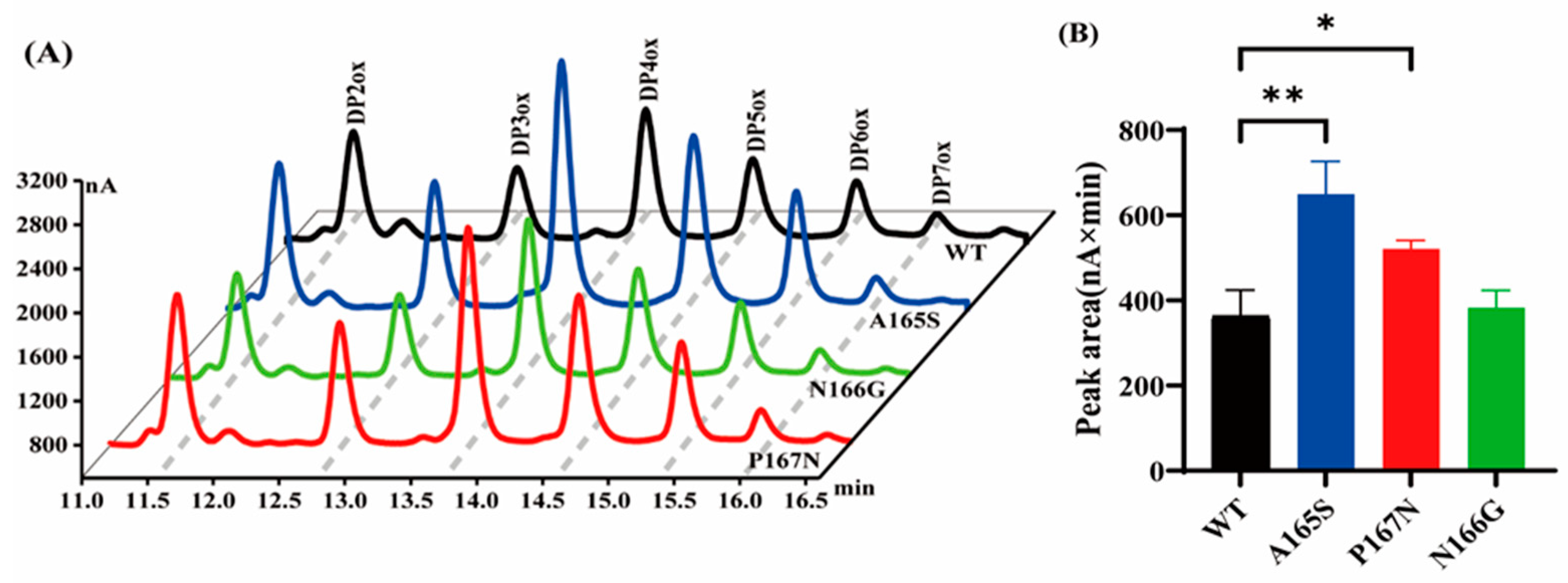
2.2. Comparison of Activities of MtLPMO9G and Its Mutants
2.3. Comparison of Thermal Stability of MtLPMO9G and Its Mutants
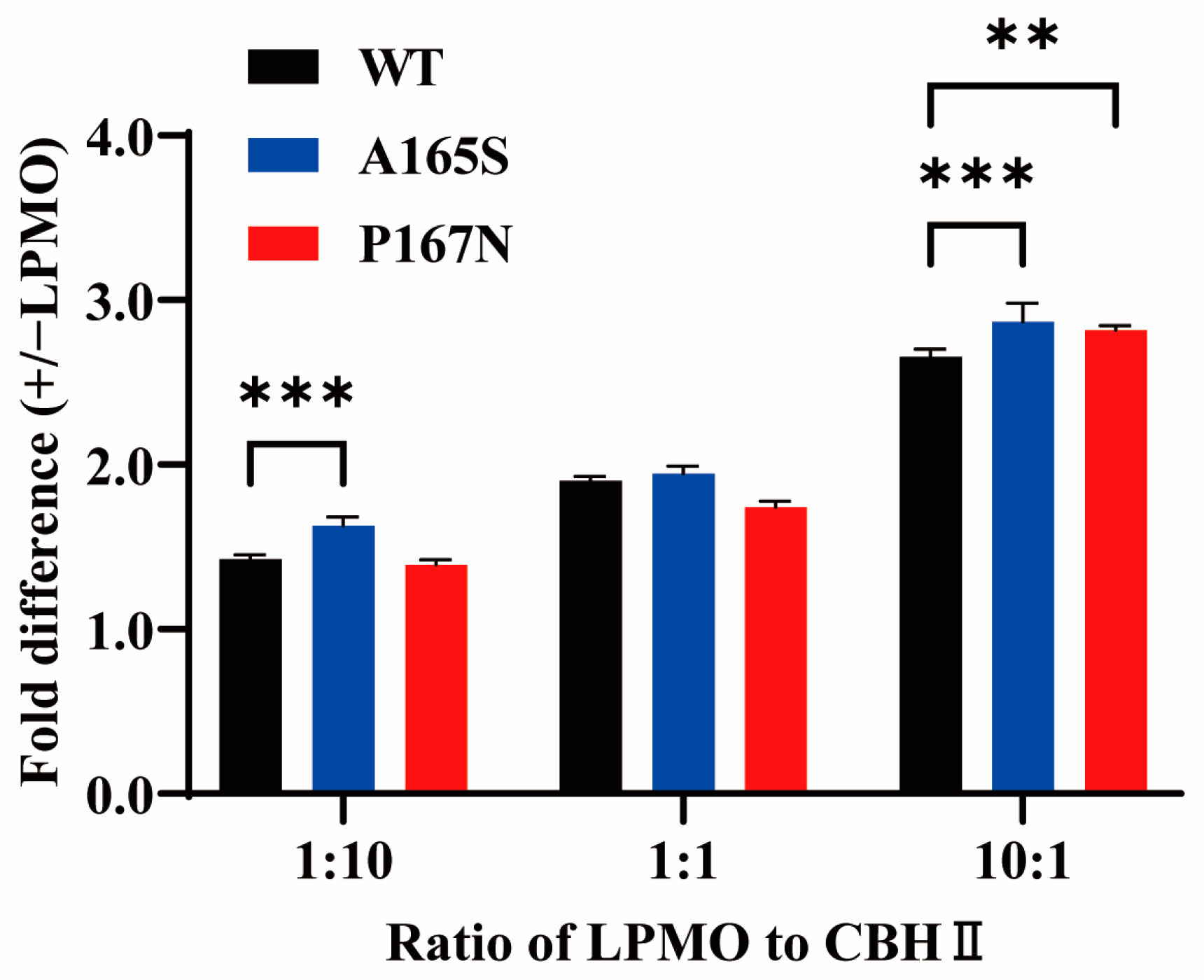
2.4. Synergistic Effect of MtLPMO9G and Its Mutants with Cellobiohydrolase
2.5. Comparison of the Substrate-Binding Ability of MtLPMO9G and Its Mutants
2.6. Comparison of the H2O2 Tolerance Ability of MtLPMO9G and Its Mutants
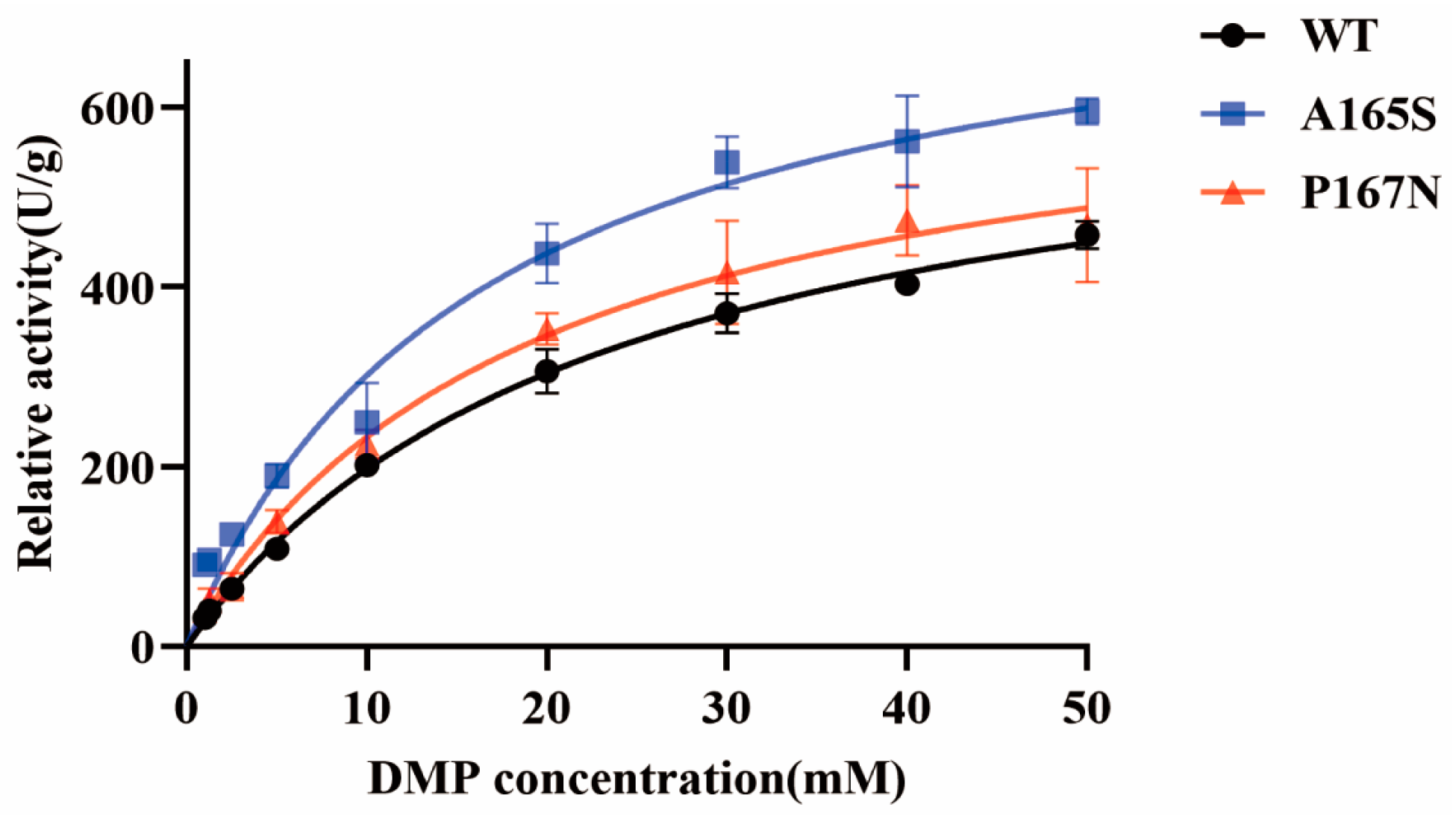
2.7. Enzyme Kinetics of MtLPMO9G and Its Mutants
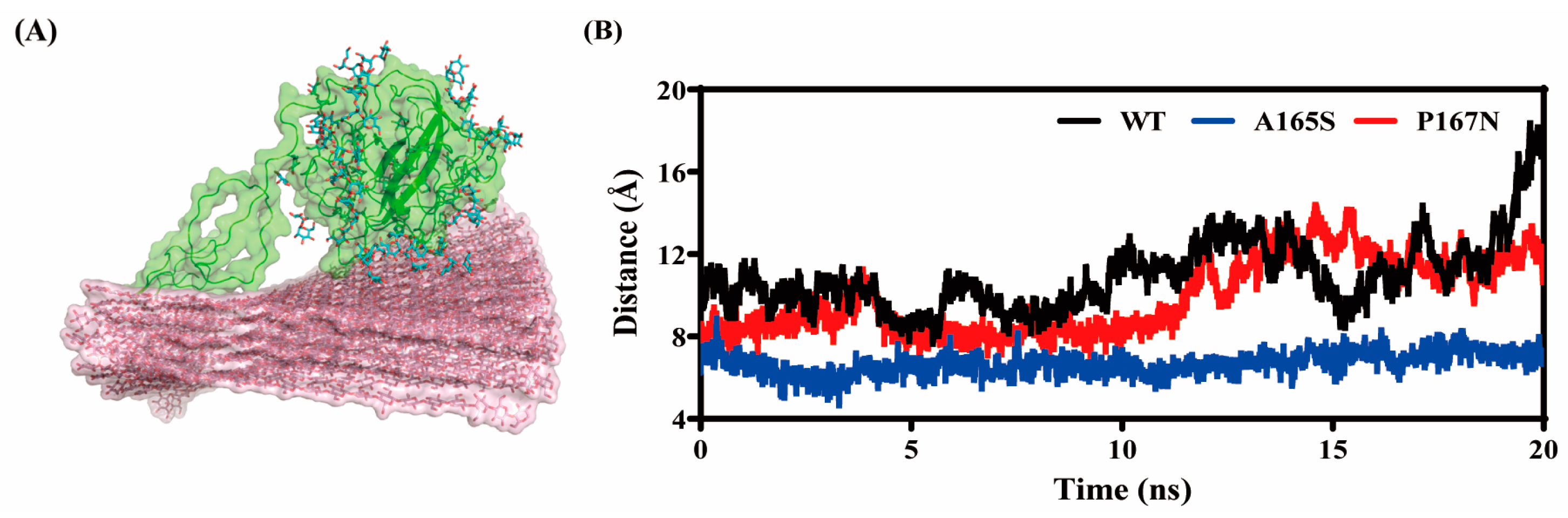
2.8. Molecular Dynamics Analysis of MtLPMO9G and Its Mutants
3. Materials and Methods
3.1. Materials
3.2. Selection of MtLPMO9G Mutation Sites
3.3. Construction of MtLPMO9G Mutants
3.4. Expression and Purification of MtLPMO9G Mutants
3.5. Assay of the Oxidative Activity of MtLPMO9G Mutants
3.6. Thermal Stability Assay of MtLPMO9G Mutants
3.7. Assay of the Synergy between MtLPMO9G Mutants and Cellobiohydrolase
3.8. Assay of the Substrate-Binding Affinity of MtLPMO9G Mutants
3.9. Assay of the H2O2 Tolerance Ability of MtLPMO9G Mutants
3.10. Assay of the Kinetics of MtLPMO9G Mutants
3.11. Molecular Dynamics Simulations
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Zhao, B.; Zhang, X.; Zhang, Y. Influence of intrinsic physicochemical properties of agroforestry waste on its pyrolysis characteristics and behavior. Materials 2022, 16, 222. [Google Scholar] [CrossRef] [PubMed]
- Agregán, R.; Lorenzo, J.M.; Kumar, M.; Shariati, M.A.; Khan, M.U.; Sarwar, A.; Sultan, M.; Rebezov, M.; Usman, M. Anaerobic digestion of lignocellulose components: Challenges and novel approaches. Energies 2022, 15, 8413. [Google Scholar] [CrossRef]
