Lignin Valorization: Production of High Value-Added Compounds by Engineered Microorganisms
Abstract
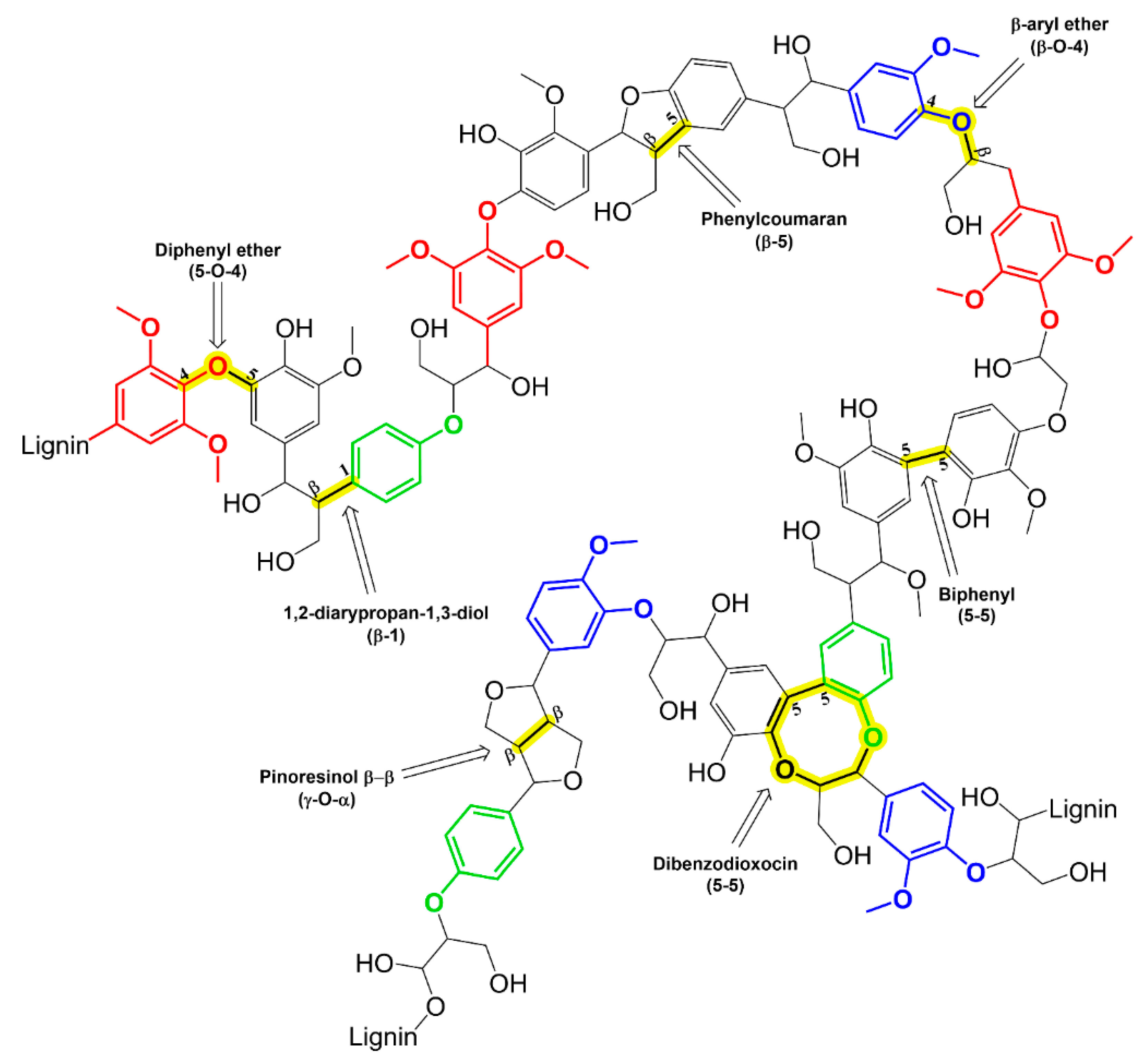
1. Introduction
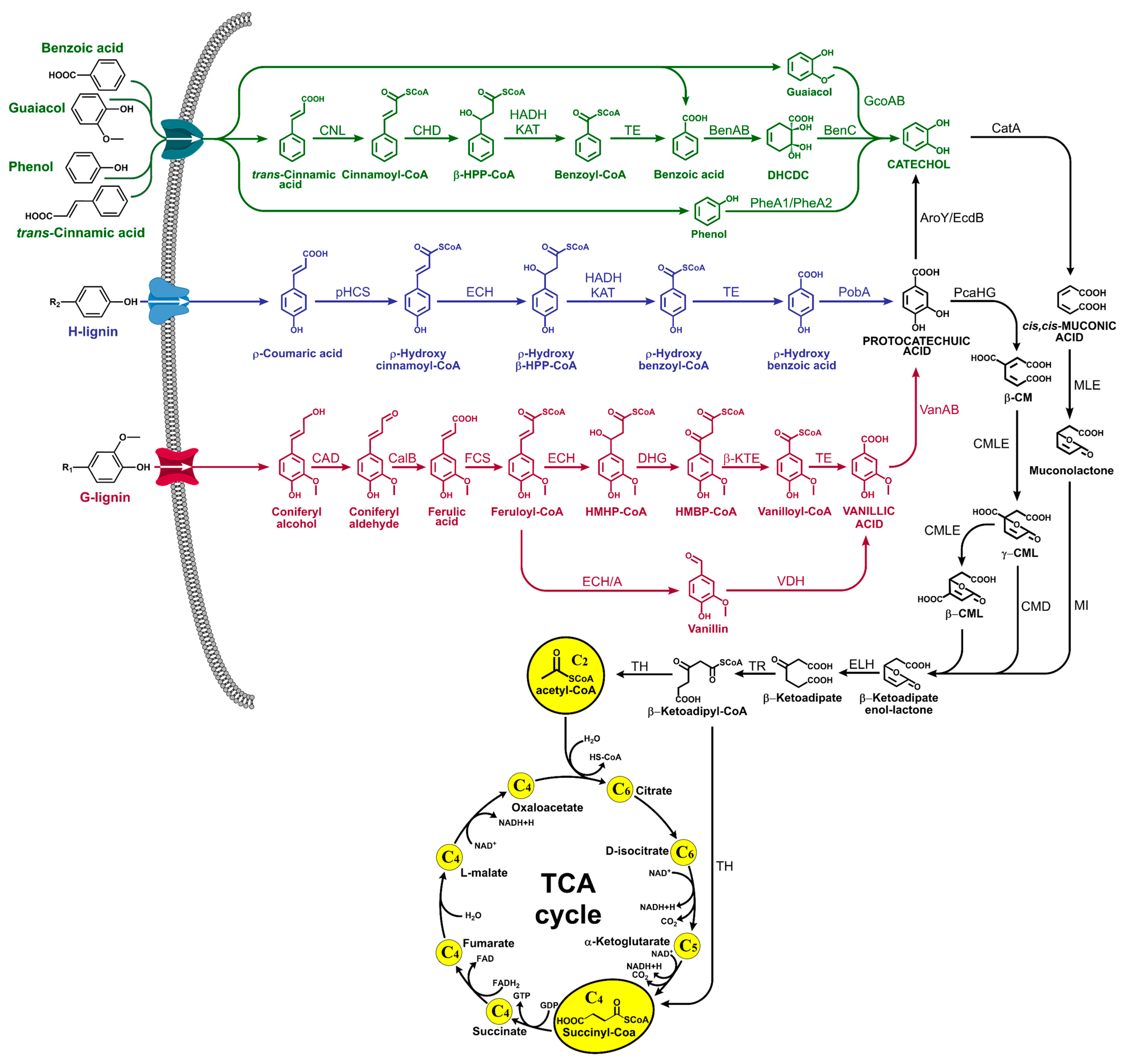
2. Microbial Lignin Depolymerization
2.1. Microbial Consortia
2.2. Extremophilic Microorganisms
3. Lignin Valorization: The Biological Routes towards Value-Added and Specialty Chemicals
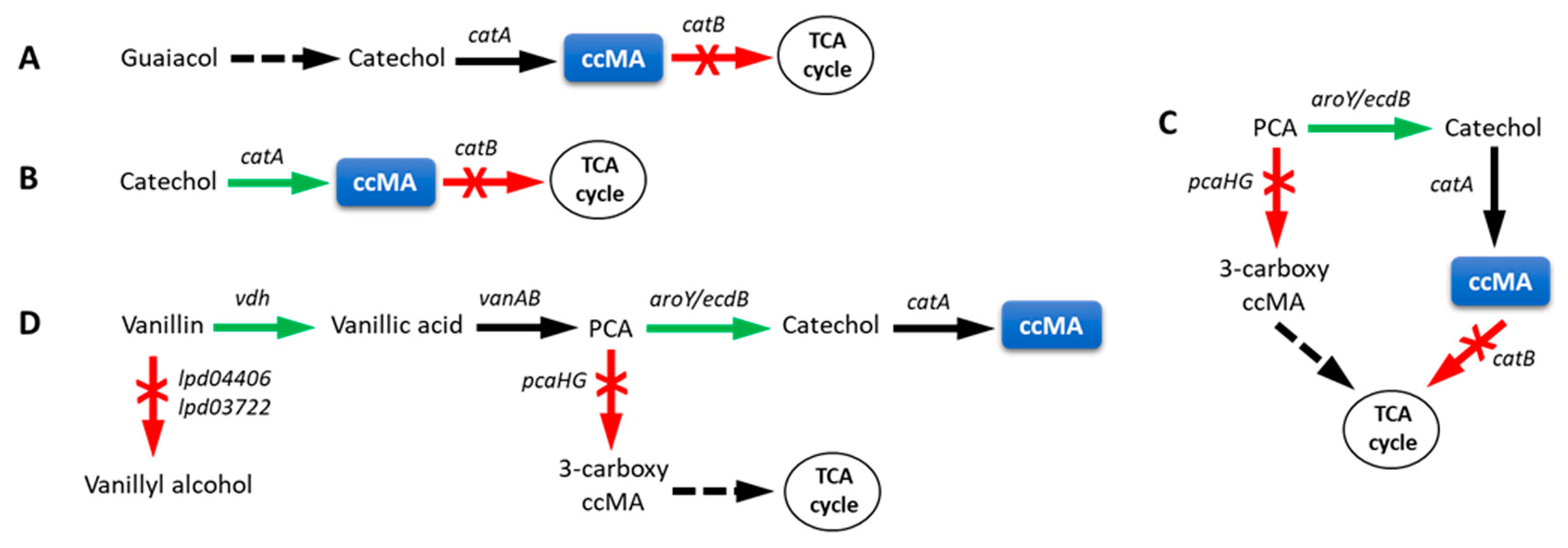
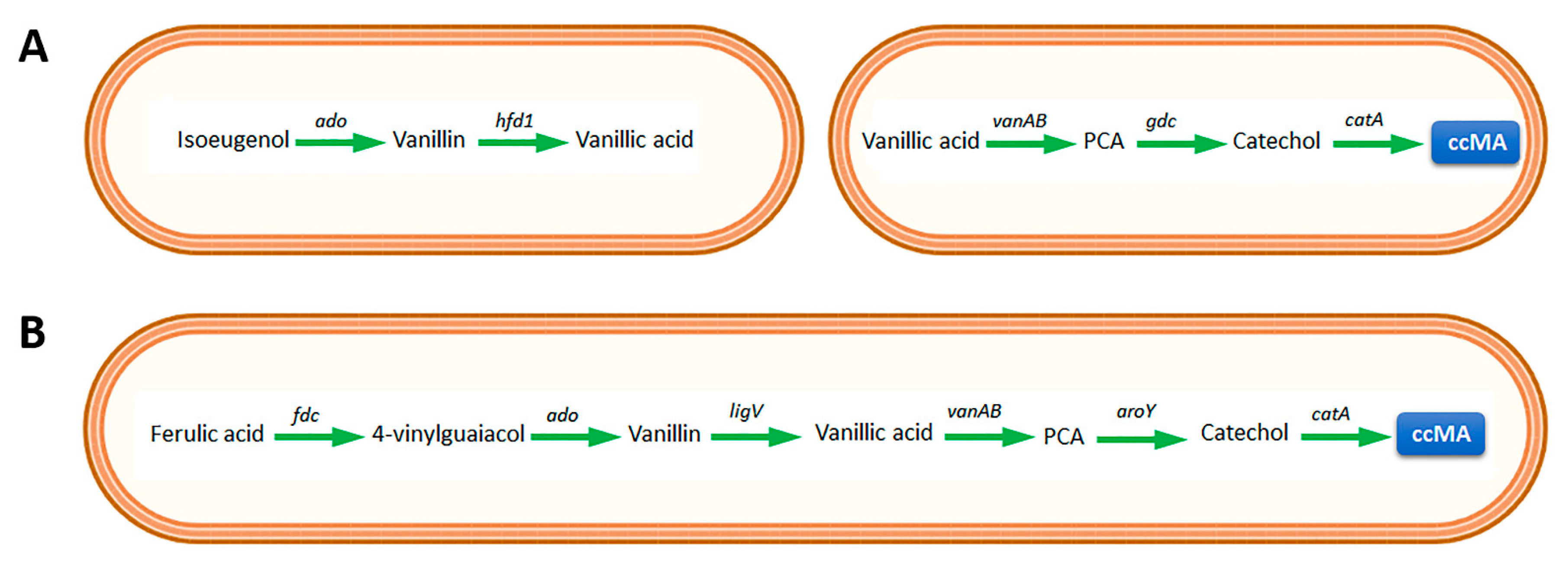
3.1. Cis,cis-Muconic Acid
3.1.1. Amycolatopsis sp. ATCC 39166
3.1.2. Corynebacterium glutamicum
3.1.3. Rhodococcus opacus
3.1.4. Escherichia coli
3.1.5. Pseudomonas putida
3.2. Bio-Based Plastics Precursors
3.3. Pharmaceuticals
4. Perspectives
- -
- the screening/use of engineered promoters with varying strength that could lead to tunable systems aimed at maximization of protein expression of selected enzymes of the relevant pathways;
- -
- the use of metabolic engineering and protein engineering approaches to remove any bottleneck of the relevant pathways;
- -
- the optimization of fermentation conditions;
- -
- the efficient supply of large quantities of lignin in a relatively uniform, purified and biocompatible form;
- -
- the optimization of lignin depolymerization to yield a substrate with high monomer concentrations;
- -
- the limitation of toxicity of lignin degradation products;
- -
- the optimization of the products’ separation process that has been estimated to account for over 60% of production costs [134].
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernández-Rodríguez, J.; Erdocia, X.; Sánchez, C.; González Alriols, M.; Labidi, J. Lignin Depolymerization for Phenolic Monomers Production by Sustainable Processes. J. Energy Chem. 2017, 26, 622–631. [Google Scholar] [CrossRef]
- van den Bosch, S.; Koelewijn, S.-F.; Renders, T.; van den Bossche, G.; Vangeel, T.; Schutyser, W.; Sels, B.F. Catalytic Strategies Towards Lignin-Derived Chemicals. Top Curr. Chem. 2018, 376, 36. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, R.; Jastrzebski, R.; Clough, M.T.; Ralph, J.; Kennema, M.; Bruijnincx, P.C.A.; Weckhuysen, B.M. Paving the Way for Lignin Valorisation: Recent Advances in Bioengineering, Biorefining and Catalysis. Angew. Chem. Int. Ed. 2016, 55, 8164–8215. [Google Scholar] [CrossRef]
- Corona, A.; Biddy, M.J.; Vardon, D.R.; Birkved, M.; Hauschild, M.Z.; Beckham, G.T. Life Cycle Assessment of Adipic Acid Production from Lignin. Green Chem. 2018, 20, 3857–3866. [Google Scholar] [CrossRef]
- Ponnusamy, V.K.; Nguyen, D.D.; Dharmaraja, J.; Shobana, S.; Banu, J.R.; Saratale, R.G.; Chang, S.W.; Kumar, G. A Review on Lignin Structure, Pretreatments, Fermentation Reactions and Biorefinery Potential. Bioresour. Technol. 2019, 271, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, K. Lignocellulose: A Chewy Problem. Nature 2011, 474, S12–S14. [Google Scholar] [CrossRef]
- Abdelaziz, O.Y.; Brink, D.P.; Prothmann, J.; Ravi, K.; Sun, M.; García-Hidalgo, J.; Sandahl, M.; Hulteberg, C.P.; Turner, C.; Lidén, G.; et al. Biological Valorization of Low Molecular Weight Lignin. Biotechnol. Adv. 2016, 34, 1318–1346. [Google Scholar] [CrossRef]
- Chio, C.; Sain, M.; Qin, W. Lignin Utilization: A Review of Lignin Depolymerization from Various Aspects. Renew. Sustain. Energy Rev. 2019, 107, 232–249. [Google Scholar] [CrossRef]
- Li, C.; Chen, C.; Wu, X.; Tsang, C.-W.; Mou, J.; Yan, J.; Liu, Y.; Lin, C.S.K. Recent Advancement in Lignin Biorefinery: With Special Focus on Enzymatic Degradation and Valorization. Bioresour. Technol. 2019, 291, 121898. [Google Scholar] [CrossRef]
- Yu, X.; Wei, Z.; Lu, Z.; Pei, H.; Wang, H. Activation of Lignin by Selective Oxidation: An Emerging Strategy for Boosting Lignin Depolymerization to Aromatics. Bioresour. Technol. 2019, 291, 121885. [Google Scholar] [CrossRef]
- Kamimura, N.; Sakamoto, S.; Mitsuda, N.; Masai, E.; Kajita, S. Advances in Microbial Lignin Degradation and Its Applications. Curr. Opin. Biotechnol. 2019, 56, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Brunow, G. Lignin Chemistry and Its Role in Biomass Conversion. In Biorefineries-Industrial Processes and Products; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2005; pp. 151–163. [Google Scholar]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Singh, R. The Emerging Role for Bacteria in Lignin Degradation and Bio-Product Formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, S.; Lukk, T.; Solbiati, J.O.; Bauer, S.; Nair, S.K.; Cronan, J.E.; Gerlt, J.A. Roles of Small Laccases from Streptomyces in Lignin Degradation. Biochemistry 2014, 53, 4047–4058. [Google Scholar] [CrossRef]
- Numata, K.; Morisaki, K. Screening of Marine Bacteria To Synthesize Polyhydroxyalkanoate from Lignin: Contribution of Lignin Derivatives to Biosynthesis by Oceanimonas doudoroffii. ACS Sustain. Chem. Eng. 2015, 3, 569–573. [Google Scholar] [CrossRef]
- Salvachúa, D.; Karp, E.M.; Nimlos, C.T.; Vardon, D.R.; Beckham, G.T. Towards Lignin Consolidated Bioprocessing: Simultaneous Lignin Depolymerization and Product Generation by Bacteria. Green Chem. 2015, 17, 4951–4967. [Google Scholar] [CrossRef]
