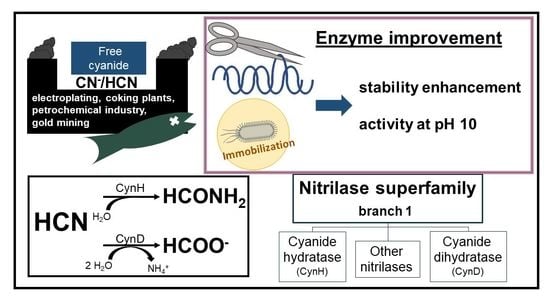
Recent Progress in the Production of Cyanide-Converting Nitrilases—Comparison with Nitrile-Hydrolyzing Enzymes
Abstract
1. Introduction
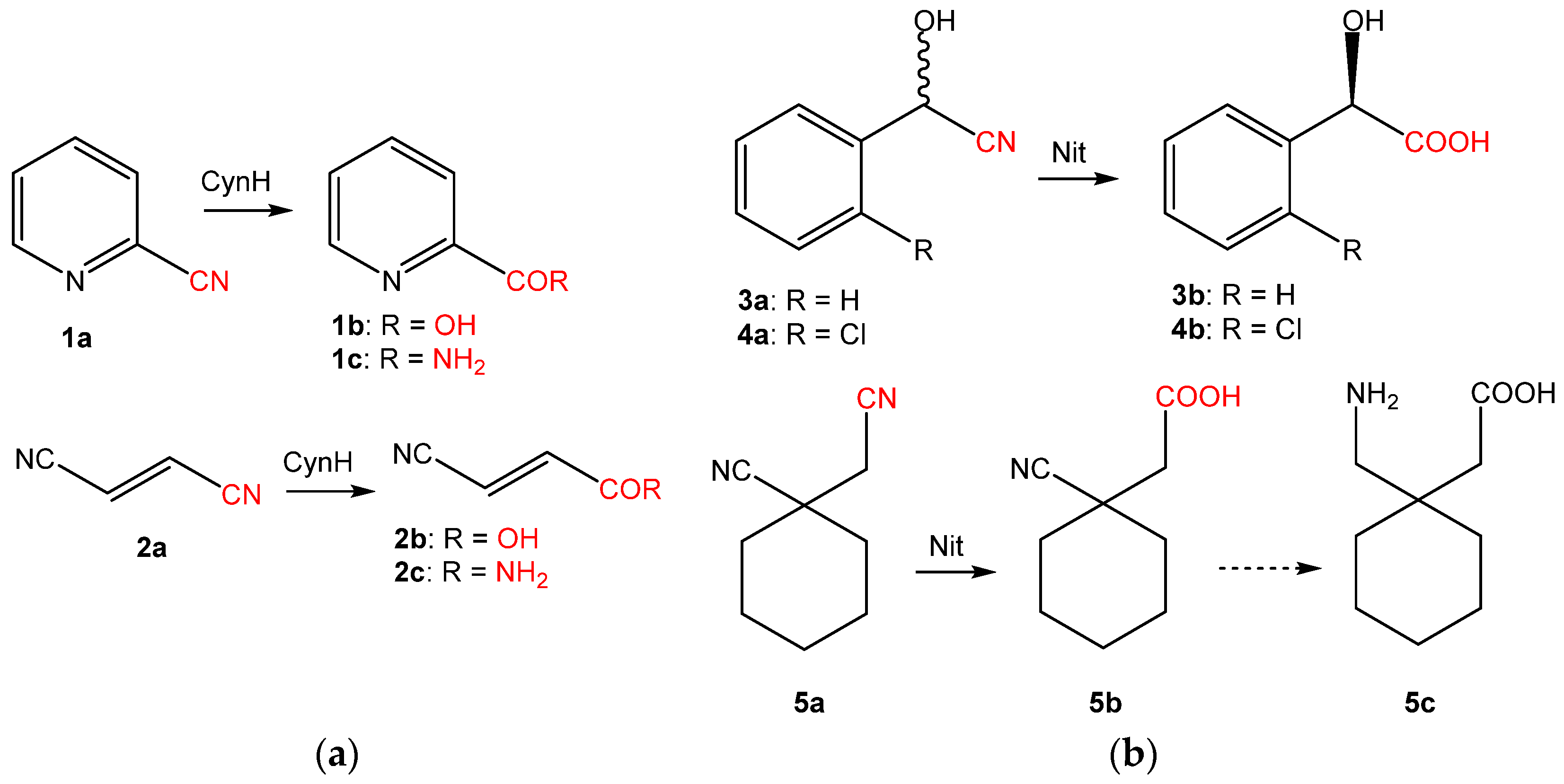
2. Origin and Distribution of the Sequences
| Phylum | Class | Genus 1 | Sequences Found 2 | Species with Confirmed CynHs (Reference) |
|---|---|---|---|---|
| Ascomycota | Dothideomycetes | Aureobasidium | 39 | |
| Alternaria | 17 | |||
| Hortaea | 12 | |||
| Pyrenophora | 4 | P. teres [29] | ||
| Leptosphaeria | 2 | L. maculans [33] | ||
| Stemphylium | 1 | S. loti [34] | ||
| Other | 46 | |||
| Lecanoromycetes | Mycoblastus | 2 | ||
| Letharia | 2 | |||
| Other | 6 | |||
| Leotiomycetes | Botrytis | 8 | ||
| Monilinia | 4 | |||
| Botryotinia | 1 | B. fuckeliana [29] | ||
| Other | 20 | |||
| Sordariomycetes | Fusarium | 43 | F. solani [35], F. lateritium [26], F. graminearum [27] 3 | |
| Colletotrichum | 21 | |||
| Monosporascus | 8 | |||
| Diaporthe | 6 | |||
| Verticillium | 6 | |||
| Neurospora | 2 | N. crassa [27] | ||
| Microdochium | 2 | M. sorghi [27] 4 | ||
| Other | 39 | |||
| Eurotiomycetes | Aspergillus | 52 | A. nidulans [27], A. niger [32] | |
| Penicillium | 29 | P. chrysogenum [28] | ||
| Other | 9 | |||
| Xylobotryomycetes | Cirrosporium | 1 | ||
| Sareomycetes | Sarea | 1 | ||
| Basidiomycota | Agaricomycetes | Auricularia | 3 | |
| Stereum | 2 | S. hirsutum [15] | ||
| Exidia | 1 | E. glandulosa [30] |
3. Overexpression of Genes
| Enzyme | Vector; His6-Tag | Medium; Cultivation Conditions | Kinetic Parameters | Reference |
|---|---|---|---|---|
| CynD | pET28a; N-terminal | LB broth; (i) 37 °C → OD 0.3; (ii) 1 mM IPTG, 30 °C, 3–4 h | Vmax 120–150 U/mg 2 KM 0.7–7.9 mM 2 | [18,19,45] |
| CynH | pET26b; C-terminal | LB broth; (i) 30 or 37 °C → OD 0.5; (ii) 1 mM IPTG, 30 °C, 3–10 h | Vmax ≈ 4400 U/mg 3 KM ≈ 90 mM 3 | [39] |
| pET28a; N-terminal pET26b; no tag | Autoinduction medium (g/L): glucose 0.5, glycerol 5, lactose 2; 30 °C, overnight | n.d. | [27] | |
| pET30a; no tag | LB broth; (i) 37 °C → OD 0.4–0.6; (ii) 0.8 mM IPTG, 26 °C, 16 h | Vmax ≈ 6800 U/mg 4 KM ≈ 109 mM 4 | [32] | |
| pET22b, pGro7 (Takara) 1; C-terminal | EnPresso (Biosilta); (i) 30 °C, 16 h; (ii) 0.02 mM IPTG, 11 mM L-arabinose; 25 °C, 24 h | n.d. | [29] | |
| pET22b; C-terminal | 2xYT; (i) 37 °C → OD 1.0; (ii) 0.02 mM IPTG, 20 °C, 20 h | Vmax ≈ 1335 U/mg 5 KM ≈ 22 mM 5 | [30] |

| Enzyme/Reaction Conditions | Compound Determined | Reagent | Method | Reference |
|---|---|---|---|---|
| CynD/4 mM KCN, pH 7.7, r.t. | Free cyanide | Picric acid, Na2CO3 | Spectrophotometric | [18,19,45] |
| CynD/4 mM KCN, pH 7.7, r.t. | Ammonia | Nessler (K2HgI4) | Spectrophotometric | [18] |
| CynH/10 mM KCN, pH 7.4, r.t. | Free cyanide | Picric acid, Na2CO3 | Spectrophotometric | [27] |
| CynH/25 mM KCN, pH 8.0, 30 °C | Formamide | - | HPLC (ion-exchange column) | [32] |
| CynH/25 mM KCN, pH 8.0, 30 °C | Formamide | - | HPLC (silica gel column) | [15] |