- Aziz, T.; Farid, A.; Haq, F.; Kiran, M.; Ullah, A.; Zhang, K.; Li, C.; Ghazanfar, S.; Sun, H.; Ullah, R.; et al. A review on the modification of cellulose and its applications. Polymers 2022, 14, 3206. [Google Scholar] [CrossRef]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Spinacé, M.A.; Lambert, C.S.; Fermoselli, K.K.; De Paoli, M.A. Characterization of lignocellulosic curaua fibres. Carbohydr. Polym. 2009, 77, 47–53. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef]
- Eibinger, M.; Ganner, T.; Bubner, P.; Rošker, S.; Kracher, D.; Haltrich, D.; Ludwig, R.; Plank, H.; Nidetzky, B. Cellulose surface degradation by a lytic polysaccharide monooxygenase and its effect on cellulase hydrolytic efficiency. J. Biol. Chem. 2014, 289, 35929–35938. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Johnston, E.M.; Davies, G.J.; Walton, P.H. Lytic polysaccharide monooxygenases in biomass conversion. Trends Biotechnol. 2015, 33, 747–761. [Google Scholar] [CrossRef]
- Quinlan, R.J.; Sweeney, M.D.; Lo Leggio, L.; Otten, H.; Poulsen, J.C.N.; Johansen, K.S.; Krogh, K.B.; Jørgensen, C.I.; Tovborg, M.; Anthonsen, A.; et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 2011, 108, 15079–15084. [Google Scholar] [CrossRef]
- Phillips, C.M.; Beeson IV, W.T.; Cate, J.H.; Marletta, M.A. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 2011, 6, 1399–1406. [Google Scholar] [CrossRef]
- Østby, H.; Hansen, L.D.; Horn, S.J.; Eijsink, V.G.; Várnai, A. Enzymatic processing of lignocellulosic biomass: Principles, recent advances and perspectives. J. Ind. Microbiol. Biot. 2020, 47, 623–657. [Google Scholar] [CrossRef] [PubMed]
- Caputo, F.; Tõlgo, M.; Naidjonoka, P.; Krogh, K.B.; Novy, V.; Olsson, L. Investigating the role of AA9 LPMOs in enzymatic hydrolysis of differentially steam-pretreated spruce. Biotechnol. Biofuels 2023, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Vandhana, T.M.; Reyre, J.L.; Sushmaa, D.; Berrin, J.G.; Bissaro, B.; Madhuprakash, J. On the expansion of biological functions of lytic polysaccharide monooxygenases. New Phytol. 2022, 233, 2380–2396. [Google Scholar] [CrossRef]