- Pollegioni, L.; Tonin, F.; Rosini, E. Lignin-Degrading Enzymes. FEBS J. 2015, 282, 1190–1213. [Google Scholar] [CrossRef] [PubMed]
- de Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial Enzymes Involved in Lignin Degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Tonin, F.; Melis, R.; Cordes, A.; Sanchez-Amat, A.; Pollegioni, L.; Rosini, E. Comparison of Different Microbial Laccases as Tools for Industrial Uses. New Biotechnol. 2016, 33, 387–398. [Google Scholar] [CrossRef]
- Vignali, E.; Tonin, F.; Pollegioni, L.; Rosini, E. Characterization and Use of a Bacterial Lignin Peroxidase with an Improved Manganese-Oxidative Activity. Appl. Microbiol. Biotechnol. 2018, 102, 10579–10588. [Google Scholar] [CrossRef]
- Masai, E.; Katayama, Y.; Fukuda, M. Genetic and Biochemical Investigations on Bacterial Catabolic Pathways for Lignin-Derived Aromatic Compounds. Biosci. Biotechnol. Biochem. 2007, 71, 1–15. [Google Scholar] [CrossRef]
- Shen, X.-H.; Zhou, N.-Y.; Liu, S.-J. Degradation and Assimilation of Aromatic Compounds by Corynebacterium Glutamicum: Another Potential for Applications for This Bacterium? Appl. Microbiol. Biotechnol. 2012, 95, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Linger, J.G.; Vardon, D.R.; Guarnieri, M.T.; Karp, E.M.; Hunsinger, G.B.; Franden, M.A.; Johnson, C.W.; Chupka, G.; Strathmann, T.J.; Pienkos, P.T.; et al. Lignin Valorization through Integrated Biological Funneling and Chemical Catalysis. Proc. Natl. Acad. Sci. USA 2014, 111, 12013–12018. [Google Scholar] [CrossRef] [PubMed]
- Bommareddy, R.R.; Chen, Z.; Rappert, S.; Zeng, A.-P. A de Novo NADPH Generation Pathway for Improving Lysine Production of Corynebacterium Glutamicum by Rational Design of the Coenzyme Specificity of Glyceraldehyde 3-Phosphate Dehydrogenase. Metab. Eng. 2014, 25, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.C.; Simmons, B.A.; Sale, K.L. Structure-Based Engineering of a Plant-Fungal Hybrid Peroxidase for Enhanced Temperature and PH Tolerance. Cell Chem. Biol. 2018, 25, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Schwille, P.; Spatz, J.; Landfester, K.; Bodenschatz, E.; Herminghaus, S.; Sourjik, V.; Erb, T.J.; Bastiaens, P.; Lipowsky, R.; Hyman, A.; et al. MaxSynBio: Avenues Towards Creating Cells from the Bottom Up. Angew. Chem. Int. Ed. 2018, 57, 13382–13392. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Gautam, A.; Dutt, D. Bio-Pulping: An Energy Saving and Environment-Friendly Approach. Phys. Sci. Rev. 2020, 5, 1–9. [Google Scholar] [CrossRef]
- Zieglowski, M.; Trosien, S.; Rohrer, J.; Mehlhase, S.; Weber, S.; Bartels, K.; Siegert, G.; Trellenkamp, T.; Albe, K.; Biesalski, M. Reactivity of Isocyanate-Functionalized Lignins: A Key Factor for the Preparation of Lignin-Based Polyurethanes. Front. Chem. 2019, 7, 562. [Google Scholar] [CrossRef]
- Kalliola, A.; Kangas, P.; Winberg, I.; Vehmas, T.; Kyllönen, H.; Heikkinen, J.; Poukka, O.; Kemppainen, K.; Sjögård, P.; Pehu-Lehtonen, L.; et al. Oxidation Process Concept to Produce Lignin Dispersants at a Kraft Pulp Mill. Nord. Pulp Pap. Res. J. 2022, 37, 394–404. [Google Scholar] [CrossRef]
- Scroccarello, A.; della Pelle, F.; Ul, Q.; Bukhari, A.; Silveri, F.; Zappi, D.; Cozzoni, E.; Compagnone, D. Eucalyptus Biochar as a Sustainable Nanomaterial for Electrochemical Sensors. Chem. Proc. 2021, 5, 13. [Google Scholar] [CrossRef]
- Scholten, P.B.V.; Figueirêdo, M.B. Back to the Future with Biorefineries: Bottom-Up and Top-Down Approaches toward Polymers and Monomers. Macromol. Chem. Phys. 2022, 223, 2200017. [Google Scholar] [CrossRef]
- Li, K.; Xu, F.; Eriksson, K.E.L. Comparison of Fungal Laccases and Redox Mediators in Oxidation of a Nonphenolic Lignin Model Compound. Appl. Env. Microbiol. 1999, 65, 2654–2660. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.C.H.; Pawar, S.V.; Hallam, S.J.; Yadav, V.G. An Improved Whole-Cell Biosensor for the Discovery of Lignin-Transforming Enzymes in Functional Metagenomic Screens. ACS Synth. Biol. 2018, 7, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.V.; Hallam, S.J.; Yadav, V.G. Metagenomic Discovery of a Novel Transaminase for Valorization of Monoaromatic Compounds. RSC Adv. 2018, 8, 22490–22497. [Google Scholar] [CrossRef] [PubMed]
- Levy-Booth, D.J.; Navas, L.E.; Fetherolf, M.M.; Liu, L.Y.; Dalhuisen, T.; Renneckar, S.; Eltis, L.D.; Mohn, W.W. Discovery of Lignin-Transforming Bacteria and Enzymes in Thermophilic Environments Using Stable Isotope Probing. ISME J. 2022, 16, 1944–1956. [Google Scholar] [CrossRef]
- Díaz-García, L.; Huang, S.; Spröer, C.; Sierra-Ramírez, R.; Bunk, B.; Overmann, J.; Jiménez, D.J.; Drake, H.L. Dilution-to-Stimulation/Extinction Method: A Combination Enrichment Strategy To Develop a Minimal and Versatile Lignocellulolytic Bacterial Consortium. Appl. Environ. Microbiol. 2021, 87, e02427-20. [Google Scholar] [CrossRef]