| CynH/25 mM KCN, pH 9.0, 30 °C | Formamide | (i) Hydroxylamine/NaOH, (ii) HCl/FeCl3 | Spectrophotometric | [30] |
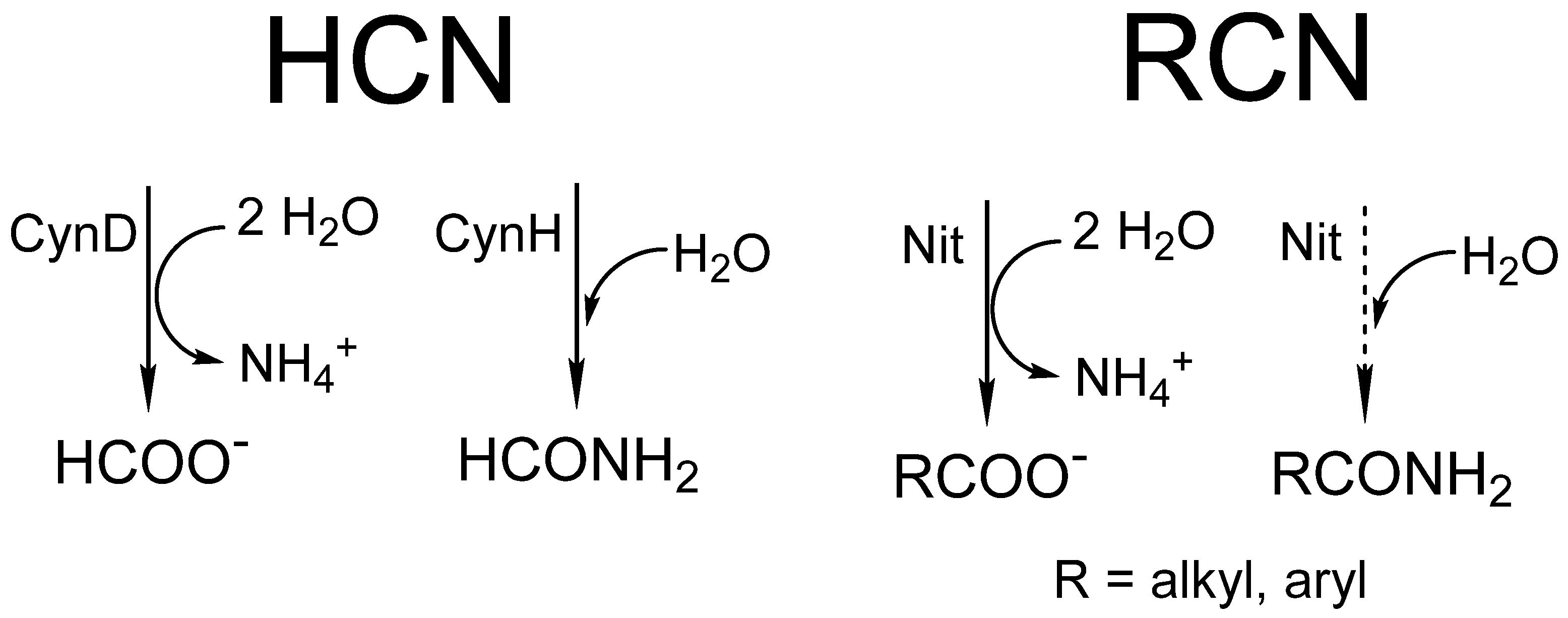
4. Activity Assays
5. Structural and Computational Studies
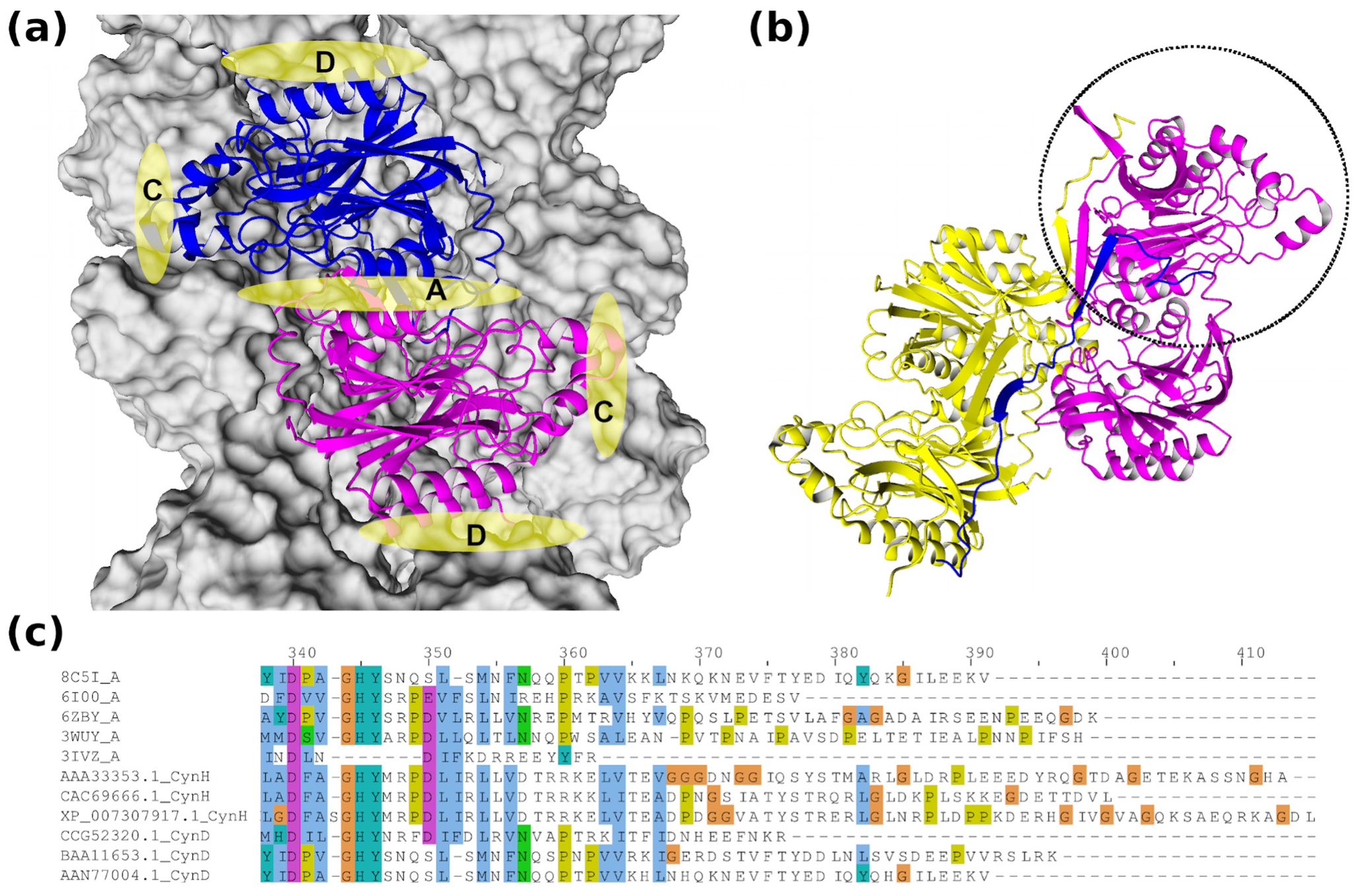
6. Engineered Enzymes
6.1. Improvement of the pH Range
6.2. Stability Enhancement
6.3. Mutants with Altered Oligomerization and Activity
7. Catalyst Forms and Uses
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Thuku, R.N.; Brady, D.; Benedik, M.J.; Sewell, B.T. Microbial nitrilases: Versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 2009, 106, 703–727. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Veselá, A.B.; Rinágelová, A.; Chmátal, M. Cyanide hydratases and cyanide dihydratases: Emerging tools in the biodegradation and biodetection of cyanide. Appl. Microbiol. Biotechnol. 2015, 99, 8875–8882. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Sewell, B.T.; Benedik, M.J. Cyanide bioremediation: The potential of engineered nitrilases. Appl. Microbiol. Biotechnol. 2017, 101, 3029–3042. [Google Scholar] [CrossRef] [PubMed]
- Howden, A.J.; Preston, G.M. Nitrilase enzymes and their role in plant-microbe interactions. Microb. Biotechnol. 2009, 2, 441–451. [Google Scholar] [CrossRef]
- Martínková, L.; Rucká, L.; Nešvera, J.; Pátek, M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J. Microbiol. Biotechnol. 2017, 33, 8. [Google Scholar] [CrossRef]
- Martínková, L.; Křen, V. Biocatalytic production of mandelic acid and analogues: A review and comparison with chemical processes. Appl. Microbiol. Biotechnol. 2018, 102, 3893–3900. [Google Scholar] [CrossRef]
- Singh, R.V.; Sambyal, K. Green synthesis aspects of (R)-(-)-mandelic acid; a potent pharmaceutically active agent and its future prospects. Crit. Rev. Biotechnol. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Shen, J.D.; Cai, X.; Liu, Z.Q.; Zheng, Y.G. Nitrilase: A promising biocatalyst in industrial applications for green chemistry. Crit. Rev. Biotechnol. 2021, 41, 72–93. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, J.; Jiang, S.Q.; Wei, D.Z. Recent research advancements on regioselective nitrilase: Fundamental and applicative aspects. Appl. Microbiol. Biotechnol. 2019, 103, 6393–6405. [Google Scholar] [CrossRef]
- Zhang, X.H.; Wang, C.Y.; Cai, X.; Xue, Y.P.; Liu, Z.Q.; Zheng, Y.G. Upscale production of (R)-mandelic acid with a stereospecific nitrilase in an aqueous system. Bioprocess Biosyst. Eng. 2020, 43, 1299–1307. [Google Scholar] [CrossRef]
- Wang, H.; Gao, W.; Sun, H.; Chen, L.; Zhang, L.; Wang, X.; Wei, D. Protein engineering of a nitrilase from Burkholderia cenocepacia J2315 for efficient and enantioselective production of (R)-o-chloromandelic acid. Appl. Environ. Microbiol. 2015, 81, 8469–8477. [Google Scholar] [CrossRef]
