The Impact of Bilirubin on 7α- and 7β-Hydroxysteroid Dehydrogenases: Spectra and Docking Analysis
Abstract
1. Introduction
2. Results and Discussion
2.1. Properties of 7α- and 7β-HSDHs
2.2. The Effects of Bilirubin on Enzymatic Activity and Kinetics
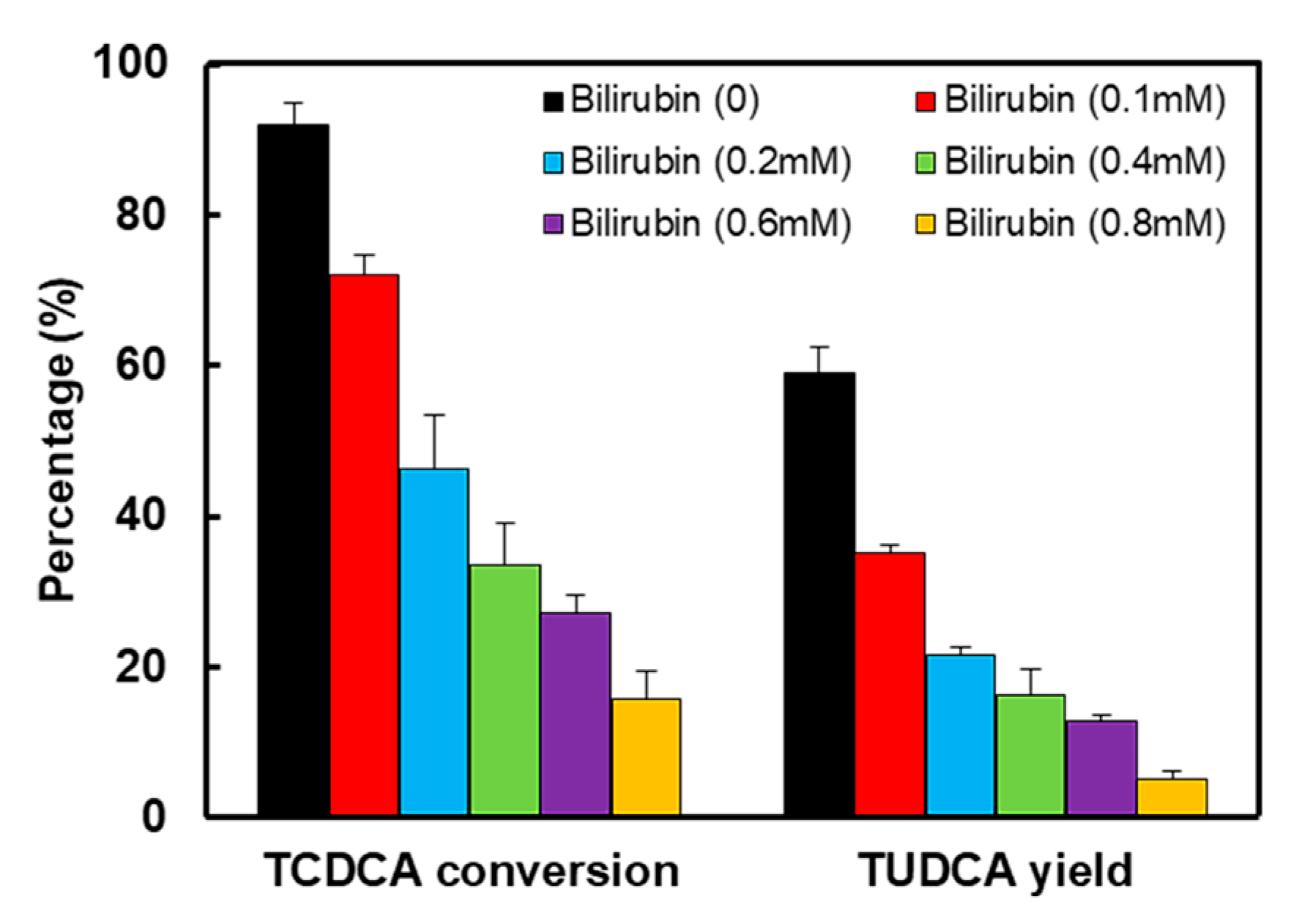
2.3. The Effect of Bilirubin on Enzymatic Reaction
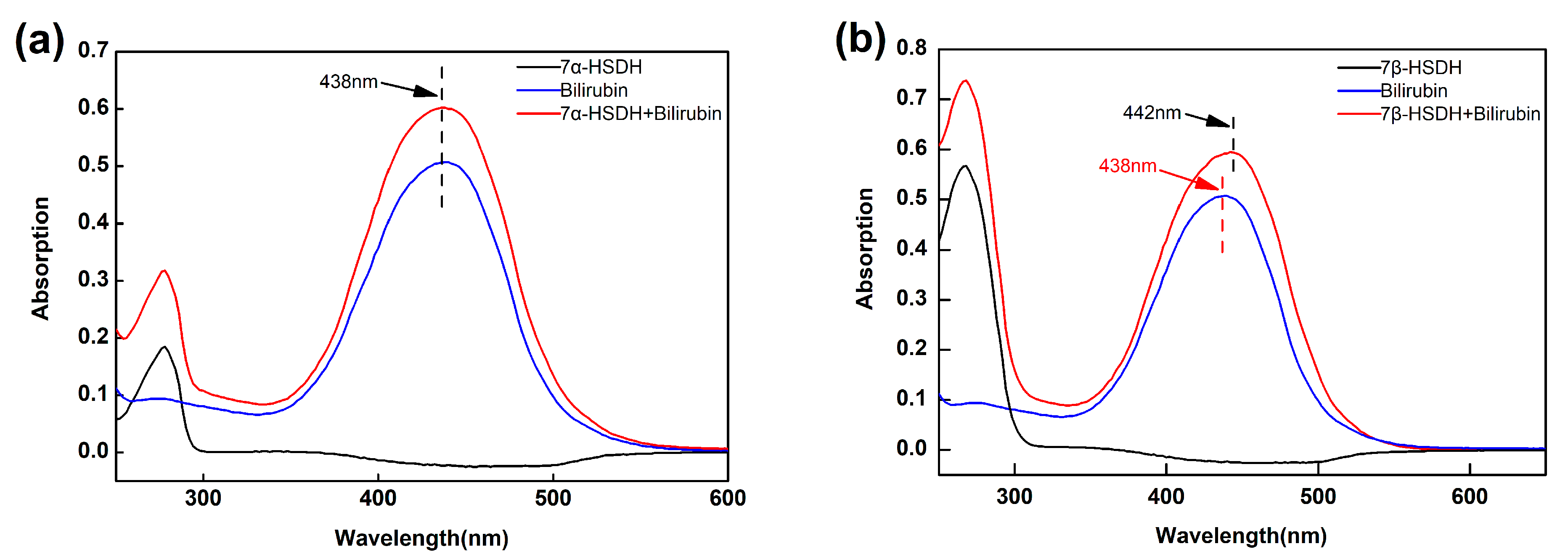
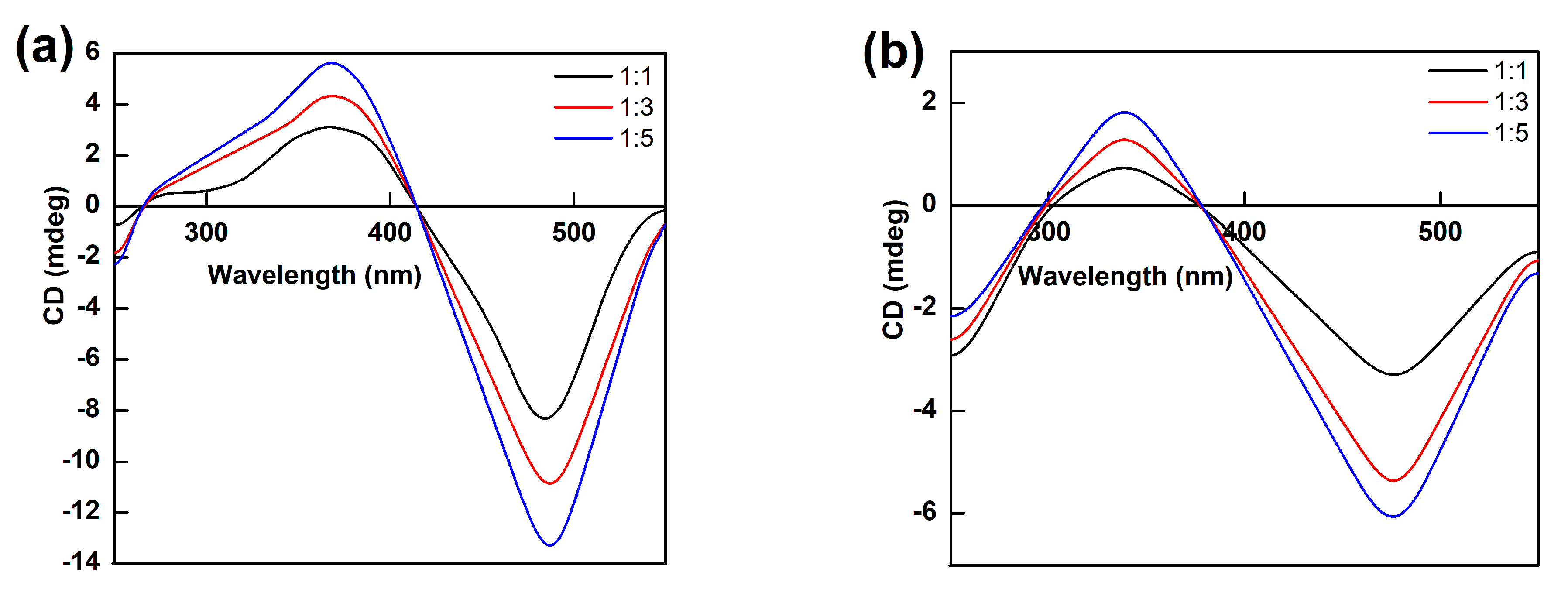
2.4. Analysis of the Binding of Enzymes and Bilirubin
2.5. Exploration of Inhibition Mechanism
3. Materials and Methods
3.1. Materials
3.2. Properties of 7α- and 7β-HSDH
3.3. Enzymatic Activity and Kinetic Characterization
3.4. Enzymatic Reaction
3.5. The Effect of Bilirubin on Enzyme Activity and TCDCA Transformation
3.6. The Combination Analysis of Bilirubin and Enzymes
3.6.1. UV–Vis
3.6.2. Fluorescence
3.6.3. CD Spectroscopy
3.7. High-Performance Liquid Chromatography (HPLC)
3.8. Molecular Docking of Bilirubin and Enzymes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Vallim, T.Q.D.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef]
- Alpini, G.; Baiocchi, L.; Glaser, S.; Ueno, Y.; Marzioni, M.; Francis, H.; Phinizy, J.L.; Angelico, M.; LeSage, G. Ursodeoxycholate and tauroursodeoxycholate inhibit cholangiocyte growth and secretion of BDL rats through activation of PKC alpha. Hepatology 2002, 35, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.H.; van Buuren, H.R.; Corpechot, C.; Thorburn, D.; Janssen, H.L.A.; Lindor, K.D.; Hirschfield, G.M.; Parés, A.; Floreani, A.; Mayo, M.J.; et al. Ursodeoxycholic acid therapy and liver transplant-free survival in patients with primary biliary cholangitis. J. Hepatol. 2019, 71, 357–365. [Google Scholar] [CrossRef]
- Wu, P.; Zhao, J.; Guo, Y.; Yu, Y.; Wu, X.; Xiao, H. Ursodeoxycholic acid alleviates nonalcoholic fatty liver disease by inhibiting apoptosis and improving autophagy via activating AMPK. Biochem. Biophys. Res. Commun. 2020, 529, 834–838. [Google Scholar] [CrossRef]