- Hu, J.; Xue, Y.; Guo, H.; Gao, M.; Li, J.; Zhang, S.; Tsang, Y.F. Design and Composition of Synthetic Fungal-Bacterial Microbial Consortia That Improve Lignocellulolytic Enzyme Activity. Bioresour. Technol. 2017, 227, 247–255. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial Interactions: From Networks to Models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Rahmanpour, R.; Rea, D.; Jamshidi, S.; Fülöp, V.; Bugg, T.D.H. Structure of Thermobifida Fusca DyP-Type Peroxidase and Activity towards Kraft Lignin and Lignin Model Compounds. Arch. Biochem. Biophys. 2016, 594, 54–60. [Google Scholar] [CrossRef]
- Akinosho, H.O.; Yoo, C.G.; Dumitrache, A.; Natzke, J.; Muchero, W.; Brown, S.D.; Ragauskas, A.J. Elucidating the Structural Changes to Populus Lignin during Consolidated Bioprocessing with Clostridium Thermocellum. ACS Sustain. Chem. Eng. 2017, 5, 7486–7491. [Google Scholar] [CrossRef]
- Welte, C.U.; de Graaf, R.; Dalcin Martins, P.; Jansen, R.S.; Jetten, M.S.M.; Kurth, J.M. A Novel Methoxydotrophic Metabolism Discovered in the Hyperthermophilic Archaeon Archaeoglobus Fulgidus. Env. Microbiol. 2021, 23, 4017–4033. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, H.; Li, L.; Shao, W. Biobleaching of Wheat Straw Pulp with Recombinant Laccase from the Hyperthermophilic Thermus Thermophilus. Biotechnol. Lett. 2012, 34, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Margesin, R.; Ludwikowski, T.M.; Kutzner, A.; Wagner, A.O. Low-Temperature Biodegradation of Lignin-Derived Aromatic Model Monomers by the Cold-Adapted Yeast Rhodosporidiobolus Colostri Isolated from Alpine Forest Soil. Microorganisms 2022, 10, 515. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.W.F.; Barato, M.B.; Nobre, F.S.; Polezel, D.A.; de Oliveira, T.B.; dos Santos, J.A.; Rodrigues, A.; Sette, L.D. Production of Cold-Adapted Enzymes by Filamentous Fungi from King George Island, Antarctica. Polar Biol. 2018, 41, 2511–2521. [Google Scholar] [CrossRef]
- Jiang, C.; Cheng, Y.; Zang, H.; Chen, X.; Wang, Y.; Zhang, Y.; Wang, J.; Shen, X.; Li, C. Biodegradation of Lignin and the Associated Degradation Pathway by Psychrotrophic Arthrobacter Sp. C2 from the Cold Region of China. Cellulose 2020, 27, 1423–1440. [Google Scholar] [CrossRef]
- Tao, X.; Feng, J.; Yang, Y.; Wang, G.; Tian, R.; Fan, F.; Ning, D.; Bates, C.T.; Hale, L.; Yuan, M.M.; et al. Winter Warming in Alaska Accelerates Lignin Decomposition Contributed by Proteobacteria. Microbiome 2020, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Bisaccia, M.; Binda, E.; Rosini, E.; Caruso, G.; Dell’Acqua, O.; Azzaro, M.; Laganà, P.; Tedeschi, G.; Maffioli, E.M.; Pollegioni, L.; et al. A Novel Promising Laccase from the Psychrotolerant and Halotolerant Antarctic Marine Halomonas Sp. M68 Strain. Front. Microbiol. 2023, 14, 1078382. [Google Scholar] [CrossRef] [PubMed]
- Boucherit, Z.; Flahaut, S.; Djoudi, B.; Mouas, T.-N.; Mechakra, A.; Ameddah, S. Potential of Halophilic Penicillium Chrysogenum Isolated from Algerian Saline Soil to Produce Laccase on Olive Oil Wastes. Curr. Microbiol. 2022, 79, 178. [Google Scholar] [CrossRef]
- Rezaei, S.; Shahverdi, A.R.; Faramarzi, M.A. Isolation, One-Step Affinity Purification, and Characterization of a Polyextremotolerant Laccase from the Halophilic Bacterium Aquisalibacillus Elongatus and Its Application in the Delignification of Sugar Beet Pulp. Bioresour. Technol. 2017, 230, 67–75. [Google Scholar] [CrossRef]
- Jafari, N.; Rezaei, S.; Rezaie, R.; Dilmaghani, H.; Khoshayand, M.R.; Faramarzi, M.A. Improved Production and Characterization of a Highly Stable Laccase from the Halophilic Bacterium Chromohalobacter Salexigens for the Efficient Delignification of Almond Shell Bio-Waste. Int. J. Biol. Macromol. 2017, 105, 489–498. [Google Scholar] [CrossRef]
- Rezaie, R.; Rezaei, S.; Jafari, N.; Forootanfar, H.; Khoshayand, M.R.; Faramarzi, M.A. Delignification and Detoxification of Peanut Shell Bio-Waste Using an Extremely Halophilic Laccase from an Aquisalibacillus Elongatus Isolate. Extremophiles 2017, 21, 993–1004. [Google Scholar] [CrossRef]
- Tonin, F.; Rosini, E.; Piubelli, L.; Sanchez-Amat, A.; Pollegioni, L. Different Recombinant Forms of Polyphenol Oxidase A, a Laccase from Marinomonas Mediterranea. Protein Expr. Purif. 2016, 123, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Kontro, J.; Maltari, R.; Mikkilä, J.; Kähkönen, M.; Mäkelä, M.R.; Hildén, K.; Nousiainen, P.; Sipilä, J. Applicability of Recombinant Laccases From the White-Rot Fungus Obba Rivulosa for Mediator-Promoted Oxidation of Biorefinery Lignin at Low PH. Front. Bioeng. Biotechnol. 2020, 8, 1387. [Google Scholar] [CrossRef] [PubMed]
- Lončar, N.; Božić, N.; Vujčić, Z. Expression and Characterization of a Thermostable Organic Solvent-Tolerant Laccase from Bacillus Licheniformis ATCC 9945a. J. Mol. Catal. B Enzym. 2016, 134, 390–395. [Google Scholar] [CrossRef]