- Xue, Y.-P.; Wang, Y.-P.; Xu, Z.; Liu, Z.-Q.; Shu, X.-R.; Jia, D.-X.; Zheng, Y.-G.; Shen, Y.-C. Chemoenzymatic synthesis of gabapentin by combining nitrilase-mediated hydrolysis with hydrogenation over Raney-nickel. Catal. Commun. 2015, 66, 121–125. [Google Scholar] [CrossRef]
- Stolz, A.; Eppinger, E.; Sosedov, O.; Kiziak, C. Comparative analysis of the conversion of mandelonitrile and 2-phenylpropionitrile by a large set of variants generated from a nitrilase originating from Pseudomonas fluorescens EBC191. Molecules 2019, 24, 4232. [Google Scholar] [CrossRef]
- Bhalla, T.C.; Kumar, V.; Kumar, V.; Thakur, N.; Savitri. Nitrile Metabolizing Enzymes in Biocatalysis and Biotransformation. Appl. Biochem. Biotechnol. 2018, 185, 925–946. [Google Scholar] [CrossRef]
- Rucká, L.; Chmátal, M.; Kulik, N.; Petrásková, L.; Pelantová, H.; Novotný, P.; Příhodová, R.; Pátek, M.; Martínková, L. Genetic and functional diversity of nitrilases in Agaricomycotina. Int. J. Mol. Sci. 2019, 20, 5990. [Google Scholar] [CrossRef]
- Martínková, L.; Bojarová, P.; Sedova, A.; Křen, V. Recent trends in the treatment of cyanide-containing effluents: Comparison of different approaches. Crit. Rev. Environ. Sci. Technol. 2023, 53, 416–434. [Google Scholar] [CrossRef]
- Sharma, M.; Akhter, Y.; Chatterjee, S. A review on remediation of cyanide containing industrial wastes using biological systems with special reference to enzymatic degradation. World J. Microbiol. Biotechnol. 2019, 35, 70. [Google Scholar] [CrossRef]
- Crum, M.A.; Park, J.M.; Mulelu, A.E.; Sewell, B.T.; Benedik, M.J. Probing C-terminal interactions of the Pseudomonas stutzeri cyanide-degrading CynD protein. Appl. Microbiol. Biotechnol. 2015, 99, 3093–3102. [Google Scholar] [CrossRef]
- Crum, M.A.; Park, J.M.; Sewell, B.T.; Benedik, M.J. C-terminal hybrid mutant of Bacillus pumilus cyanide dihydratase dramatically enhances thermal stability and pH tolerance by reinforcing oligomerization. J. Appl. Microbiol. 2015, 118, 881–889. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, V.; Bhalla, T.C. Alkaline active cyanide dihydratase of Flavobacterium indicum MTCC 6936: Growth optimization, purification, characterization and in silico analysis. Int. J. Biol. Macromol. 2018, 116, 591–598. [Google Scholar] [CrossRef]
- Arevalo, S.J.; Sifuentes, D.Z.; Portocarrero, A.C.; Reategui, M.B.; Pimentel, C.M.; Martins, L.F.; Pierry, P.M.; Piroupo, C.M.; Santa Cruz, A.G.; Aguilar, M.Q.; et al. Genomic characterization of Bacillus safensis isolated from mine tailings in Peru and evaluation of its cyanide-degrading enzyme CynD. Appl. Environ. Microbiol. 2022, 88, e00916-22. [Google Scholar] [CrossRef]