- Fernandez-Sanchez, L.; Lax, P.; Noailles, A.; Angulo, A.; Maneu, V.; Cuenca, N. Natural Compounds from Saffron and Bear Bile Prevent Vision Loss and Retinal Degeneration. Molecules 2015, 20, 13875–13893. [Google Scholar] [CrossRef]
- Hu, J.; Xu, M.; Yuan, J.; Li, B.; Entenman, S.; Yu, H.; Zheng, Q.Y. Tauroursodeoxycholic Acid Prevents Hearing Loss and Hair Cell Death in Cdh23erl/Erl Mice. Neuroscience 2016, 316, 311–320. [Google Scholar] [CrossRef]
- Qi, H.; Shen, D.; Jiang, C.; Wang, H.; Chang, M. Ursodeoxycholic acid protects dopaminergic neurons from oxidative stress via regulating mitochondrial function, autophagy, and apoptosis in MPTP/MPP+-induced Parkinson’s disease. Neurosci. Lett. 2021, 741, 135493. [Google Scholar] [CrossRef]
- Rosa, A.I.; Duarte-Silva, S.; Silva-Fernandes, A.; Nunes, M.J.; Carvalho, A.N.; Rodrigues, E.; Gama, M.J.; Rodrigues, C.M.P.; Maciel, P.; Castro-Caldas, M. Tauroursodeoxycholic Acid Improves Motor Symptoms in a Mouse Model of Parkinson’s Disease. Mol. Neurobiol. 2018, 55, 9139–9155. [Google Scholar] [CrossRef]
- Cho, J.G.; Lee, J.H.; Hong, S.H.; Lee, H.N.; Kim, C.M.; Kim, S.Y.; Yoon, K.J.; Oh, B.J.; Kim, J.H.; Jung, S.Y.; et al. Tauroursodeoxycholic Acid, a Bile Acid, Promotes Blood Vessel Repair by Recruiting Vasculogenic Progenitor Cells. Stem Cells 2015, 33, 792–805. [Google Scholar] [CrossRef]
- Guo, Q.; Shi, Q.; Li, H.; Liu, J.; Wu, S.; Sun, H.; Zhou, B. Glycolipid Metabolism Disorder in the Liver of Obese Mice Is Improved by TUDCA via the Restoration of Defective Hepatic Autophagy. Int. J. Endocrinol. 2015, 2015, 11. [Google Scholar] [CrossRef]
- Nabil, T.M.; Khalil, A.H.; Gamal, K. Effect of oral ursodeoxycholic acid on cholelithiasis following laparoscopic sleeve gastrectomy for morbid obesity. Surg. Obes. Relat. Dis. 2019, 15, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.Y.; Hong, S.H.; Lee, Y.J.; Chung, S.S.; Jung, H.S.; Park, S.G.; Park, K.S. Tauroursodeoxycholate (TUDCA), chemical chaperone, enhances function of islets by reducing ER stress. Biochem. Biophys. Res. Commun. 