- Johansen, K.S. Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 2016, 44, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Filiatrault-Chastel, C.; Navarro, D.; Haon, M.; Grisel, S.; Herpoël-Gimbert, I.; Chevret, D.; Fanuel, M.; Henrissat, B.; Heiss-Blanquet, S.; Margeot, A.; et al. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol. Biofuels 2019, 12, 55. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Henrissat, B.; Davies, G.J.; Walton, P.H. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat. Chem. Biol. 2014, 10, 122–126. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Lo Leggio, L.; Simmons, T.J.; Poulsen, J.-C.N.; Frandsen, K.E.; Hemsworth, G.R.; Stringer, M.A.; Von Freiesleben, P.; Tovborg, M.; Johansen, K.S.; De Maria, L.; et al. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat. Commun. 2015, 6, 5961. [Google Scholar] [CrossRef]
- Sabbadin, F.; Hemsworth, G.R.; Ciano, L.; Henrissat, B.; Dupree, P.; Tryfona, T.; Marques, R.D.; Sweeney, S.T.; Besser, K.; Elias, L.; et al. An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 2018, 9, 756. [Google Scholar] [CrossRef]
- Sabbadin, F.; Urresti, S.; Henrissat, B.; Avrova, A.O.; Welsh, L.R.; Lindley, P.J.; Csukai, M.; Squires, J.N.; Walton, P.H.; Davies, G.J.; et al. Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Science 2021, 373, 774–779. [Google Scholar] [CrossRef]
- Voshol, G.P.; Vijgenboom, E.; Punt, P.J. The discovery of novel LPMO families with a new Hidden Markov model. BMC Res. Notes 2017, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.V.; Welner, D.; McFarland, K.; Re, E.; Navarro Poulsen, J.-C.