- Overhage, J.; Steinbüchel, A.; Priefert, H. Harnessing Eugenol as a Substrate for Production of Aromatic Compounds with Recombinant Strains of Amycolatopsis Sp. HR167. J. Biotechnol. 2006, 125, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Steinbüchel, A.; Priefert, H. Highly Efficient Biotransformation of Eugenol to Ferulic Acid and Further Conversion to Vanillin in Recombinant Strains of Escherichia Coli. Appl. Env. Microbiol. 2003, 69, 6569–6576. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.W.; Beckham, G.T. Aromatic Catabolic Pathway Selection for Optimal Production of Pyruvate and Lactate from Lignin. Metab. Eng. 2015, 28, 240–247. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Tran, M.H.; Lee, E.Y. Co-Upgrading of Ethanol-Assisted Depolymerized Lignin: A New Biological Lignin Valorization Approach for the Production of Protocatechuic Acid and Polyhydroxyalkanoic Acid. Bioresour. Technol. 2021, 338, 125563. [Google Scholar] [CrossRef]
- Hong, C.-Y.; Ryu, S.-H.; Jeong, H.; Lee, S.-S.; Kim, M.; Choi, I.-G. Phanerochaete Chrysosporium Multienzyme Catabolic System for in Vivo Modification of Synthetic Lignin to Succinic Acid. ACS Chem. Biol. 2017, 12, 1749–1759. [Google Scholar] [CrossRef]
- Sainsbury, P.D.; Hardiman, E.M.; Ahmad, M.; Otani, H.; Seghezzi, N.; Eltis, L.D.; Bugg, T.D.H. Breaking Down Lignin to High-Value Chemicals: The Conversion of Lignocellulose to Vanillin in a Gene Deletion Mutant of Rhodococcus Jostii RHA1. ACS Chem. Biol. 2013, 8, 2151–2156. [Google Scholar] [CrossRef]
- Sharma, R.K.; Mukhopadhyay, D.; Gupta, P. Microbial Fuel Cell-Mediated Lignin Depolymerization: A Sustainable Approach. J. Chem. Technol. Biotechnol. 2019, 94, 927–932. [Google Scholar] [CrossRef]
- Overhage, J.; Priefert, H.; Steinbüchel, A. Biochemical and Genetic Analyses of Ferulic Acid Catabolism in Pseudomonas sp. Strain HR199. Appl. Env. Microbiol. 1999, 65, 4837–4847. [Google Scholar] [CrossRef] [PubMed]
- Yaguchi, A.L.; Lee, S.J.; Blenner, M.A. Synthetic Biology towards Engineering Microbial Lignin Biotransformation. Trends Biotechnol. 2021, 39, 1037–1064. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. A Field of Dreams: Lignin Valorization into Chemicals, Materials, Fuels, and Health-Care Products. Biotechnol. Adv. 2019, 37, 107360. [Google Scholar] [CrossRef]
- Rosini, E.; Allegretti, C.; Melis, R.; Cerioli, L.; Conti, G.; Pollegioni, L.; D’Arrigo, P. Cascade Enzymatic Cleavage of the β-O-4 Linkage in a Lignin Model Compound. Catal. Sci. Technol. 2016, 6, 2195–2205. [Google Scholar] [CrossRef]
- Vignali, E.; Pollegioni, L.; di Nardo, G.; Valetti, F.; Gazzola, S.; Gilardi, G.; Rosini, E. Multi-Enzymatic Cascade Reactions for the Synthesis of Cis,Cis -Muconic Acid. Adv. Synth. Catal. 2022, 364, 114–123. [Google Scholar] [CrossRef]
- Muheim, A.; Lerch, K. Towards a High-Yield Bioconversion of Ferulic Acid to Vanillin. Appl. Microbiol. Biotechnol. 1999, 51, 456–461. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Z.; Pereira, B.; Stephanopoulos, G. Engineering E. Coli–E. Coli Cocultures for Production of Muconic Acid from Glycerol. Microb. Cell Fact. 2015, 14, 134. [Google Scholar] [CrossRef]
- Matthiesen, J.E.; Carraher, J.M.; Vasiliu, M.; Dixon, D.A.; Tessonnier, J.-P. Electrochemical Conversion of Muconic Acid to Biobased Diacid Monomers. ACS Sustain. Chem. Eng. 2016, 4, 3575–3585. [Google Scholar] [CrossRef]
- Xie, N.-Z.; Liang, H.; Huang, R.-B.; Xu, P. Biotechnological Production of Muconic Acid: Current Status and Future Prospects. Biotechnol. Adv. 2014, 32, 615–622. [Google Scholar] [CrossRef]
- Kruyer, N.S.; Peralta-Yahya, P. Metabolic Engineering Strategies to Bio-Adipic Acid Production. Curr. Opin. Biotechnol. 2017, 45, 136–143. [Google Scholar] [CrossRef]
- Khalil, I.; Quintens, G.; Junkers, T.; Dusselier, M. Muconic Acid Isomers as Platform Chemicals and Monomers in the Biobased Economy. Green Chem. 2020, 22, 1517–1541. [Google Scholar] [CrossRef]
- Curran, K.A.; Leavitt, J.M.; Karim, A.S.; Alper, H.S. Metabolic Engineering of Muconic Acid Production in Saccharomyces Cerevisiae. Metab. Eng. 2013, 15, 55–66. [Google Scholar] [CrossRef]
- Wu, W.; Dutta, T.; Varman, A.M.; Eudes, A.; Manalansan, B.; Loqué, D.; Singh, S. Lignin Valorization: Two Hybrid Biochemical Routes for the Conversion of Polymeric Lignin into Value-Added Chemicals. Sci. Rep. 2017, 7, 8420. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Sun, X.; Yuan, Q.; Yan, Y. Extending Shikimate Pathway for the Production of Muconic Acid and Its Precursor Salicylic Acid in Escherichia Coli. Metab. Eng. 2014, 23, 62–69. [Google Scholar] [CrossRef]
- Weber, C.; Brückner, C.; Weinreb, S.; Lehr, C.; Essl, C.; Boles, E. Biosynthesis of Cis, Cis -Muconic Acid and Its Aromatic Precursors, Catechol and Protocatechuic Acid, from Renewable Feedstocks by Saccharomyces Cerevisiae. Appl. Env. Microbiol. 2012, 78, 8421–8430. [Google Scholar] [CrossRef] [PubMed]
- van Duuren, J.B.J.H.; Wijte, D.; Karge, B.; Martins dos Santos, V.A.P.; Yang, Y.; Mars, A.E.; Eggink, G. PH-Stat Fed-Batch Process to Enhance the Production of Cis, Cis-Muconate from Benzoate by Pseudomonas Putida KT2440-JD1. Biotechnol. Prog. 2012, 28, 85–92. [Google Scholar] [CrossRef]
- Polen, T.; Spelberg, M.; Bott, M. Toward Biotechnological Production of Adipic Acid and Precursors from Biorenewables. J. Biotechnol. 2013, 167, 75–84. [Google Scholar] [CrossRef]
- van Duuren, J.B.J.H.; Wijte, D.; Leprince, A.; Karge, B.; Puchałka, J.; Wery, J.; dos Santos, V.A.P.M.; Eggink, G.; Mars, A.E. Generation of a CatR Deficient Mutant of P. Putida KT2440 That Produces Cis, Cis-Muconate from Benzoate at High Rate and Yield. J. Biotechnol. 2011, 156, 163–172. [Google Scholar] [CrossRef]