2010, 397, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Brevini, T.; Maes, M.; Webb, G.J.; John, B.V.; Fuchs, C.D.; Buescher, G.; Wang, L.; Griffiths, C.; Brown, M.L.; Scott, W.E.; et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2022, 615, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.M.; Wang, R.F.; Li, C.X.; Xu, J.H. Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7 beta-hydroxysteroid dehydrogenase from Ruminococcus torques. Process Biochem. 2015, 50, 598–604. [Google Scholar] [CrossRef]
- Ji, Q.Z.; Tan, J.; Zhu, L.C.; Lou, D.S.; Wang, B.C. Preparing tauroursodeoxycholic acid (TUDCA) using a double-enzyme-coupled system. Biochem. Eng. J. 2016, 105, 1–9. [Google Scholar] [CrossRef]
- Ferrandi, E.E.; Bertolesi, G.M.; Polentini, F.; Negri, A.; Riva, S.; Monti, D. In search of sustainable chemical processes: Cloning, recombinant expression, and functional characterization of the 7 alpha- and 7 beta-hydroxysteroid dehydrogenases from Clostridium absonum. Appl. Microbiol. Biotechnol. 2012, 95, 1221–1233. [Google Scholar] [CrossRef]
- Bennett, M.J.; McKnight, S.L.; Coleman, J.P. Cloning and characterization of the NAD-dependent 7 alpha-hydroxysteroid dehydrogenase from Bacteroides fragilis. Curr. Microbiol. 2003, 47, 475–484. [Google Scholar] [CrossRef]
- Pedrini, P.; Andreotti, E.; Guerrini, A.; Dean, M.; Fantin, G.; Giovannini, P.P. Xanthomonas maltophilia CBS 897.97 as a source of new 7 beta- and 7 alpha-hydroxysteroid dehydrogenases and cholylglycine hydrolase: Improved biotransformations of bile acids. Steroids 2006, 71, 189–198. [Google Scholar] [CrossRef]
- Huang, B.; Yang, K.; Amanze, C.; Yan, Z.; Zhou, H.; Liu, X.; Qiu, G.; Zeng, W. Sequence and structure-guided discovery of a novel NADH-dependent 7β-hydroxysteroid dehydrogenase for efficient biosynthesis of ursodeoxycholic acid. Bioorg. Chem. 2023, 131, 106340. [Google Scholar] [CrossRef]