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, H.; Xu, F. Identifying and engineering a critical amino acid residue to enhance the catalytic efficiency of Pseudomonas sp. methyl parathion hydrolase. Appl. Microbiol. Biotechnol. 2018, 102, 6537–6545. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; An, Y.; Chai, C.; Sang, J.; Jiang, L.; Lu, F.; Dai, Y.; Liu, F. Construction of the R17L mutant of MtC1LPMO for improved lignocellulosic biomass conversion by rational point mutation and investigation of the mechanism by molecular dynamics simulations. Bioresour. Technol. 2020, 317, 124024. [Google Scholar] [CrossRef]
- Kruer-Zerhusen, N.; Alahuhta, M.; Lunin, V.V.; Himmel, M.E.; Bomble, Y.J.; Wilson, D.B. Structure of a Thermobifida fusca lytic polysaccharide monooxygenase and mutagenesis of key residues. Biotechnol. Biofuels 2017, 10, 243. [Google Scholar] [CrossRef]
- Yang, Y.J.; Pei, X.Q.; Liu, Y.; Wu, Z.L. Thermostabilizing ketoreductase ChKRED20 by consensus mutagenesis at dimeric interfaces. Enzyme Microb. Technol. 2022, 158, 110052. [Google Scholar] [CrossRef]
- Porebski, B.T.; Buckle, A.M. Consensus protein design. Protein Eng. Des. Sel. 2016, 29, 245–251. [Google Scholar] [CrossRef]
- Sternke, M.; Tripp, K.W.; Barrick, D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 11275–11284. [Google Scholar] [CrossRef]
- Gumulya, Y.; Baek, J.M.; Wun, S.J.; Thomson, R.E.; Harris, K.L.; Hunter, D.J.; Behrendorff, J.B.; Kulig, J.; Zheng, S.; Wu, X.; et al. Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nat. Catal. 2018, 1, 878–888. [Google Scholar] [CrossRef]
- Berka, R.M.; Grigoriev, I.V.; Otillar, R.; Salamov, A.; Grimwood, J.; Reid, I.; Ishmael, N.; John, T.; Darmond, C.; Moisan, M.C.; et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 2011, 29, 922–927. [Google Scholar] [CrossRef]