- Barton, N.; Horbal, L.; Starck, S.; Kohlstedt, M.; Luzhetskyy, A.; Wittmann, C. Enabling the Valorization of Guaiacol-Based Lignin: Integrated Chemical and Biochemical Production of Cis,Cis-Muconic Acid Using Metabolically Engineered Amycolatopsis sp ATCC 39116. Metab. Eng. 2018, 45, 200–210. [Google Scholar] [CrossRef]
- Becker, J.; Klopprogge, C.; Zelder, O.; Heinzle, E.; Wittmann, C. Amplified Expression of Fructose 1,6-Bisphosphatase in Corynebacterium Glutamicum Increases In Vivo Flux through the Pentose Phosphate Pathway and Lysine Production on Different Carbon Sources. Appl. Env. Microbiol. 2005, 71, 8587–8596. [Google Scholar] [CrossRef]
- Becker, J.; Kuhl, M.; Kohlstedt, M.; Starck, S.; Wittmann, C. Metabolic Engineering of Corynebacterium Glutamicum for the Production of Cis, Cis-Muconic Acid from Lignin. Microb. Cell Fact. 2018, 17, 115. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Xu, Z.; Xu, M.; Cai, M.; Jin, M. Development of a Rhodococcus Opacus Cell Factory for Valorizing Lignin to Muconate. ACS Sustain. Chem. Eng. 2020, 8, 2016–2031. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, Z.; Cai, C.; Li, J.; Jin, M. Deciphering the Metabolic Distribution of Vanillin in Rhodococcus Opacus during Lignin Valorization. Bioresour. Technol. 2022, 347, 126348. [Google Scholar] [CrossRef]
- Sonoki, T.; Morooka, M.; Sakamoto, K.; Otsuka, Y.; Nakamura, M.; Jellison, J.; Goodell, B. Enhancement of Protocatechuate Decarboxylase Activity for the Effective Production of Muconate from Lignin-Related Aromatic Compounds. J. Biotechnol. 2014, 192, 71–77. [Google Scholar] [CrossRef]
- Molinari, F.; Pollegioni, L.; Rosini, E. Whole-Cell Bioconversion of Renewable Biomasses-Related Aromatics to Cis,Cis -Muconic Acid. ACS Sustain. Chem. Eng. 2023, 11, 2476–2485. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, B.; Xiao, G.; Ko, L.-Y.; Kao, T.-Y.; Fan, C.; Yuan, J. Bioconversion of Lignin-Derived Feedstocks to Muconic Acid by Whole-Cell Biocatalysis. ACS Food Sci. Technol. 2021, 1, 382–387. [Google Scholar] [CrossRef]
- Vardon, D.R.; Franden, M.A.; Johnson, C.W.; Karp, E.M.; Guarnieri, M.T.; Linger, J.G.; Salm, M.J.; Strathmann, T.J.; Beckham, G.T. Adipic Acid Production from Lignin. Energy Env. Sci. 2015, 8, 617–628. [Google Scholar] [CrossRef]
- Hernández-Arranz, S.; Moreno, R.; Rojo, F. The Translational Repressor Crc Controls the Pseudomonas Putida Benzoate and Alkane Catabolic Pathways Using a Multi-Tier Regulation Strategy. Env. Microbiol. 2013, 15, 227–241. [Google Scholar] [CrossRef]
- Johnson, C.W.; Abraham, P.E.; Linger, J.G.; Khanna, P.; Hettich, R.L.; Beckham, G.T. Eliminating a Global Regulator of Carbon Catabolite Repression Enhances the Conversion of Aromatic Lignin Monomers to Muconate in Pseudomonas Putida KT2440. Metab. Eng. Commun. 2017, 5, 19–25. [Google Scholar] [CrossRef]
- Kuatsjah, E.; Johnson, C.W.; Salvachúa, D.; Werner, A.Z.; Zahn, M.; Szostkiewicz, C.J.; Singer, C.A.; Dominick, G.; Okekeogbu, I.; Haugen, S.J.; et al. Debottlenecking 4-Hydroxybenzoate Hydroxylation in Pseudomonas Putida KT2440 Improves Muconate Productivity from p-Coumarate. Metab. Eng. 2022, 70, 31–42. [Google Scholar] [CrossRef]
- Kohlstedt, M.; Starck, S.; Barton, N.; Stolzenberger, J.; Selzer, M.; Mehlmann, K.; Schneider, R.; Pleissner, D.; Rinkel, J.; Dickschat, J.S.; et al. From Lignin to Nylon: Cascaded Chemical and Biochemical Conversion Using Metabolically Engineered Pseudomonas Putida. Metab. Eng. 2018, 47, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Akutsu, M.; Abe, N.; Sakamoto, C.; Kurimoto, Y.; Sugita, H.; Tanaka, M.; Higuchi, Y.; Sakamoto, K.; Kamimura, N.; Kurihara, H.; et al. Pseudomonas sp. NGC7 as a Microbial Chassis for Glucose-Free Muconate Production from a Variety of Lignin-Derived Aromatics and Its Application to the Production from Sugar Cane Bagasse Alkaline Extract. Bioresour. Technol. 2022, 359, 127479. [Google Scholar] [CrossRef] [PubMed]
- Sonoki, T.; Takahashi, K.; Sugita, H.; Hatamura, M.; Azuma, Y.; Sato, T.; Suzuki, S.; Kamimura, N.; Masai, E. Glucose-Free Cis, Cis-Muconic Acid Production via New Metabolic Designs Corresponding to the Heterogeneity of Lignin. ACS Sustain. Chem. Eng. 2018, 6, 1256–1264. [Google Scholar] [CrossRef]
- Pometto III, A.L.; Sutherland, J.B.; Crawford, D.L. Streptomyces Setonii: Catabolism of Vanillic Acid via Guaiacol and Catechol. Can. J. Microbiol. 1981, 27, 636–638. [Google Scholar] [CrossRef] [PubMed]
- Rosini, E.; D’Arrigo, P.; Pollegioni, L. Demethylation of Vanillic Acid by Recombinant LigM in a One-Pot Cofactor Regeneration System. Catal. Sci. Technol. 2016, 6, 7729–7737. [Google Scholar] [CrossRef]
- Wu, W.; Liu, F.; Singh, S. Toward Engineering E. Coli with an Autoregulatory System for Lignin Valorization. Proc. Natl. Acad. Sci. USA 2018, 115, 2970–2975. [Google Scholar] [CrossRef]
- Nguyen, T.T.M.; Iwaki, A.; Izawa, S. The ADH7 Promoter of Saccharomyces Cerevisiae Is Vanillin-Inducible and Enables MRNA Translation Under Severe Vanillin Stress. Front. Microbiol. 2015, 6, 1390. [Google Scholar] [CrossRef]