- Ji, S.; Pan, Y.; Zhu, L.; Tan, J.; Tang, S.; Yang, Q.; Zhang, Z.; Lou, D.; Wang, B. A novel 7α-hydroxysteroid dehydrogenase: Magnesium ion significantly enhances its activity and thermostability. Int. J. Biol. Macromol. 2021, 177, 111–118. [Google Scholar] [CrossRef]
- Ji, Q.; Wang, B.; Li, C.; Hao, J.; Feng, W. Co-immobilised 7α- and 7β-HSDH as recyclable biocatalyst: High-performance production of TUDCA from waste chicken bile. RSC Adv. 2018, 8, 34192–34201. [Google Scholar] [CrossRef] [PubMed]
- Stocker, R.; Yamamoto, Y.; Mcdonagh, A.; Glazer, A.; Ames, B.J.S. Bilirubin is an antioxidant of possible physiological importance. Science 1987, 235, 1043–1046. [Google Scholar]
- Baraano, D.E.; Rao, M.; Ferris, C.D.; Snyder, S.H. Biliverdin reductase: A major physiologic cytoprotectant. Proc. Natl. Acad. Sci. USA 2002, 99, 16093–16098. [Google Scholar] [CrossRef] [PubMed]
- Creeden, J.F.; Gordon, D.M.; Stec, D.E.; Hinds, T.D. Bilirubin as a metabolic hormone: The physiological relevance of low levels. Am. J. Physiol.-Endocrinol. Metab. 2020, 320, E191–E207. [Google Scholar] [CrossRef]
- Vitek, L.; Hinds, T.D.; Stec, D.E.; Tiribelli, C. The physiology of bilirubin: Health and disease equilibrium. Trends Mol. Med. 2023, 29, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-W.; Gong, L.-N.; Lai, K.; Yu, X.-F.; Liu, Z.-Q.; Li, M.-X.; Yin, X.-L.; Liang, M.; Shi, H.-S.; Jiang, L.-H.; et al. Bilirubin gates the TRPM2 channel as a direct agonist to exacerbate ischemic brain damage. Neuron 2023, 111, 1609–1625. [Google Scholar] [CrossRef]
- Zhou, S.; Wei, Y. Kaleidoscope megamolecules synthesis and application using self-assembly technology. Biotechnol. Adv. 2023, 65, 108147. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guo, Y.; Xianyu, Y. Applications of self-assembly strategies in immunoassays: A review. Coord. Chem. Rev. 2023, 478, 214974. [Google Scholar] [CrossRef]
- Girelli, A.M.; Chiappini, V. Renewable, sustainable, and natural lignocellulosic carriers for lipase immobilization: A review. J. Biotechnol. 2023, 365, 29–47. [Google Scholar] [CrossRef]