- Dos Santos, H.B.; Bezerra, T.M.S.; Pradella, J.G.; Delabona, P.; Lima, D.; Gomes, E.; Hartson, S.D.; Rogers, J.; Couger, B.; Prade, R. Myceliophthora thermophila M77 utilizes hydrolytic and oxidative mechanisms to deconstruct biomass. AMB Express 2016, 6, 103. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, M.A.S.; Godoy, M.O.d.; Kumagai, P.S.; Costa-Filho, A.J.d.; Mort, A.; Prade, R.A.; Polikarpov, I. Characterization of a new glyoxal oxidase from the thermophilic fungus Myceliophthora thermophila M77: Hydrogen peroxide production retained in 5-hydroxymethylfurfural oxidation. Catalysts 2018, 8, 476. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, H.; Li, T.; Ju, J.; Zhou, H.; Zong, X.; Yin, H. Controlled depolymerization of cellulose by photoelectrochemical bioreactor using a lytic polysaccharide monooxygenase. Biochem. Eng. J. 2022, 187, 108597. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Davies, G.J.; Walton, P.H. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr. Opin. Struct. Biol. 2013, 23, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Forsberg, Z.; Loose, J.S.; Bissaro, B.; Eijsink, V.G. Structural diversity of lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 2017, 44, 67–76. [Google Scholar] [CrossRef]
- Tandrup, T.; Frandsen, K.E.; Johansen, K.S.; Berrin, J.G.; Lo Leggio, L. Recent insights into lytic polysaccharide monooxygenases (LPMOs). Biochem. Soc. Trans. 2018, 46, 1431–1447. [Google Scholar] [CrossRef]
- Xu, P.; Ni, Z.F.; Zong, M.H.; Ou, X.Y.; Yang, J.G.; Lou, W.Y. Improving the thermostability and activity of Paenibacillus pasadenensis chitinase through semi-rational design. Int. J. Biol. Macromol. 2020, 150, 9–15. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, Z.; Li, Y.; He, J.; Zhu, H. Improvement of the stability and activity of an LPMO through rational disulfide bonds design. Front. Bioeng. Biotechnol. 2022, 9, 815990. [Google Scholar] [CrossRef]