- Salmon, R.C.; Cliff, M.J.; Rafferty, J.B.; Kelly, D.J. The CouPSTU and TarPQM Transporters in Rhodopseudomonas Palustris: Redundant, Promiscuous Uptake Systems for Lignin-Derived Aromatic Substrates. PLoS ONE 2013, 8, e59844. [Google Scholar] [CrossRef]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; de Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of Substrate Toxicity by IPTG in Escherichia Coli BL21 (DE3) Carrying a Synthetic Metabolic Pathway. Microb. Cell Fact. 2015, 14, 201. [Google Scholar] [CrossRef]
- Bai, Z.; Gao, Z.; He, B.; Wu, B. Effect of Lignocellulose-Derived Inhibitors on the Growth and d-Lactic Acid Production of Sporolactobacillus Inulinus YBS1-5. Bioprocess Biosyst. Eng. 2015, 38, 1993–2001. [Google Scholar] [CrossRef]
- Sakai, S.; Tsuchida, Y.; Okino, S.; Ichihashi, O.; Kawaguchi, H.; Watanabe, T.; Inui, M.; Yukawa, H. Effect of Lignocellulose-Derived Inhibitors on Growth of and Ethanol Production by Growth-Arrested Corynebacterium Glutamicum R. Appl. Env. Microbiol. 2007, 73, 2349–2353. [Google Scholar] [CrossRef] [PubMed]
- Kunjapur, A.M.; Tarasova, Y.; Prather, K.L.J. Synthesis and Accumulation of Aromatic Aldehydes in an Engineered Strain of Escherichia coli. J. Am. Chem. Soc. 2014, 136, 11644–11654. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, Y.-T.; Tao, F.; Peng, Y.; Xu, P. A Coenzyme-Free Biocatalyst for the Value-Added Utilization of Lignin-Derived Aromatics. J. Am. Chem. Soc. 2018, 140, 16001–16005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, P.; Ko, L.-Y.; Kao, T.-Y.; Liu, L.; Zhang, Y.; Yuan, J. High-Yielding Protocatechuic Acid Synthesis from l-Tyrosine in Escherichia coli. ACS Sustain. Chem. Eng. 2020, 8, 14949–14954. [Google Scholar] [CrossRef]
- Hibi, M.; Sonoki, T.; Mori, H. Functional Coupling between Vanillate- O -Demethylase and Formaldehyde Detoxification Pathway. FEMS Microbiol. Lett. 2005, 253, 237–242. [Google Scholar] [CrossRef]
- Nordlund, I.; Powlowski, J.; Hagstrom, A.; Shingler, V. Conservation of Regulatory and Structural Genes for a Multi-Component Phenol Hydroxylase within Phenol-Catabolizing Bacteria That Utilize a Meta-Cleavage Pathway. J. Gen. Microbiol. 1993, 139, 2695–2703. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Salvachúa, D.; Johnson, C.W.; Singer, C.A.; Rohrer, H.; Peterson, D.J.; Black, B.A.; Knapp, A.; Beckham, G.T. Bioprocess Development for Muconic Acid Production from Aromatic Compounds and Lignin. Green Chem. 2018, 20, 5007–5019. [Google Scholar] [CrossRef]
- Shinoda, E.; Takahashi, K.; Abe, N.; Kamimura, N.; Sonoki, T.; Masai, E. Isolation of a Novel Platform Bacterium for Lignin Valorization and Its Application in Glucose-Free Cis, Cis-Muconate Production. J. Ind. Microbiol. Biotechnol. 2019, 46, 1071–1080. [Google Scholar] [CrossRef]
- Qian, Y.; Otsuka, Y.; Sonoki, T.; Mukhopadhyay, B.; Nakamura, M.; Jellison, J.; Goodell, B. Engineered Microbial Production of 2-Pyrone-4,6-Dicarboxylic Acid from Lignin Residues for Use as an Industrial Platform Chemical. Bioresources 2016, 11, 6097–6109. [Google Scholar] [CrossRef]
- Mycroft, Z.; Gomis, M.; Mines, P.; Law, P.; Bugg, T.D.H. Biocatalytic Conversion of Lignin to Aromatic Dicarboxylic Acids in Rhodococcus Jostii RHA1 by Re-Routing Aromatic Degradation Pathways. Green Chem. 2015, 17, 4974–4979. [Google Scholar] [CrossRef]
- Skoog, E.; Shin, J.H.; Saez-jimenez, V.; Mapelli, V.; Olsson, L. Biobased Adipic Acid—The Challenge of Developing the Production Host. Biotechnol. Adv. 2018, 36, 2248–2263. [Google Scholar] [CrossRef] [PubMed]
- Suitor, J.T.; Varzandeh, S.; Wallace, S. One-Pot Synthesis of Adipic Acid from Guaiacol in Escherichia coli. ACS Synht. Biol. 2020, 9, 2472–2476. [Google Scholar] [CrossRef] [PubMed]
- Kruyer, N.S.; Wauldron, N.; Bommarius, A.S.; Yahya, P.P. Fully Biological Production of Adipic Acid Analogs from Branched Catechols. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Niu, W.; Willett, H.; Mueller, J.; He, X.; Kramer, L.; Ma, B.; Guo, J. Direct Biosynthesis of Adipic Acid from Lignin-Derived Aromatics Using Engineered Pseudomonas Putida KT2440. Metab. Eng. 2020, 59, 151–161. [Google Scholar] [CrossRef]
- Notonier, S.; Werner, A.Z.; Kuatsjah, E.; Dumalo, L.; Abraham, P.E.; Hatmaker, E.A.; Hoyt, C.B.; Amore, A.; Ramirez, K.J.; Woodworth, S.P.; et al. Metabolism of Syringyl Lignin-Derived Compounds in Pseudomonas Putida Enables Convergent Production of 2-Pyrone-4,6-Dicarboxylic Acid. Metab. Eng. 2021, 65, 111–122. [Google Scholar] [CrossRef]
- Lee, S.; Jean, Y.; Jae, S.; Ryu, M.; Eon, J.; Min, H.; Hee, K.; Keun, B.; Hyun, B.; Hwan, Y.; et al. Microbial Production of 2-Pyrone-4,6-Dicarboxylic Acid from Lignin Derivatives in an Engineered Pseudomonas Putida and Its Application for the Synthesis of Bio-Based Polyester. Bioresour. Technol. 2022, 352, 127106. [Google Scholar] [CrossRef]
- Perez, J.M.; Kontur, W.S.; Alherech, M.; Coplien, J.; Karlen, S.D.; Stahl, S.S.; Donohue, T.J.; Noguera, D.R. Funneling Aromatic Products of Chemically Depolymerized Lignin into 2-Pyrone-4-6-Dicarboxylic Acid with Novosphingobium Aromaticivorans. Green Chem. 2019, 21, 1340–1350. [Google Scholar] [CrossRef]