- Zdarta, J.; Kołodziejczak-Radzimska, A.; Bachosz, K.; Rybarczyk, A.; Bilal, M.; Iqbal, H.M.N.; Buszewski, B.; Jesionowski, T. Nanostructured supports for multienzyme co-immobilization for biotechnological applications: Achievements, challenges and prospects. Adv. Colloid Interface Sci. 2023, 315, 102889. [Google Scholar] [CrossRef]
- Odell, G.B. Studies in Kernicterus. I. The Protein Binding of Bilirubin. J. Clin. Investig. 1959, 38, 823–833. [Google Scholar] [CrossRef]
- Hulzebos, C.V.; Dijk, P.H. Bilirubin–albumin binding, bilirubin/albumin ratios, and free bilirubin levels: Where do we stand? Semin. Perinatol. 2014, 38, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Vítek, L.; Tiribelli, C. Bilirubin: The yellow hormone? J. Hepatol. 2021, 75, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Morioka, I.; Iwatani, S.; Koda, T.; Iijima, K.; Nakamura, H. Disorders of bilirubin binding to albumin and bilirubin-induced neurologic dysfunction. Semin. Fetal Neonatal Med. 2015, 20, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.B. Bilirubin Binding Capacity in the Preterm Neonate. Clin. Perinatol. 2016, 43, 241–257. [Google Scholar] [CrossRef]
- Arriaga, S.M.; Basiglio, C.L.; Mottino, A.D.; Almará, A.M. Unconjugated bilirubin inhibits C1 esterase activity. Clin. Biochem. 2009, 42, 919–921. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, U.; Gladstone Christopher, J.; Jonnalagadda, R.R.; Chandrasekaran, B.; Balachandran, U.N. Studies on the chemico-biological characteristics of bilirubin binding with collagen. Mater. Sci. Eng. C 2013, 33, 4965–4971. [Google Scholar] [CrossRef]
- Gligorijević, N.; Minić, S.; Robajac, D.; Nikolić, M.; Ćirković Veličković, T.; Nedić, O. Characterisation and the effects of bilirubin binding to human fibrinogen. Int. J. Biol. Macromol. 2019, 128, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Lear, S.; Cobb, S.L. Pep-Calc.com: A set of web utilities for the calculation of peptide and peptoid properties and automatic mass spectral peak assignment. J. Comput. Aid. Mol. Des. 2016, 30, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Michelin, M.; Ximenes, E.; Maria de Lourdes, T.M.; Ladisch, M.R. Inhibition of enzyme hydrolysis of cellulose by phenols from hydrothermally pretreated sugarcane straw. Enzym. Microb. Technol. 2023, 166, 110227. [Google Scholar] [CrossRef]