- Zong, Z.; Gao, L.; Cai, W.; Yu, L.; Cui, C.; Chen, S.; Zhang, D. Computer-assisted rational modifications to improve the thermostability of β-glucosidase from Penicillium piceum H16. Bioenergy Res. 2015, 8, 1384–1390. [Google Scholar] [CrossRef]
- Szilágyi, A.; Závodszky, P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: Results of a comprehensive survey. Structure 2000, 8, 493–504. [Google Scholar] [CrossRef]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Shirley, B.A.; Hendricks, M.M.; Iimura, S.; Gajiwala, K.; Scholtz, J.M.; et al. Contribution of hydrophobic interactions to protein stability. J. Mol. Biol. 2011, 408, 514–528. [Google Scholar] [CrossRef]
- Nguyen, H.; Kondo, K.; Yagi, Y.; Iseki, Y.; Okuoka, N.; Watanabe, T.; Mikami, B.; Nagata, T.; Katahira, M. Functional and structural characterizations of lytic polysaccharide monooxygenase, which cooperates synergistically with cellulases, from Ceriporiopsis subvermispora. ACS Sustain. Chem. Eng. 2022, 10, 923–934. [Google Scholar] [CrossRef]
- Hansson, H.; Karkehabadi, S.; Mikkelsen, N.; Douglas, N.R.; Kim, S.; Lam, A.; Kaper, T.; Kelemen, B.; Meier, K.K.; Jones, S.M.; et al. High-resolution structure of a lytic polysaccharide monooxygenase from Hypocrea jecorina reveals a predicted linker as an integral part of the catalytic domain. J. Biol. Chem. 2017, 292, 19099–19109. [Google Scholar] [CrossRef] [PubMed]
- de Gouvêa, P.F.; Gerolamo, L.E.; Bernardi, A.V.; Pereira, L.; Uyemura, S.A.; Dinamarco, T.M. Lytic polysaccharide monooxygenase from Aspergillus fumigatus can improve enzymatic cocktail activity during sugarcane bagasse hydrolysis. Protein Pept. Lett. 2019, 26, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Haider, J.; Liu, P.; Yang, J.; Tan, Z.; Huang, T.; Lin, J.; Jiang, M.; Liu, H.; Zhu, L. Engineered LPMO significantly boosting cellulase-catalyzed depolymerization of cellulose. J. Agric. Food. Chem. 2020, 68, 15257–15266. [Google Scholar] [CrossRef]