- Terada, A.; Komatsu, D.; Ogawa, T.; Flamandita, D.; Sahlan, M.; Nishimura, M.; Yohda, M. Isolation of cyanide-degrading bacteria and molecular characterization of its cyanide-degrading nitrilase. Biotechnol. Appl. Biochem. 2022, 69, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Ponder, C.M.; Sewell, B.T.; Benedik, M.J. Residue Y70 of the nitrilase cyanide dihydratase from Bacillus pumilus is critical for formation and activity of the spiral oligomer. J. Microbiol. Biotechnol. 2016, 26, 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Mulelu, A.; Sewell, B.T.; Benedik, M.J. Probing an interfacial surface in the cyanide dihydratase from Bacillus pumilus, a spiral forming nitrilase. Front. Microbiol. 2015, 6, 1479. [Google Scholar] [CrossRef] [PubMed]
- Sewell, B.T.; Frangakis, A.; Mulelu, A.; Reitz, J. The structure of the cyanide dihydratase (CynD) from Bacillus pumilus. Acta Crystallogr. Sect. A 2017, 73, C1296. [Google Scholar] [CrossRef]
- Nolan, L.M.; Harnedy, P.A.; Turner, P.; Hearne, A.B.; O’Reilly, C. The cyanide hydratase enzyme of Fusarium lateritium also has nitrilase activity. FEMS Microbiol. Lett. 2003, 221, 161–165. [Google Scholar] [CrossRef]
- Basile, L.J.; Willson, R.C.; Sewell, B.T.; Benedik, M.J. Genome mining of cyanide-degrading nitrilases from filamentous fungi. Appl. Microbiol. Biotechnol. 2008, 80, 427–435. [Google Scholar] [CrossRef]
- Kaplan, O.; Veselá, A.B.; Petříčková, A.; Pasquarelli, F.; Pičmanová, M.; Rinágelová, A.; Bhalla, T.C.; Pátek, M.; Martínková, L. A comparative study of nitrilases identified by genome mining. Mol. Biotechnol. 2013, 54, 996–1003. [Google Scholar] [CrossRef]
- Veselá, A.B.; Rucká, L.; Kaplan, O.; Pelantová, H.; Nešvera, J.; Pátek, M.; Martínková, L. Bringing nitrilase sequences from databases to life: The search for novel substrate specificities with a focus on dinitriles. Appl. Microbiol. Biotechnol. 2016, 100, 2193–2202. [Google Scholar] [CrossRef]
- Sedova, A.; Rucká, L.; Bojarová, P.; Glozlová, M.; Novotný, P.; Křístková, B.; Pátek, M.; Martínková, L. Application potential of cyanide hydratase from Exidia glandulosa: Free cyanide removal from simulated industrial effluents. Catalysts 2021, 11, 1410. [Google Scholar] [CrossRef]
- O’Reilly, C.; Turner, P.D. The nitrilase family of CN hydrolysing enzymes—A comparative study. J. Appl. Microbiol. 2003, 95, 1161–1174. [Google Scholar] [CrossRef]
- Rinágelová, A.; Kaplan, O.; Veselá, A.B.; Chmátal, M.; Křenková, A.; Plíhal, O.; Pasquarelli, F.; Cantarella, M.; Martínková, L. Cyanide hydratase from Aspergillus niger K10: Overproduction in Escherichia coli, purification, characterization and use in continuous cyanide degradation. Process Biochem. 2014, 49, 445–450. [Google Scholar] [CrossRef]
- Sexton, A.C.; Howlett, B.J. Characterisation of a cyanide hydratase gene in the phytopathogenic fungus Leptosphaeria maculans. Mol. Gen. Genet. 2000, 263, 463–470. [Google Scholar] [CrossRef]
- Fry, W.E.; Millar, R.L. Cyanide degradation by an enzyme from Stemphylium loti. Arch. Biochem. Biophys. 1972, 151, 468–474. [Google Scholar] [CrossRef]
- Barclay, M.; Day, J.C.; Thompson, I.P.; Knowles, C.J.; Bailey, M.J. Substrate-Regulated cyanide hydratase (chy) gene expression in Fusarium solani: The potential of a transcription-based assay for monitoring the biotransformation of cyanide complexes. Environ. Microbiol. 2002, 4, 183–189. [Google Scholar] [CrossRef]