- Baksi, S.; Sarkar, U.; Villa, R.; Basu, D.; Sengupta, D. Conversion of biomass to biofuels through sugar platform: A review of enzymatic hydrolysis highlighting the trade-off between product and substrate inhibitions. Sustain. Energy Technol. Assess. 2023, 55, 102963. [Google Scholar] [CrossRef]
- Haldar, D.; Gayen, K.; Sen, D. Enumeration of monosugars’ inhibition characteristics on the kinetics of enzymatic hydrolysis of cellulose. Process Biochem. 2018, 72, 130–136. [Google Scholar] [CrossRef]
- Orlov, S.; Goncharova, I.; Urbanova, M. Circular dichroism study of the interaction between mutagens and bilirubin bound to different binding sites of serum albumins. Spectrochim. Acta A 2014, 126, 68–75. [Google Scholar] [CrossRef]
- Rodriguez Galdón, B.; Pinto Corraliza, C.; Cestero Carrillo, J.J.; Macias Laso, P. Spectroscopic study of the interaction between lycopene and bovine serum albumin. Luminescence 2013, 28, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Huang, J.; Lin, J.; Li, Z.; Liu, F.; Jiang, Z.; Sun, Y. A study of the binding of C.I. Mordant Red 3 with bovine serum albumin using fluorescence spectroscopy. Dyes Pigments 2009, 82, 65–70. [Google Scholar] [CrossRef]
- Yuan, T.; Weljie, A.M.; Vogel, H.J. Tryptophan Fluorescence Quenching by Methionine and Selenomethionine Residues of Calmodulin: Orientation of Peptide and Protein Binding. Biochemistry 1998, 37, 3187–3195. [Google Scholar] [CrossRef]