- Spence, E.M.; Bado, L.C.; Mines, P.; Bugg, T.D.H. Metabolic Engineering of Rhodococcus Jostii RHA1 for Production of Pyridine—Dicarboxylic Acids from Lignin. Microb. Cell Fact. 2021, 20, 15. [Google Scholar] [CrossRef]
- Fu, B.; Xiao, G.; Zhang, Y.; Yuan, J. One-Pot Bioconversion of Lignin-Derived Substrates into Gallic Acid. Agric. Food Chem. 2021, 69, 11336–11341. [Google Scholar] [CrossRef]
- Cai, C.; Xu, Z.; Zhou, H.; Chen, S.; Jin, M. Valorization of Lignin Components into Gallate by Integrated Biological Hydroxylation, O-Demethylation, and Aryl Side-Chain Oxidation. Sci. Adv. 2021, 7, eabg4585. [Google Scholar] [CrossRef] [PubMed]
- Galman, J.L.; Parmeggiani, F.; Seibt, L.; Birmingham, W.R.; Turner, N.J. One-Pot Biocatalytic In Vivo Methylation-Hydroamination of Bioderived Lignin Monomers to Generate a Key Precursor to L-DOPA. Angew. Chem. Int. Ed. 2022, 61, e202112855. [Google Scholar] [CrossRef] [PubMed]
- Raza, Z.A.; Abid, S.; Banat, I.M. Polyhydroxyalkanoates: Characteristics, Production, Recent Developments and Applications. Int. Biodeterior. Biodegrad. 2018, 126, 45–56. [Google Scholar] [CrossRef]
- Rydzak, T.; De Capite, A.; Black, B.A.; Bouvier, J.T.; Cleveland, N.S.; Joshua, R.; Huenemann, J.D.; Katahira, R.; Michener, E.; Peterson, D.J.; et al. Metabolic Engineering of Pseudomonas Putida for Increased Polyhydroxyalkanoate Production from Lignin. Microb. Biotechnol. 2020, 13, 290–298. [Google Scholar] [CrossRef]
- Ichinobu, B.T.M.; Ito, M.B.; Amada, Y.Y.; Animura, M.T.; Atayama, Y.K.; Asai, E.M.; Akamura, M.N.; Tsuka, Y.O.; Hara, S.O.; Higehara, K.S. Fusible, Elastic, and Biodegradable Polyesters of 2-Pyrone-4,6-Dicarboxylic Acid (PDC). Polym. J. 2009, 41, 1111–1116. [Google Scholar] [CrossRef]
- Kaiser, J.; Feng, Y.; Bollag, J.-M. Microbial Metabolism of Pyridine, Quinoline, Acridine, and Their Derivatives under Aerobic and Anaerobic Conditions. Microbiol. Rev. 1996, 60, 483–498. [Google Scholar] [CrossRef]
- Domìnguez-Robles, J.; Càrcamo-Màrtinez, A.; Stewart, S.A.; Donnelly, R.F. Lignin for Pharmaceutical and Biomedical Applications—Could This Become a Reality? Sustain. Chem. Pharm. 2020, 18, 100320. [Google Scholar] [CrossRef]
- Bai, J.; Zhang, Y.; Tang, C.; Hou, Y.; Ai, X.; Chen, X.; Zhang, Y.; Wang, X.; Meng, X. Gallic Acid: Pharmacological Activities and Molecular Mechanisms Involved in Inflammation-Related Diseases. Biomed. Pharmacother. 2021, 133, 110985. [Google Scholar] [CrossRef]
- Badhani, B.; Sharma, N.; Kakkar, R. Gallic Acid: A Versatile Antioxidant with Promising Therapeutic and Industrial Applications. RSC Adv. 2015, 5, 27540–27557. [Google Scholar] [CrossRef]
- Kambourakis, S.; Draths, K.M.; Frost, J.W. Synthesis of Gallic Acid and Pyrogallol from Glucose: Replacing Natural Product Isolation with Microbial Catalysis. J. Am. Chem. Soc. 2000, 122, 9042–9043. [Google Scholar] [CrossRef]
- Min, K.; Park, K.; Park, D. Overview on the Biotechnological Production of L-DOPA. Appl. Microbiol. Biotechnol. 2015, 99, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Tang, X.; Suo, H.; Feng, L.; Liu, X.; Yang, J. Biochemical Characterization of a Novel Tyrosine Phenol-Lyase from Fusobacterium Nucleatum for Highly Efficient Biosynthesis of L -DOPA. Enzym. Microb. Technol. 2018, 112, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Bechthold, I.; Bretz, K.; Kabasci, S.; Kopitzky, R.; Springer, A. Succinic Acid: A New Platform Chemical for Biobased Polymers from Renewable Resources. Chem. Eng. Technol. 2008, 31, 647–654. [Google Scholar] [CrossRef]
- Liu, Z.-H.; Hao, N.; Wang, Y.-Y.; Dou, C.; Lin, F.; Shen, R.; Bura, R.; Hodge, D.B.; Dale, B.E.; Ragauskas, A.J.; et al. Transforming Biorefinery Designs with ‘Plug-In Processes of Lignin’ to Enable Economic Waste Valorization. Nat. Commun. 2021, 12, 3912. [Google Scholar] [CrossRef] [PubMed]
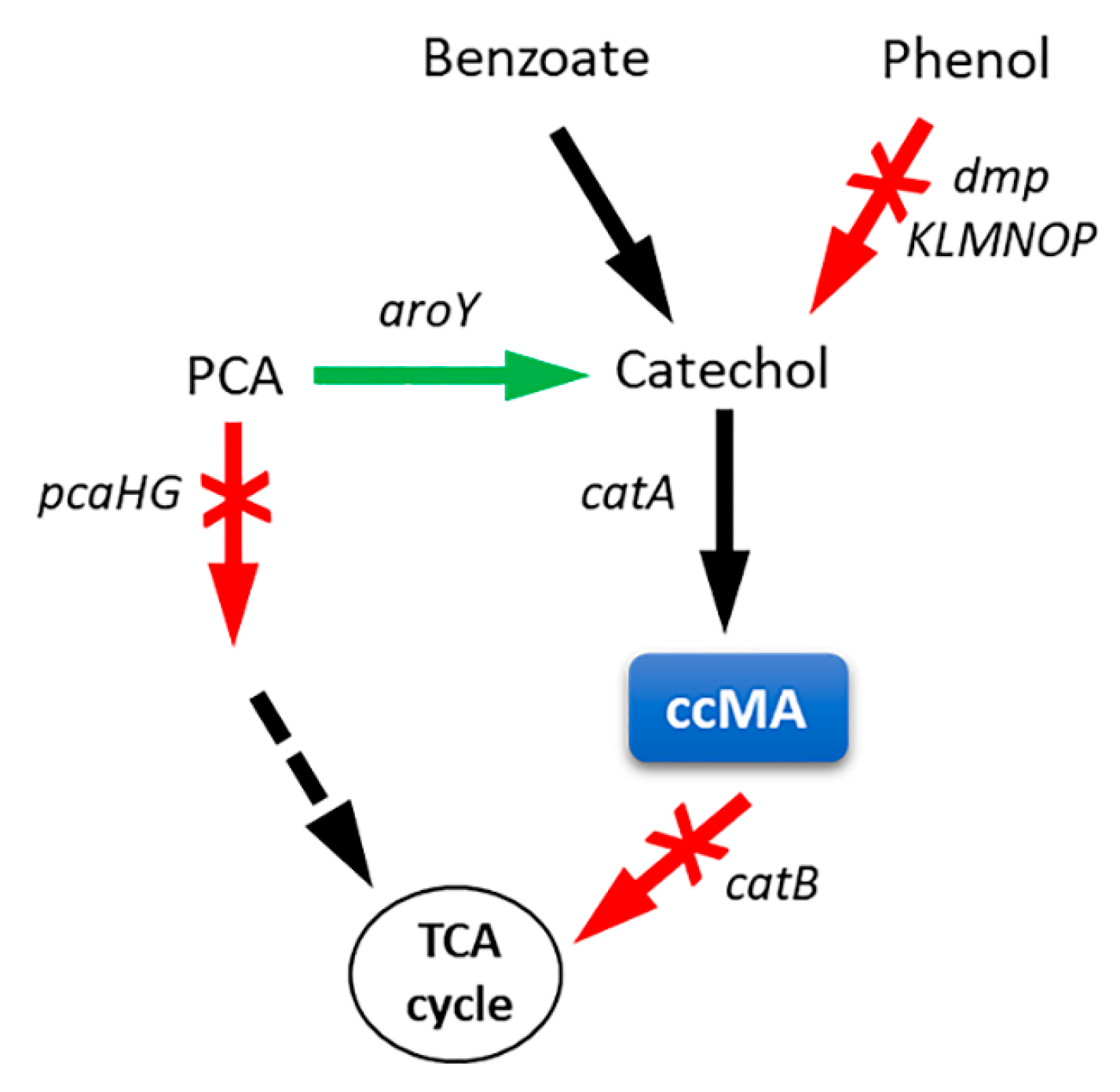
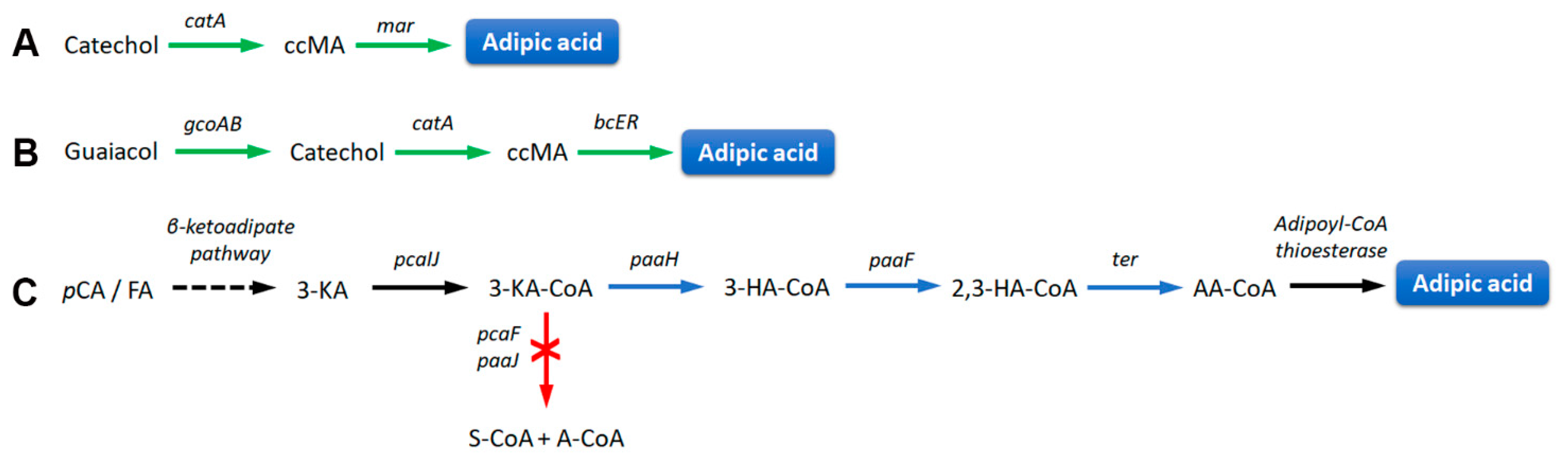
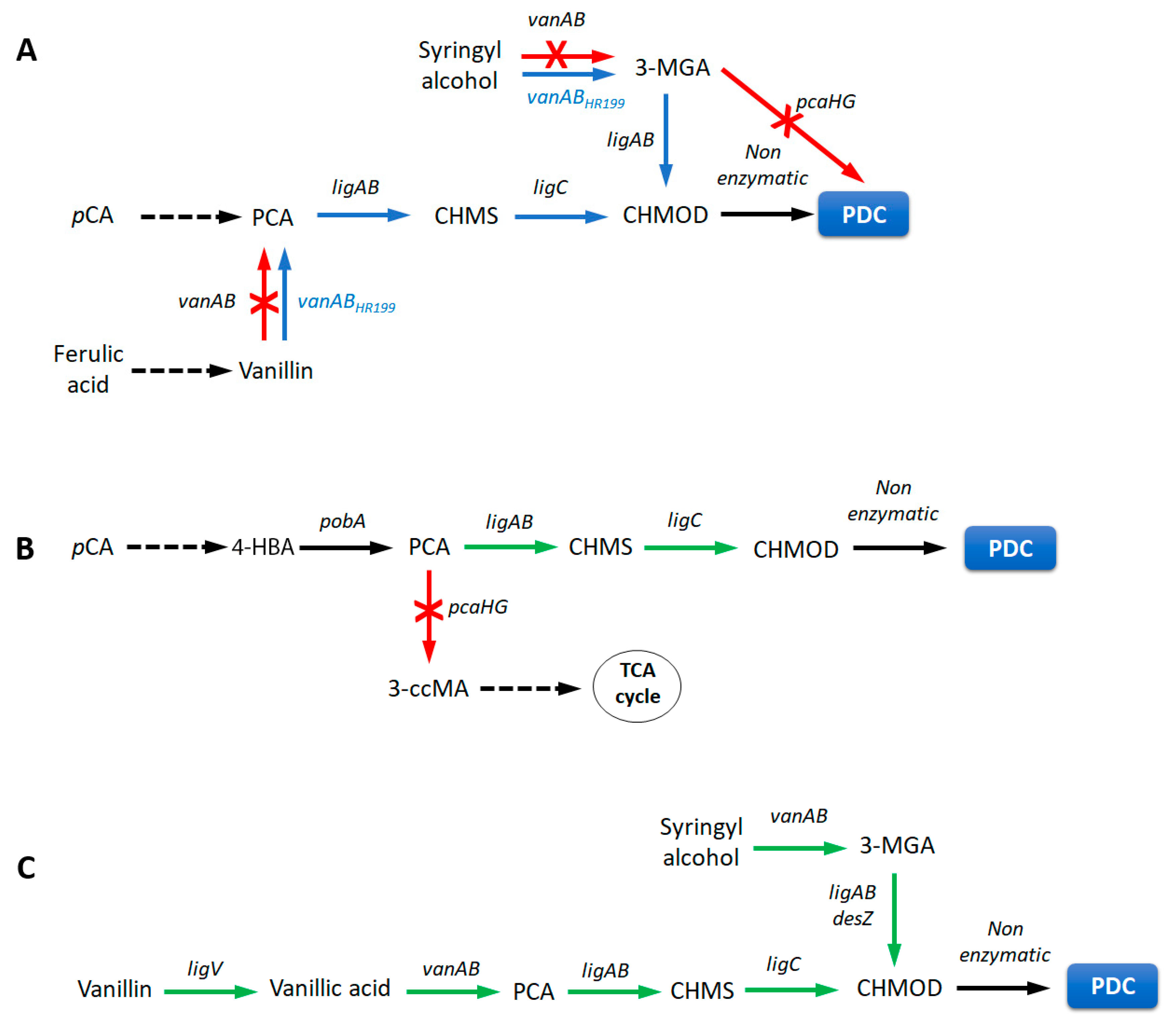
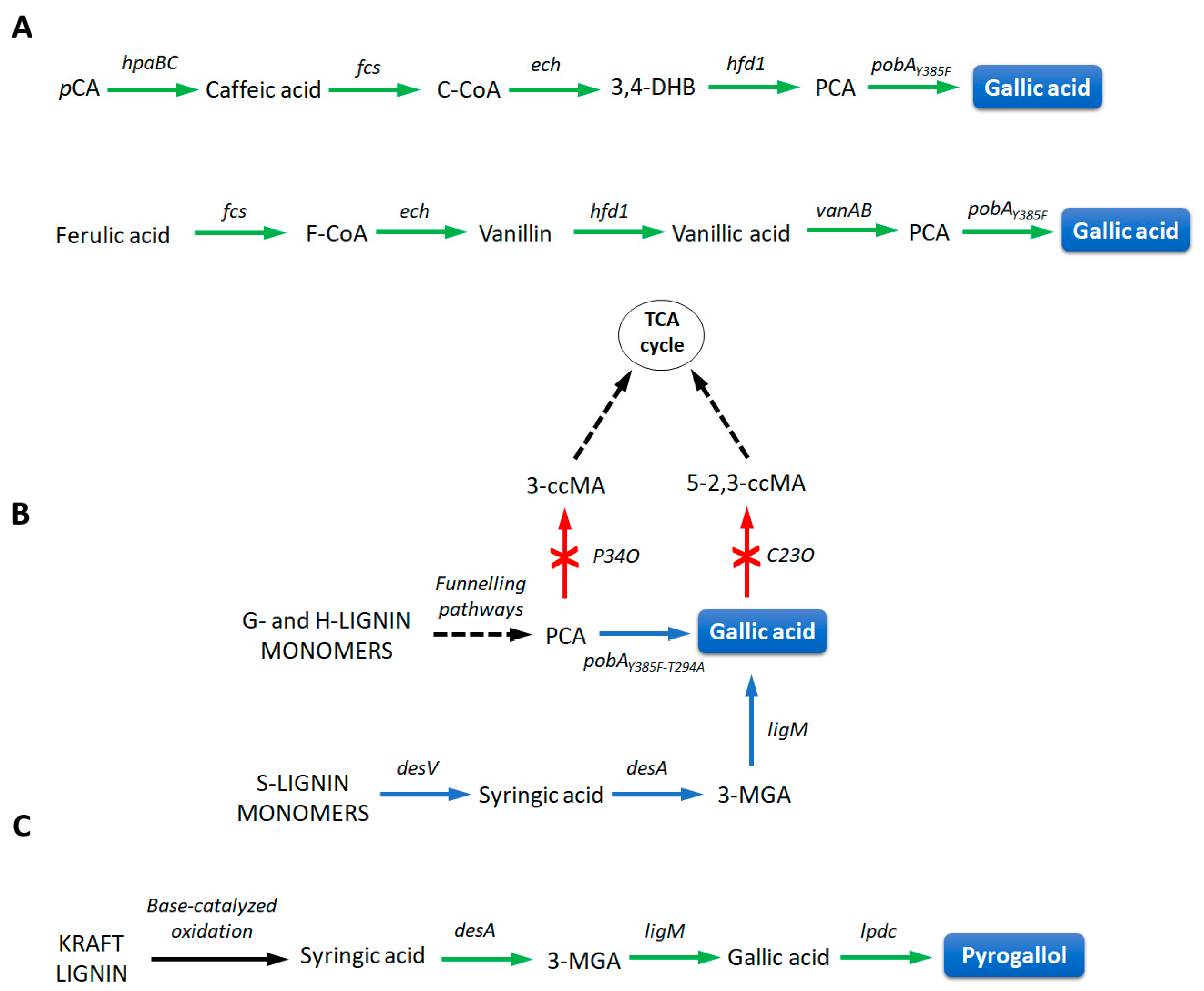
| Methods | Advantages | Limitations |
|---|---|---|
| Chemical treatments | ||
| Acid catalysts Base catalysts Metallic catalysts Ionic liquids assisted catalysis Supercritical fluids assisted catalysis Oxidative catalysis | Effective lignin degradation High stability Low substrate/product inhibition effects | Low selectivity Environmental concern Costly Difficult recovery of products from the mixture Harsh operational conditions |
| Biological treatments | ||
| Bacteria Fungi In vitro enzymatic depolymerization | Mild operational conditions Environmentally friendly Optimized by metabolic engineering Optimized by protein engineering Funneling pathways to key intermediates High selectivity and catalytic efficiency | Low conversion yield Long culture/biotransformation time Substrate/product inhibition Difficult to scale up |
| Product | Host | Starting Material | Yield | Reference |
|---|---|---|---|---|
| Coniferyl alcohol | Amycolatopsis sp. HR167 pRLE6SKvaom | Eugenol | 4.7 g/L | [55] |
| Ferulate | E. coli XL-1 Blue pSKvaomPcalAmcalB | Eugenol | 14.7 g/L | [56] |