- Makumire, S.; Su, S.Y.; Weber, B.W.; Woodward, J.D.; Kimani, S.W.; Hunter, R.; Sewell, B.T. The structures of the C146A variant of the amidase from Pyrococcus horikoshii bound to glutaramide and acetamide suggest the basis of amide recognition. J. Struct. Biol. 2022, 214, 107859. [Google Scholar] [CrossRef]
- Weber, B.W.; Kimani, S.W.; Varsani, A.; Cowan, D.A.; Hunter, R.; Venter, G.A.; Gumbart, J.C.; Sewell, B.T. The mechanism of the amidases mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning. J. Biol. Chem. 2013, 288, 28514–28523. [Google Scholar] [CrossRef]
- Křístková, B.; Rädisch, R.; Kulik, N.; Horvat, M.; Rucká, L.; Grulich, M.; Rudroff, F.; Kádek, A.; Pátek, M.; Winkler, M.; et al. Scanning aldoxime dehydratase sequence space and characterization of a new aldoxime dehydratase from Fusarium vanettenii. Enzyme Microb. Technol. 2023, 164, 110187. [Google Scholar] [CrossRef]
- Jandhyala, D.M.; Willson, R.C.; Sewell, B.T.; Benedik, M.J. Comparison of cyanide-degrading nitrilases. Appl. Microbiol. Biotechnol. 2005, 68, 327–335. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Gong, J.S.; Zhang, Q.; Gu, B.C.; Dong, T.T.; Li, H.; Li, H.; Lu, Z.M.; Shi, J.S.; Xu, Z.H. Efficient biocatalytic synthesis of nicotinic acid by recombinant nitrilase via high density culture. Bioresour. Technol. 2018, 260, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Tang, L.; Liang, Y.; Jiao, S.; Yu, H.; Luo, H. Novel chaperones RrGroEL and RrGroES for activity and stability enhancement of nitrilase in Escherichia coli and Rhodococcus ruber. Molecules 2020, 25, 1002. [Google Scholar] [CrossRef] [PubMed]
- Petříčková, A.; Veselá, A.B.; Kaplan, O.; Kubáč, D.; Uhnáková, B.; Malandra, A.; Felsberg, J.; Rinágelová, A.; Weyrauch, P.; Křen, V.; et al. Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl. Microbiol. Biotechnol. 2012, 93, 1553–1561, Erratum in Appl. Microbiol. Biotechnol. 2013, 97, 9263–9264. [Google Scholar] [CrossRef] [PubMed]
- Lévy-Schil, S.; Soubrier, F.; Crutz-Le Coq, A.M.; Faucher, D.; Crouzet, J.; Pétré, D. Aliphatic nitrilase from a soil-isolated Comamonas testosteroni sp.: Gene cloning and overexpression, purification and primary structure. Gene 1995, 161, 15–20. [Google Scholar] [CrossRef]
- Crum, M.A.; Sewell, B.T.; Benedik, M.J. Bacillus pumilus cyanide dihydratase mutants with higher catalytic activity. Front. Microbiol. 2016, 7, 1264. [Google Scholar] [CrossRef]
- Shen, Q.; Yu, Z.; Lv, P.J.; Li, Q.; Zou, S.P.; Xiong, N.; Liu, Z.Q.; Xue, Y.P.; Zheng, Y.G. Engineering a Pichia pastoris nitrilase whole cell catalyst through the increased nitrilase gene copy number and co-expressing of ER oxidoreductin 1. Appl. Microbiol. Biotechnol. 2020, 104, 2489–2500. [Google Scholar] [CrossRef]