- Guo, Y.; Yue, Q.; Gao, B.; Zhong, Q. Spectroscopic studies on the interaction between disperse blue SBL and bovine serum albumin. J. Lumin. 2010, 130, 1384–1389. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, S.; Tang, Y.; Hu, Z.; Shi, L.; Lu, J.; Li, H.; Wang, Y.; Zhu, Y.; Lin, H.; et al. Direct inhibition of bisphenols on human and rat 11β-hydroxysteroid dehydrogenase 2: Structure-activity relationship and docking analysis. Ecotoxicol. Environ. Saf. 2023, 254, 114715. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tian, F.; Tang, Y.; Shi, L.; Wang, S.; Hu, Z.; Zhu, Y.; Wang, Y.; Li, H.; Ge, R.-s.; et al. Inhibition of human and rat placental 3β-hydroxysteroid dehydrogenase/Δ5,4-isomerase activities by insecticides and fungicides: Mode action by docking analysis. Chem. Biol. Interact. 2023, 369, 110292. [Google Scholar] [CrossRef]
- Lou, D.; Wang, B.; Tan, J.; Zhu, L.; Cen, X.; Ji, Q.; Wang, Y. The three-dimensional structure of Clostridium absonum 7α-hydroxysteroid dehydrogenase: New insights into the conserved arginines for NADP(H) recognition. Sci. Rep. 2016, 6, 22885. [Google Scholar] [CrossRef]
- Wang, R.; Wu, J.; Jin, D.K.; Chen, Y.; Lv, Z.; Chen, Q.; Miao, Q.; Huo, X.; Wang, F. Structure of NADP+-bound 7 beta-hydroxysteroid dehydrogenase reveals two cofactor-binding modes. Acta Cryst. 2017, F73, 246–252. [Google Scholar] [CrossRef]
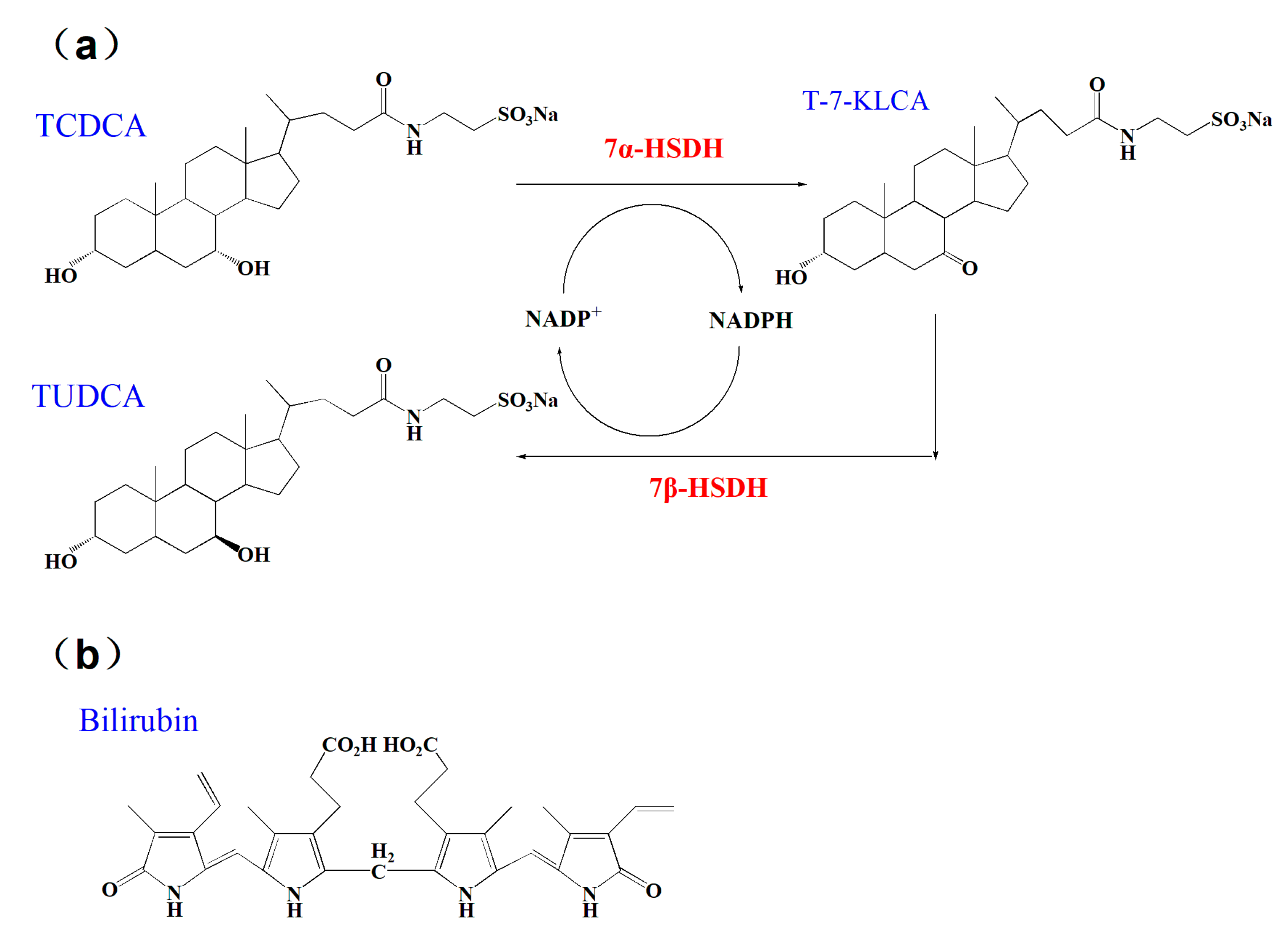
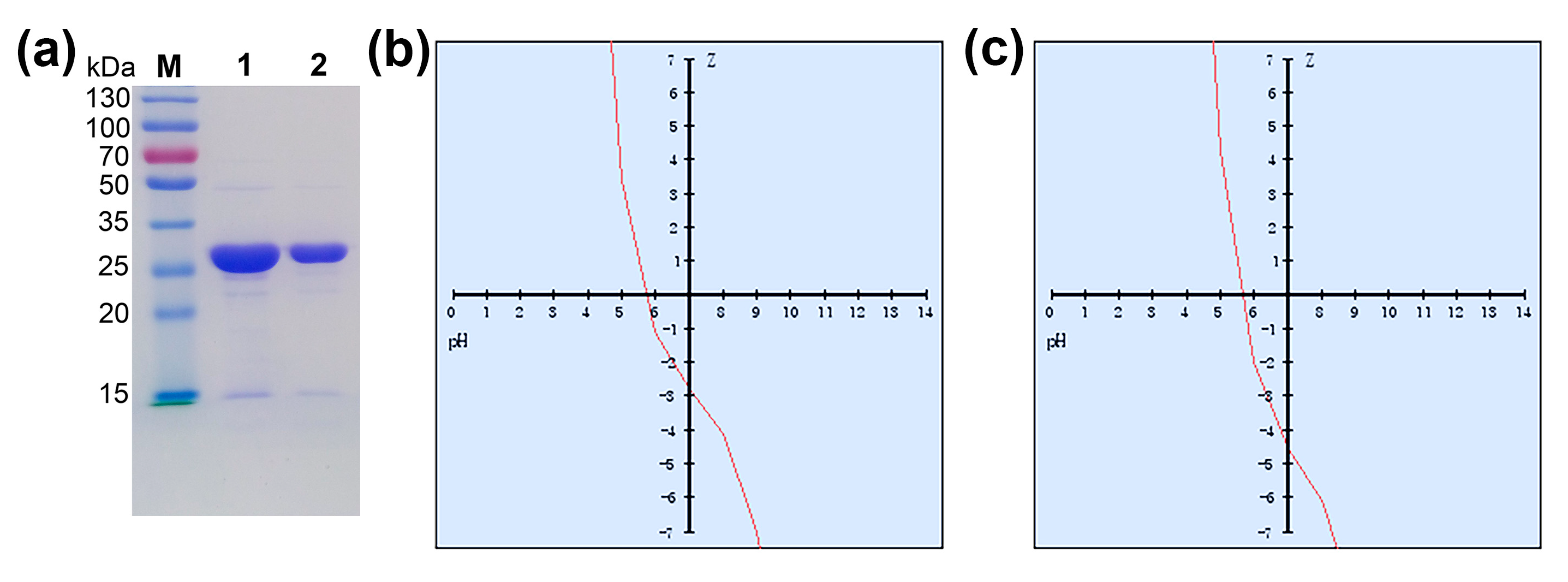




| Molecular Weight (g/mol) | Isoelectric Point | Charge at pH = 7.0 | Average Hydrophilicity | Ratio of Hydrophilic Residues | |
|---|---|---|---|---|---|
| 7α-HSDH | 28,289.16 | 5.7 | −2.8 | 0.0 | 37% |
| 7β-HSDH | 29,382.85 | 5.5 | −4.6 | 0.1 | 37% |
| Enzyme | Substrate | Vmax (μmol·min−1·mg−1) | Km (mM) | kcat (s−1) | kcat/Km (s−1·mM−1) |
|---|---|---|---|---|---|
| Free 7α-HSDH | TCDCA | 22.47 | 0.24 | 6.25 × 103 | 2.60 × 104 |
| NADP+ | 20.24 | 0.28 | 5.63 × 103 | 2.01 × 104 | |
| Free 7β-HSDH | TUDCA | 52.80 | 1.14 | 7.28 × 104 | 6.39 × 104 |
| NADP+ | 53.61 | 1.37 | 7.39 × 104 | 5.39 × 104 | |
| 7α-HSDH + bilirubin | TCDCA | 6.78 | 0.63 | 1.02 × 103 | 1.62 × 103 |
| NADP+ | 18.76 | 0.59 | 2.82 × 103 | 4.78 × 103 | |
| 7β-HSDH + bilirubin | TUDCA | 4.32 | 1.87 | 2.64 × 104 | 1.41 × 104 |
| NADP+ | 9.65 | 1.32 | 5.90 × 104 | 4.47 × 104 |