- Wood, T. The cellulase of Fusarium solani. Purification and specificity of the β-(1→4)-glucanase and the β-D-glucosidase components. Biochem. J. 1971, 121, 353–362. [Google Scholar] [CrossRef]
- Jia, X.; Chen, Y.; Shi, C.; Ye, Y.; Wang, P.; Zeng, X.; Wu, T. Preparation and characterization of cellulose regenerated from phosphoric acid. J. Agric. Food. Chem. 2013, 61, 12405–12414. [Google Scholar] [CrossRef]
- Butera, G.; De Pasquale, C.; Maccotta, A.; Alonzo, G.; Conte, P. Thermal transformation of micro-crystalline cellulose in phosphoric acid. Cellulose 2011, 18, 1499–1507. [Google Scholar] [CrossRef]
- Van Den Ent, F.; Löwe, J. RF cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Bioph. Meth. 2006, 67, 67–74. [Google Scholar] [CrossRef]
- Westereng, B.; Agger, J.W.; Horn, S.J.; Vaaje-Kolstad, G.; Aachmann, F.L.; Stenstrøm, Y.H.; Eijsink, V.G. Efficient separation of oxidized cello-oligosaccharides generated by cellulose degrading lytic polysaccharide monooxygenases. J. Chromatogr. A 2013, 1271, 144–152. [Google Scholar] [CrossRef]
- Westereng, B.; Arntzen, M.Ø.; Aachmann, F.L.; Várnai, A.; Eijsink, V.G.; Agger, J.W. Simultaneous analysis of C1 and C4 oxidized oligosaccharides, the products of lytic polysaccharide monooxygenases acting on cellulose. J. Chromatogr. A 2016, 1445, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Breslmayr, E.; Hanžek, M.; Hanrahan, A.; Leitner, C.; Kittl, R.; Šantek, B.; Oostenbrink, C.; Ludwig, R. A fast and sensitive activity assay for lytic polysaccharide monooxygenase. Biotechnol. Biofuels 2018, 11, 79. [Google Scholar] [CrossRef] [PubMed]

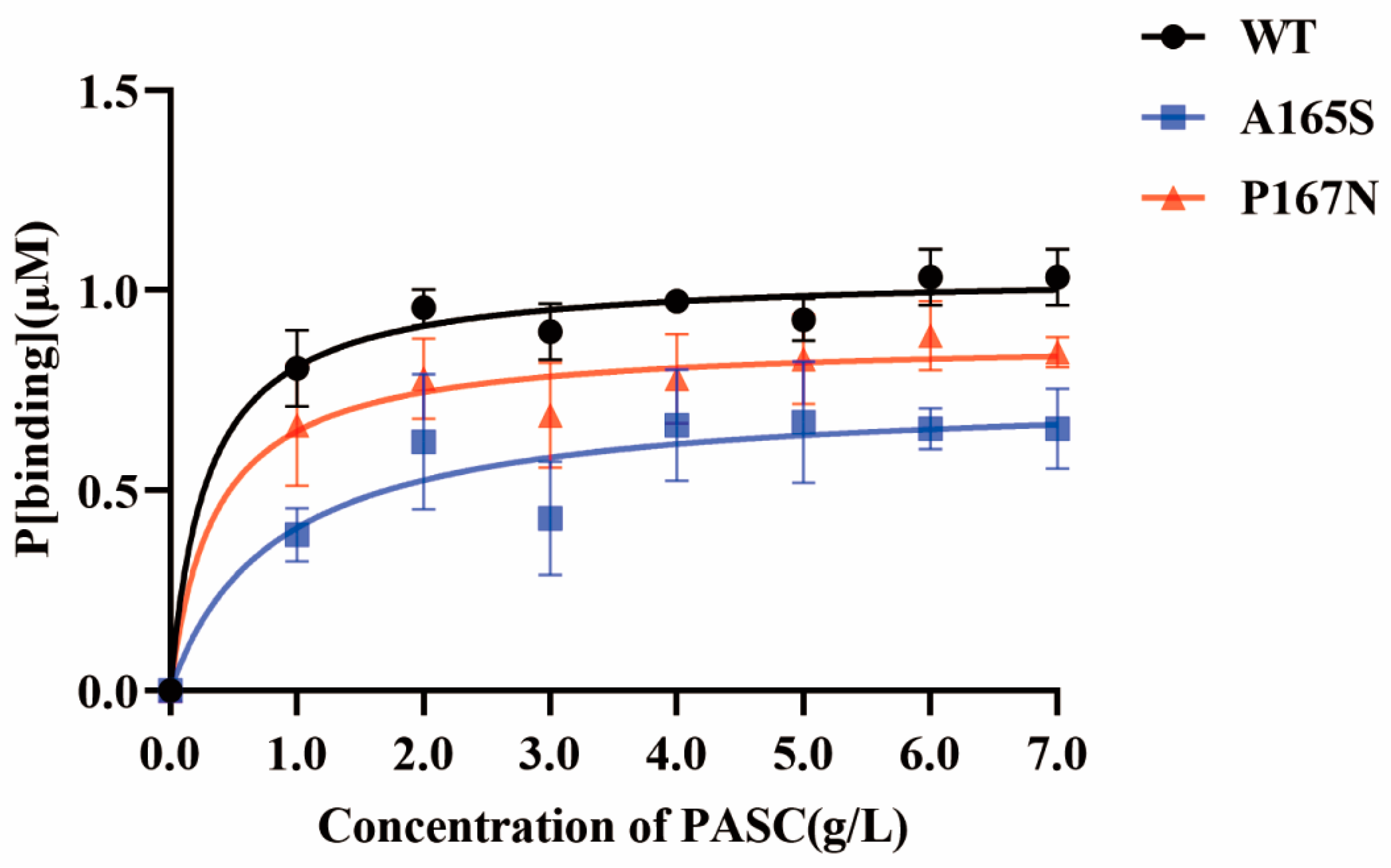

| Kd (μM) | Bmax (μM) | Kr (Bmax/Kd) | |