- Rustler, S.; Motejadded, H.; Altenbuchner, J.; Stolz, A. Simultaneous expression of an arylacetonitrilase from Pseudomonas fluorescens and a (S)-oxynitrilase from Manihot esculenta in Pichia pastoris for the synthesis of (S)-mandelic acid. Appl. Microbiol. Biotechnol. 2008, 80, 87–97. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Ge, Y.; Meng, Q.; Zhang, Y. Constitutive secretory expression and characterization of nitrilase from Alcaligenes faecalis in Pichia pastoris for production of R-mandelic acid. Biotechnol. Appl. Biochem. 2021, 69, 587–595. [Google Scholar] [CrossRef]
- Kuyucak, N.; Akcil, A. Cyanide and removal options from effluents in gold mining and metallurgical processes. Miner. Eng. 2013, 50–51, 13–29. [Google Scholar] [CrossRef]
- Cluness, M.J.; Turner, P.D.; Clements, E.; Brown, D.T.; Oreilly, C. Purification and properties of cyanide hydratase from Fusarium lateritium and analysis of the corresponding chy1 gene. J. Gen. Microbiol. 1993, 139, 1807–1815. [Google Scholar] [CrossRef]
- Olsen, B.A. Hydrophilic interaction chromatography using amino and silica columns for the determination of polar pharmaceuticals and impurities. J. Chromatogr. A 2001, 913, 113–122. [Google Scholar] [CrossRef]
- Jandhyala, D.; Berman, M.; Meyers, P.R.; Sewell, B.T.; Willson, R.C.; Benedik, M.J. CynD, the cyanide dihydratase from Bacillus pumilus: Gene cloning and structural studies. Appl. Environ. Microbiol. 2003, 69, 4794–4805. [Google Scholar] [CrossRef]
- Sewell, B.T.; Berman, M.N.; Meyers, P.R.; Jandhyala, D.; Benedik, M.J. The cyanide degrading nitrilase from Pseudomonas stutzeri AK61 is a two-fold symmetric, 14-subunit spiral. Structure 2003, 11, 1413–1422. [Google Scholar] [CrossRef]
- Mulelu, A.E.; Kirykowicz, A.M.; Woodward, J.D. Cryo-EM and directed evolution reveal how Arabidopsis nitrilase specificity is influenced by its quaternary structure. Commun. Biol. 2019, 2, 260. [Google Scholar] [CrossRef]
- Sewell, B.T.; Thuku, R.N.; Zhang, X.; Benedik, M.J. Oligomeric structure of nitrilases—Effect of mutating interfacial residues on activity. In Natural Products and Molecular Therapy; Kotwal, G.J., Lahiri, D.K., Eds.; Annals of the New York Academy of Sciences: Wiley, Hoboken, NJ, USA, 2005; Volume 1056, pp. 153–159. [Google Scholar]
- Zhang, L.; Yin, B.; Wang, C.; Jiang, S.; Wang, H.; Yuan, Y.A.; Wei, D. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803. J. Struct. Biol. 2014, 188, 93–101. [Google Scholar] [CrossRef]
- Xu, Z.; Cai, T.; Xiong, N.; Zou, S.P.; Xue, Y.P.; Zheng, Y.G. Engineering the residues on “A” surface and C-terminal region to improve thermostability of nitrilase. Enzyme Microb. Technol. 2018, 113, 52–58. [Google Scholar] [CrossRef]
- Jiang, S.Q.; Zhang, L.J.; Yao, Z.Q.; Gao, B.; Wang, H.L.; Mao, X.Z.; Wei, D.Z. Switching a nitrilase from Syechocystis sp. PCC6803 to a nitrile hydratase by rationally regulating reaction pathways. Catal. Sci. Technol. 2017, 7, 1122–1128. [Google Scholar] [CrossRef]