| Number | 7α-HSDH Binding with Bilirubin | 7β-HSDH Binding with Bilirubin | ||
|---|---|---|---|---|
| Binding Energy (kcal/mol) | Structures | Binding Energy (kcal/mol) | Structures | |
| 1 | −4.71 |  | −5.79 |  |
| 2 | −4.67 |  | −5.62 |  |
| 3 | −4.6 | 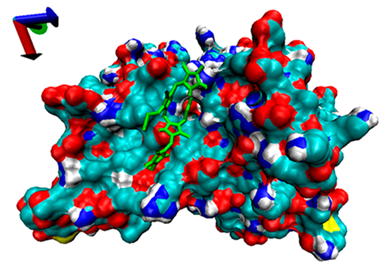 | −5.39 | 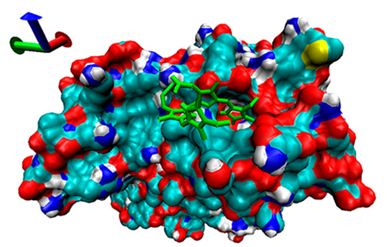 |
| 4 | −4.55 | 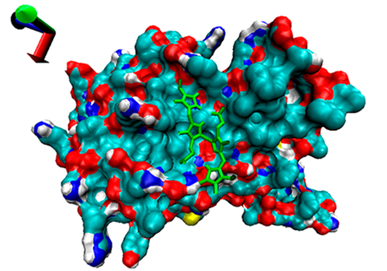 | −4.97 | 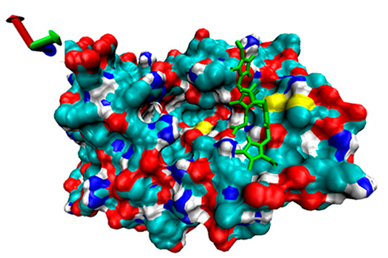 |
| 5 | −4.39 | 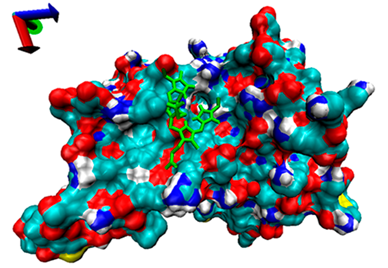 | −4.91 | 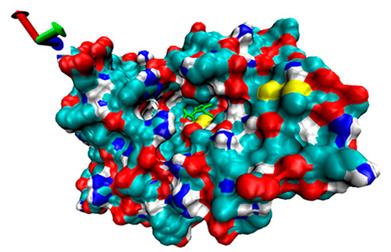 |
| 6 | −4.35 | 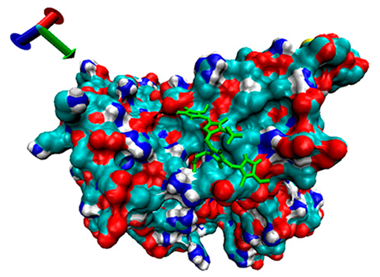 | −4.68 | 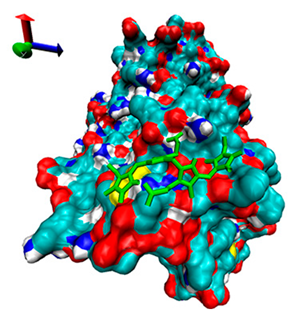 |
| 7 | −4.31 |  | −4.83 | 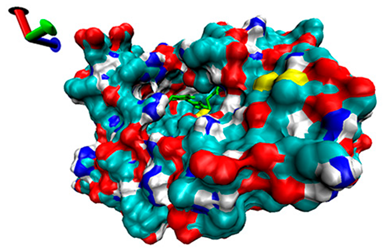 |
| 8 | −4.27 | 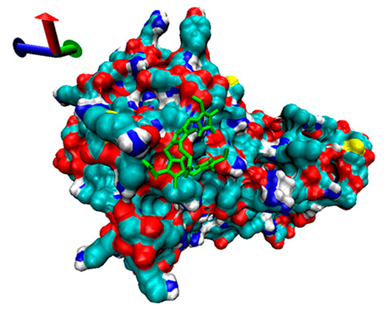 | −4.8 | 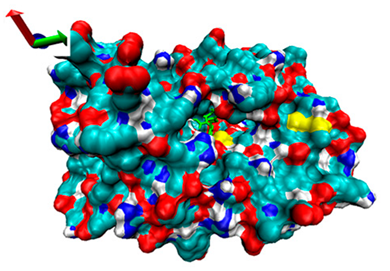 |
| 9 | −4.16 | 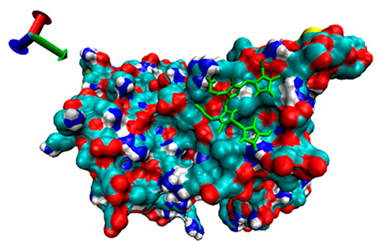 | −4.74 | 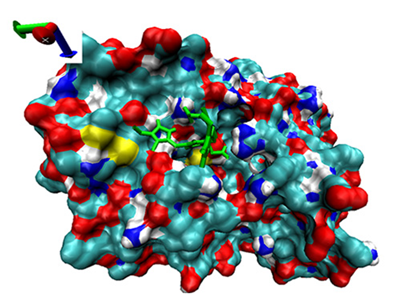 |
| 10 | −4.07 | 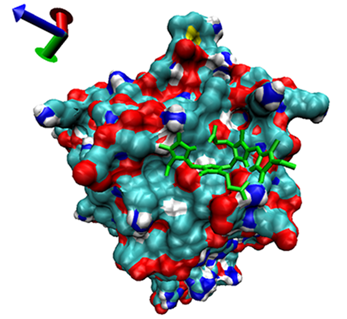 | −4.32 | 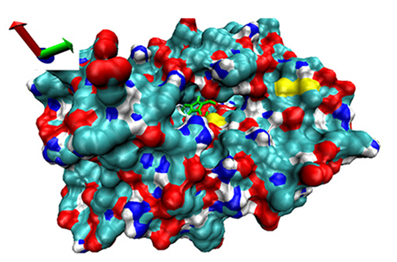 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Q.; Chen, J.; Zhu, L.; Wang, R.; Wang, B. The Impact of Bilirubin on 7α- and 7β-Hydroxysteroid Dehydrogenases: Spectra and Docking Analysis. Catalysts 2023, 13, 965. https://doi.org/10.3390/catal13060965
Ji Q, Chen J, Zhu L, Wang R, Wang B. The Impact of Bilirubin on 7α- and 7β-Hydroxysteroid Dehydrogenases: Spectra and Docking Analysis. Catalysts. 2023; 13(6):965. https://doi.org/10.3390/catal13060965
Chicago/Turabian StyleJi, Qingzhi, Jiamin Chen, Luping Zhu, Ruiyao Wang, and Bochu Wang. 2023. "The Impact of Bilirubin on 7α- and 7β-Hydroxysteroid Dehydrogenases: Spectra and Docking Analysis" Catalysts 13, no. 6: 965. https://doi.org/10.3390/catal13060965
APA StyleJi, Q., Chen, J., Zhu, L., Wang, R., & Wang, B. (2023). The Impact of Bilirubin on 7α- and 7β-Hydroxysteroid Dehydrogenases: Spectra and Docking Analysis. Catalysts, 13(6), 965. https://doi.org/10.3390/catal13060965