| Lactate | P. putida KT2440-CJ127 P. putida KT2440-CJ124 | Benzoate p-Coumarate | 0.5 g/L 0.5 g/L | [57] [57] |
| PCA | P. putida KT2440 | Ethanol-assisted depolymerized lignin | 6.7 mg/L | [58] |
| Pyruvate | P. putida KT2440-CJ112 P. putida KT2440-CJ116 | Benzoate p-Coumarate | 0.7 g/L 2 g/L | [57] [57] |
| Succinate | Phanerochaete chrysosporium | Lignin | 20 mg/L | [59] |
| Vanillin | R. jostii RHA045 Shewanella putrefaciens Pseudomonas sp. HR199 Δvdh E. coli pSKechE/Hfcs Streptomyces setonii | Wheat straw lignocellulose Lignin from wheat straw Eugenol Ferulate Ferulate | 96 mg/L 275 mg/L 400 mg/L 300 mg/L 6.4 g/L | [60] [61] [62] [56] [67] |
| Host | Starting Material | Deleted Genes | Overexpressed Genes | Productivity (g/L/h) | Reference |
|---|---|---|---|---|---|
| Amycolatopsis sp. ATCC 39166 | Guaiacol | G10GW-3575, G10GW-1735 (putative cycloisomerases) | - | 0.13 | [80] |
| Hydrothermal depolymerized Kraft lignin | 0.26 | ||||
| C. glutamicum | Catechol | catB | catA | 2.4 | [82] |
| Hydrothermal depolymerized Kraft lignin | 0.07 | ||||
| R. opacus | PCA Lignin from corn stover | catB, pcaHG | aroY, ecdB | 0.56 0.02 | [83] |
| Vanillin | LPD03722, LPD04406 (vanillin reductases) pcaHG | vdh, aroY, ecdB | 0.22 | [84] | |
| E. coli | Vanillin | - | vdh, vanAB, catA, aroY, kpdB | 0.04 | [85] |
| ligV, ligM, aroY, catA | 0.01 | [74] | |||
| ligV, vanAB, aroY, catA | 0.7 | [86] | |||
| Vanillin from lignin | - | ligV, vanAB, aroY, catA | 0.3 | ||
| Isoeugenol | - | ado, hfd1, vanAB, gdc, catA | 0.14 | [87] | |
| Ferulic acid | - | fdc, ado, ligV, vanAB, aroY, catA | 0.14 | [86] | |
| Ferulic acid from wheat bran | 0.04 | ||||
| P. putida | p-Coumarate | pcaHG, catBC, dmpKMLNOP | aroY | 0.17 | [88] |
| Alkaline pretreated liquor | 0.03 | ||||
| p-Coumarate | pcaHG, catRBCA, crc | catA, aroy | 0.08 | [90] | |
| Ferulic acid | 0.01 | ||||
| p-Coumarate | pobA, catRBCA, crc | praI, ecdBD, vanAB | 0.4 | [91] | |
| Catechol | catBC, endA-1, endA-2 | catA | 4.3 | [92] | |
| Hydrothermal depolymerized Kraft lignin | catA, dmpKLMOP | 0.24 | |||
| Vanillic acid | pcaHG, catB | pdc | 0.03 | [94] | |
| Vanillic acid | catB | pcaHG, aroY, catA. vanAB | 0.21 | [93] | |
| Sugar cane bagasse alkaline extract | 0.01 |
| Product | Host | Starting Material | Deleted Genes | Chromosome Insertion | Overexpressed Genes | Productivity (g/L/h) | Reference |
|---|---|---|---|---|---|---|---|
| Adipic acid | E. coli | Catechol | - | - | catA, mar | 0.6 × 10−4 | [115] |
| Guaiacol | - | - | gcoAB, catA, bcER | 0.02 | [114] | ||
| P. putida KT2440 | p-Coumarate Ferulic acid | pcaF | paaH, paaF, 2,3-dehydroadipoyil-CoA reductase | - | 0.01 | [116] | |
| PHA | P. putida KT2440 | Ethanol-assisted depolymerized lignin | - | - | hps, phi, ada | 0.3 × 10−2 | [58] |
| p-Coumarate from lignin | fadAB | phaG, alkK, phaC1, phaC2 | - | 1.2 | [58] | ||
| PDC | P. putida KT2440 | Syringic acid Ferulic acid p-Coumarate | pcaHG, vanAB | vanABHR199, ligAB, ligC | - | 0.02 | [117] |
| p-Coumarate | pcaHG | - | ligAB, ligC | 0.19 | [118] | ||
| P. putida PDH | Depolymerized lignin | - | - | vanAB, ligV, ligAb, ligC, desZ | 0.03 | [111] | |
| N. aromaticivorans DSM12444 | Vanillin Vanillic acid | desC, desD, ligI | - | - | 0.1 | [119] | |
| Depolymerized lignin | 0.1 × 10−2 | [119] | |||||
| 2,4-PDCA | R. jostii RHA1 | Wheat straw Kraft lignin | - | - | praA | 0.5 × 10−6 | [112] |
| Wheat straw Protobind lignin | pcaHG | praA | dyp2 | 0.007 | [120] | ||
| 2,5-PDCA | R. jostii RHA1 | Wheat straw | - | - | ligAB | 0.5 × 10−3 | [112] |
| Gallic acid | E. coli MG1655 RARE | p-Coumarate | - | - | hpaBC, fcs, ech, hfd1, pobA | 0.1 | [121] |
| Ferulic acid | - | - | fcs, ech, hfd1, vanAB, pobA | 0.1 | [121] | ||
| R. opacus PD630 | Base-depolymerized AFEX lignin | protocatechuate 3,4-dioxygenase, putative catechol 2,3-dioxygenase | pobA, desV, desA, ligM, metF, ligH | - | 0.6 × 10−2 | [122] | |
| Pyrogallol | E. coli DH1 | Base-depolymerized Kraft lignin | - | - | ligM, desA, lpdc | 0.3 × 10−3 | [74] |
| L-veratrylglycine | E. coli BL21(DE3) | Ferulic acid | metJ | - | ejomt, mntN, luxS, metk, AL-11 | 0.01 | [123] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosini, E.; Molinari, F.; Miani, D.; Pollegioni, L. Lignin Valorization: Production of High Value-Added Compounds by Engineered Microorganisms. Catalysts 2023, 13, 555. https://doi.org/10.3390/catal13030555
Rosini E, Molinari F, Miani D, Pollegioni L. Lignin Valorization: Production of High Value-Added Compounds by Engineered Microorganisms. Catalysts. 2023; 13(3):555. https://doi.org/10.3390/catal13030555
Chicago/Turabian StyleRosini, Elena, Filippo Molinari, Davide Miani, and Loredano Pollegioni. 2023. "Lignin Valorization: Production of High Value-Added Compounds by Engineered Microorganisms" Catalysts 13, no. 3: 555. https://doi.org/10.3390/catal13030555
APA StyleRosini, E., Molinari, F., Miani, D., & Pollegioni, L. (2023). Lignin Valorization: Production of High Value-Added Compounds by Engineered Microorganisms. Catalysts, 13(3), 555. https://doi.org/10.3390/catal13030555