- Wang, L.; Watermeyer, J.M.; Mulelu, A.E.; Sewell, B.T.; Benedik, M.J. Engineering pH-tolerant mutants of a cyanide dihydratase. Appl. Microbiol. Biotechnol. 2012, 94, 131–140. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, X.F.; Zhang, Y.; Tang, X.L.; Zheng, R.C.; Zheng, Y.G. Development of a robust nitrilase by fragment swapping and semi-rational design for efficient biosynthesis of pregabalin precursor. Biotechnol. Bioeng. 2020, 117, 318–329. [Google Scholar] [CrossRef]
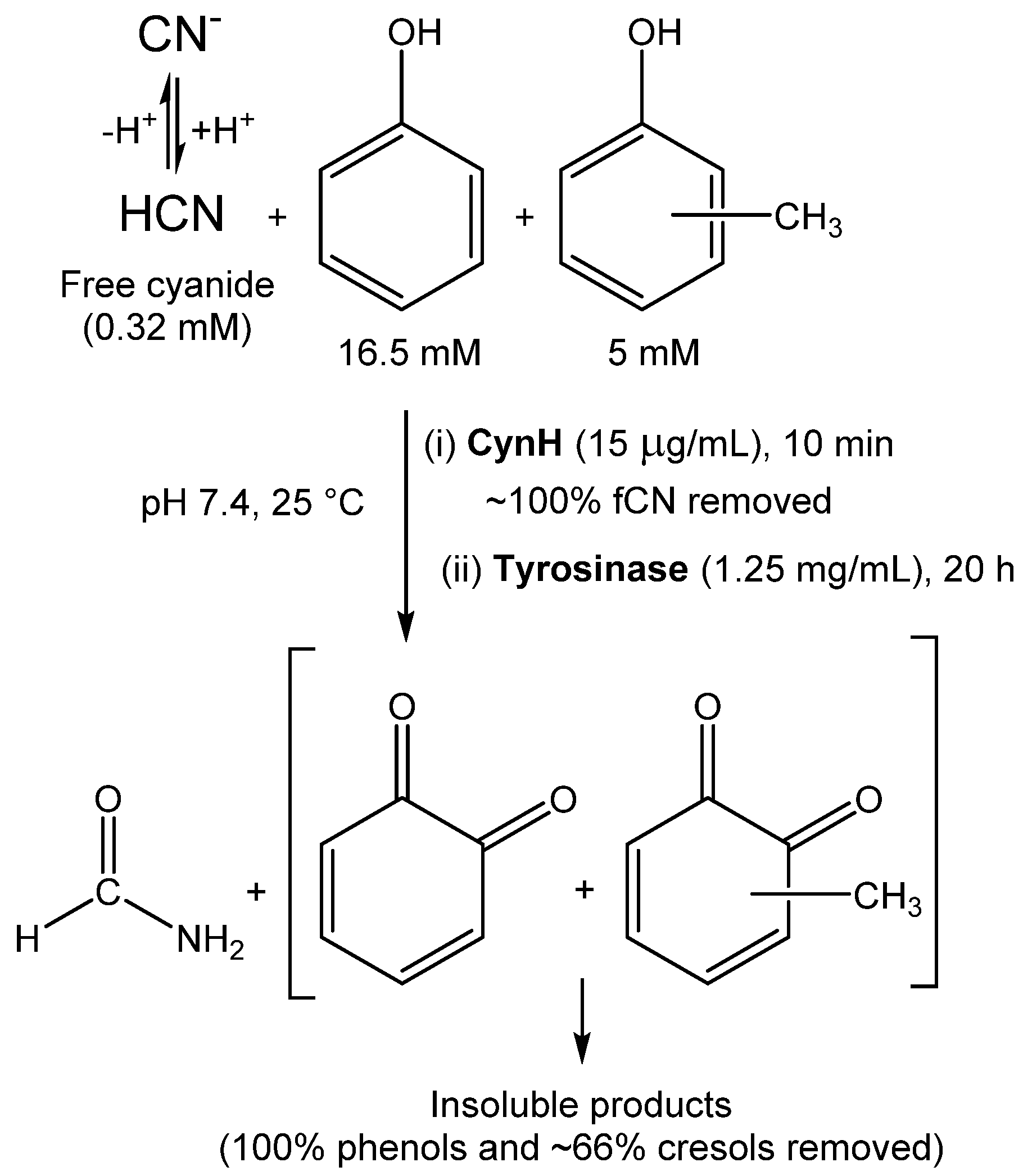
- Martínková, L.; Chmátal, M. The integration of cyanide hydratase and tyrosinase catalysts enables effective degradation of cyanide and phenol in coking wastewaters. Water Res. 2016, 102, 90–95. [Google Scholar] [CrossRef]
- Carmona-Orozco, M.L.; Panay, A.J. Immobilization of E. coli expressing Bacillus pumilus CynD in three organic polymer matrices. Appl. Microbiol. Biotechnol. 2019, 103, 5401–5410. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, V.; Singh, A.K.; Verma, N.; Bhalla, T.C. A potentiometric biosensor for cyanide detection using immobilized whole cell cyanide dihydratase of Flavobacterium indicum MTCC 6936. J. Anal. Chem. 2018, 73, 1014–1019. [Google Scholar] [CrossRef]
- Wang, X.C.; Davis, I.; Liu, A.M.; Miller, A.; Shamsi, S.A. Improved separation and detection of picolinic acid and quinolinic acid by capillary electrophoresis-mass spectrometry: Application to analysis of human cerebrospinal fluid. J. Chromatogr. A 2013, 1316, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Huang, J.W.; Xia, C.L.; Zou, S.P.; Xue, Y.P.; Zheng, Y.G. Enhanced catalytic stability and reusability of nitrilase encapsulated in ethyleneamine-mediated biosilica for regioselective hydrolysis of 1-cyanocycloalkaneacetonitrile. Int. J. Biol. Macromol. 2019, 130, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, C.; Yip, T.W.S.; Patwardhan, S.V. CO2 sequestration by enzyme immobilized onto bioinspired silica. Chem. Commun. 2013, 49, 3191–3193. [Google Scholar] [CrossRef]
- Khatik, A.G.; Jain, A.K.; Muley, A.B. Preparation, characterization and stability of cross linked nitrilase aggregates (nitrilase-CLEAs) for hydroxylation of 2-chloroisonicotinonitrile to 2-chloroisonicotinic acid. Bioprocess Biosyst. Eng. 2022, 45, 1559–1579. [Google Scholar] [CrossRef]
- Sheldon, R.A. Cross-linked enzyme aggregates as industrial biocatalysts. Org. Process Res. Dev. 2011, 15, 213–223. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Schoevaart, R.; Van Langen, L.M. Cross-linked enzyme aggregates (CLEAs): A novel and versatile method for enzyme immobilization (a review). Biocatal. Biotransform. 2005, 23, 141–147. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]


| Phylum | Class | Genus | Sequences Found 1 | Species with Confirmed CynD Activity (Reference) |
|---|---|---|---|---|
| Firmicutes | Bacilli | Bacillus | 52 2 | B. pumilus [23,24,25] |
| Paenibacillus | 12 3 | |||
| Other | 4 | |||
| Clostridia | Clostridium | 6 | ||
| Lacrimispora | 1 | |||