|---|---|---|---|
| WT | 9.15 ± 4.94 | 1.05 ± 0.06 | 1.1 × 10−1 |
| A165S | 34.37 ± 27.97 | 0.78 ± 0.17 | 2.3 × 10−2 |
| P167N | 12.60 ± 10.34 | 0.89 ± 0.10 | 7.0 × 10−2 |
| Vmax (U/g) | Km (mM) | |
|---|---|---|
| WT | 662.2 ± 44.3 | 23.7 ± 3.5 |
| A165S | 808.5 ± 94.4 | 17.3 ± 5.1 |
| P167N | 683.5 ± 82.9 | 19.9 ± 5.7 |
| Name | Sequence (5′-3′) | Usage |
|---|---|---|
| 5′AOX | GACTGGTTCCAATTGACAAGC | Transformant verification |
| 3′AOX | GCAAATGGCATTCTGACATCC | |
| pPICZαA-A165S-F | CAGTTCCAACCCCGGCCC | Construction of mutant A165S |
| pPICZαA-A165S-R | TTGGAACTGCCGCCACCAGT | |
| pPICZαA-N166G-F | TGCCGGCCCCGGCCCGACCGTCT | Construction of mutant N166G |
| pPICZαA-N166G-R | GGGGCCGGCACTGCCGCCACCA | |
| pPICZαA-P167N-F | CAACAACGGCCCGACCGT | Construction of mutant P167N |
| pPICZαA-P167N-R | GCCGTTGTTGGCACTGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, Y.; Gao, W.; Liu, X.; Li, T.; Li, K.; Yin, H. Improving the Catalytic Efficiency of an AA9 Lytic Polysaccharide Monooxygenase MtLPMO9G by Consensus Mutagenesis. Catalysts 2024, 14, 614. https://doi.org/10.3390/catal14090614
Meng Y, Gao W, Liu X, Li T, Li K, Yin H. Improving the Catalytic Efficiency of an AA9 Lytic Polysaccharide Monooxygenase MtLPMO9G by Consensus Mutagenesis. Catalysts. 2024; 14(9):614. https://doi.org/10.3390/catal14090614
Chicago/Turabian StyleMeng, Yao, Wa Gao, Xiaohua Liu, Tang Li, Kuikui Li, and Heng Yin. 2024. "Improving the Catalytic Efficiency of an AA9 Lytic Polysaccharide Monooxygenase MtLPMO9G by Consensus Mutagenesis" Catalysts 14, no. 9: 614. https://doi.org/10.3390/catal14090614
APA StyleMeng, Y., Gao, W., Liu, X., Li, T., Li, K., & Yin, H. (2024). Improving the Catalytic Efficiency of an AA9 Lytic Polysaccharide Monooxygenase MtLPMO9G by Consensus Mutagenesis. Catalysts, 14(9), 614. https://doi.org/10.3390/catal14090614