| Proteobacteria | Gammaproteobacteria | Acinetobacter | 1 | |
| Stutzerimonas | 1 | S. stutzeri [18] | ||
| Betaproteobacteria | Burkholderia | 2 | ||
| Actinobacteria | Actinomycetia | Brevibacterium | 1 | |
| Bacteroidota | Flavobacteria | Flavobacterium | 147 | F. indicum [20] |
| Other | 5 | |||
| Ascomycota | Saccharomycetes | Scheffersomyces | 1 |
| Variant | Increased Thermostability | Activity at pH ≥ 9.0 | Reference |
|---|---|---|---|
| K39R | + | (+) | [45] |
| Q86R | ++ | + | [59] |
| D172N | (+) | - | [45] |
| E327K | (+) | (+) | [45] |
| E327G | - | ++ | [59] |
| E35/Q322R/E327G | - | ++ | [59] |
| Q86R/E96G/D254E | + | ++ | [59] |
| Q86R/E96G/D254E/E327G | + | ++ | [59] |
| K39R/D172N/E327K | ++ | - | [45] |
| 307NHQKNE312 replaced with GERDST 1 | + | (+) | [19] |
| CynDpum-stut hybrid 2 | ++ | ++ | [19] |
| CynDpum-stut K39R | +++ | ++ | [19] |
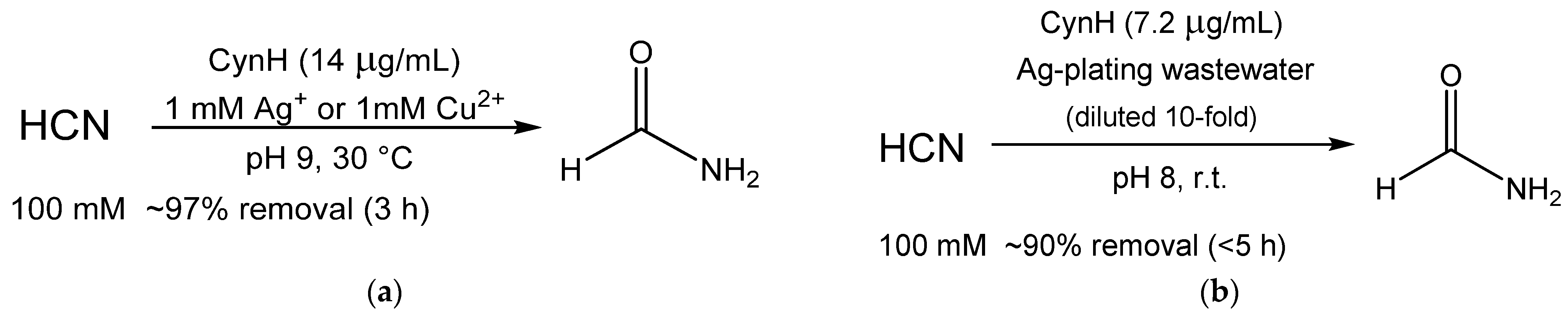
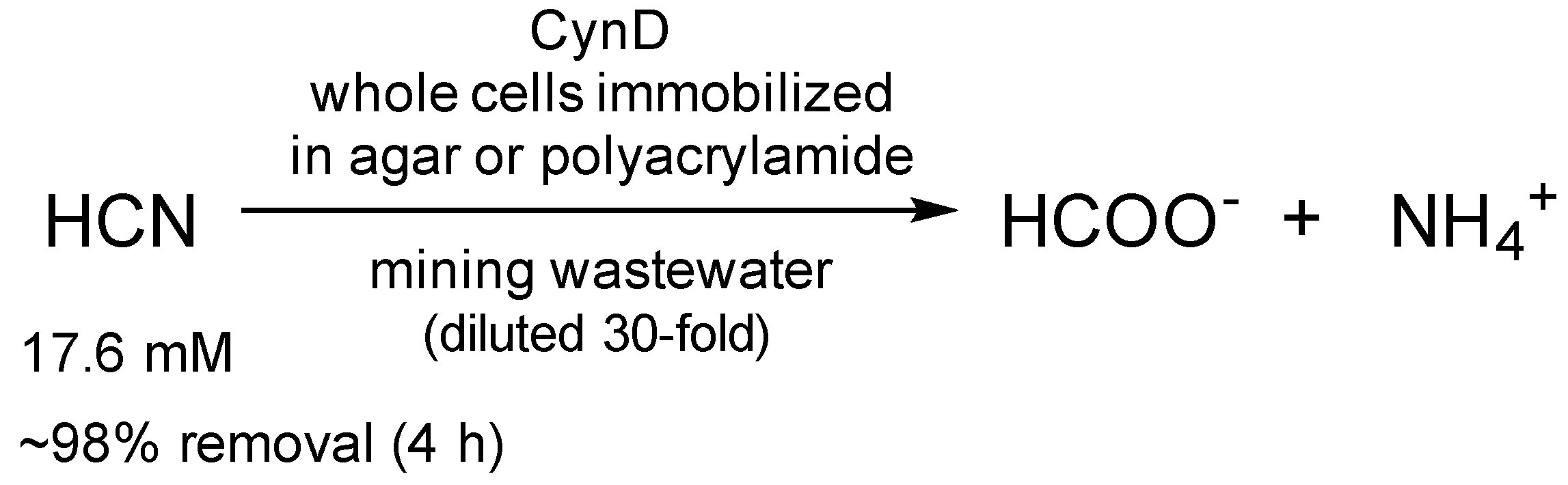
| Enzyme | Catalyst Form | Properties | Application | Cyanide Concentration; Removal | Reference |
|---|---|---|---|---|---|
| CynD | Whole cells immobilized in agar | Recyclability (>20-fold); Activity at pH 9–10 | Elimination of cyanide from mining waste | 17.6 mM, 98% 1 | [62] |
| Whole cells immobilized in polyacrylamide | Recyclability (5-fold); Activity at pH 9–10 | 17.6 mM, 98% 1; 528 mM, 43% 2 | |||
| CynD | Whole cells immobilized in agar | Optimal activity at pH 8.0, 35 °C Half-life 4 h at 40 °C | Cyanide sensor | n.a. | [63] |
| CynH | Whole cells in a flow reactor | Operational stability (3 days) | Elimination of cyanide 1 | 25 mM, >80% 3 | [32] |
| CynH | Purified | Activity at up to pH 10; Prolonged shelf-life (>3 months) | Elimination of cyanide | 0.6–100 mM; 96–100% 3 | [30] |
| Purified | Activity at up to pH 10 | Elimination of cyanide from electroplating waste | 100 mM, 30–100% 4 | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínková, L.; Kulik, N.; Sedova, A.; Křístková, B.; Bojarová, P. Recent Progress in the Production of Cyanide-Converting Nitrilases—Comparison with Nitrile-Hydrolyzing Enzymes. Catalysts 2023, 13, 500. https://doi.org/10.3390/catal13030500
Martínková L, Kulik N, Sedova A, Křístková B, Bojarová P. Recent Progress in the Production of Cyanide-Converting Nitrilases—Comparison with Nitrile-Hydrolyzing Enzymes. Catalysts. 2023; 13(3):500. https://doi.org/10.3390/catal13030500
Chicago/Turabian StyleMartínková, Ludmila, Natalia Kulik, Anastasia Sedova, Barbora Křístková, and Pavla Bojarová. 2023. "Recent Progress in the Production of Cyanide-Converting Nitrilases—Comparison with Nitrile-Hydrolyzing Enzymes" Catalysts 13, no. 3: 500. https://doi.org/10.3390/catal13030500
APA StyleMartínková, L., Kulik, N., Sedova, A., Křístková, B., & Bojarová, P. (2023). Recent Progress in the Production of Cyanide-Converting Nitrilases—Comparison with Nitrile-Hydrolyzing Enzymes. Catalysts, 13(3), 500. https://doi.org/10.3390/catal13030500