Abstract
Chemical reactions can be performed to covalently modify specific residues in proteins. When applied to native enzymes, these chemical modifications can greatly expand the available set of building blocks for the development of biocatalysts. Nucleophilic canonical amino acid sidechains are the most readily accessible targets for such endeavors. A rich history of attempts to design enhanced or novel enzymes, from various protein scaffolds, has paved the way for a rapidly developing field with growing scientific, industrial, and biomedical applications. A major challenge is to devise reactions that are compatible with native proteins and can selectively modify specific residues. Cysteine, lysine, N-terminus, and carboxylate residues comprise the most widespread naturally occurring targets for enzyme modifications. In this review, chemical methods for selective modification of enzymes will be discussed, alongside with examples of reported applications. We aim to highlight the potential of such strategies to enhance enzyme function and create novel semisynthetic biocatalysts, as well as provide a perspective in a fast-evolving topic.
1. Introduction
Biocatalysis is defined as the use of enzymes in chemical synthesis [1]. Enzymes offer synthetic advantages such as: the potential for regio-, chemo and stereo control, reactivity in aqueous media under mild conditions (temperature and pressure), and non-toxic and biodegradable components [2]. Enzyme-mediated reactions have been explored for thousands of years (e.g., fermentation of foods and beverages), however in the last two decades they have become increasingly more widespread in the chemical and pharmaceutical sectors [3,4,5]. However, despite their great potential, employment of enzymes in an industrial setting poses some issues related to protein features, such as stability, catalytic efficiency, and specificity [6]. Strategies to discover novel biocatalysts, or to improve the existing ones, include screening for natural enzyme repertoires, protein engineering through directed evolution and rational design or a combination of these approaches [7,8]. Within this framework a particular category of protein engineering, the chemical modification of native enzymes, has been around for decades [9,10,11]. The covalent alteration of amino acid side chains of an enzyme could enhance function or even create a novel biocatalyst, by expanding beyond the 20 naturally available building blocks [11,12].
The various described methodologies for chemical modification of proteins can be categorized in three main classes: (1) modification of naturally occurring amino acids; (2) modification by bioorthogonal reactions using non-canonical amino acids (nCAAs); (3) ligand directed modifications [13]. Regarding this subject, a thorough review by Pagar and co-workers has been recently published covering the application of these methods in the development of new or enhanced enzymes for biocatalysis, with a particular interest in the insertion and modification of nCAAs [14]. Here we focused on protein modification strategies targeting canonical amino acids, including cysteines, lysines, N-terminus and carboxylate residues for selective enzyme modification and functionalization. Despite great potential and increasing interest, aromatic residues (tyrosine, tryptophan, and histidine) were excluded, due to the lack of more general labeling methods and reduced number of applications. We also highlighted the chemistry behind selective modifications and their potential to create novel biocatalysts with a vast array of possible applications.
2. Historical Background
The pivotal example of protein chemical modification was published independently by Koshland Jr. and Bender, more than 50 years ago [15,16]. Both groups described the conversion of the active site serine from subtilisin (a serine protease from Bacillus subtilis) into a cysteine residue, to form a new enzyme, thiosubtilisin. The three-step reaction included the activation of the nucleophilic serine residue with phenylmethylsulfonyl fluoride (PMSF), followed by the SN2 displacement of the resulting sulfonate ester by thioacetate. The final step involves the spontaneous hydrolysis of the thioacetyl group resulting in a cysteine residue inside the active site. A decade later Ishii group reported the first successful application of the same method to a disulfide containing serine-protease, trypsin form Streptomyces griseus [17]. In a complementary work, Lowe’s group expanded this proof-of-concept to specifically modify a catalytic cysteine residue in papain [18]. Their approach was based on the successive alkylation of the active-site cysteine by a phenacyl bromide and photolysis (λ > 320 nm) of the thioether derivative. Spontaneous hydrolysis of the resulting thioaldehyde, followed by loss of hydrogen sulfide (H2S) resulted in the corresponding aldehyde, dehydroserine (formylglycine). The reduction of the photolysate with sodium borohydride (NaBH4) yielded either serine or glycine (if performed under alkaline conditions, pH 9.0).
Despite validating that is feasible to chemically modify enzymes to create new semisynthetic catalysts, these early examples lacked novel functionalities and were limited to only a few protein scaffolds. In this context, the work from E.T. Kaiser research group has provided interesting examples of enzyme scaffold functionalization [19]. First, the production of a semisynthetic protein by alkylating the active site cysteine of papain with brominated flavin coenzyme analogs [20]. The resulting flavopapain was able to catalyze de oxidation of dehydronicotinamides and dithiols, which represented the chemical conversion of a hydrolase into an oxidoreductase. Following the same strategy, E.T. Kaiser’s group also created a flavohemoglobin, containing a conjugated flavin in the vicinity of the heme group from hemoglobin [21]. The modified protein was capable of hydroxylating aniline using a 2 electrons donor (NADPH), in the absence of a reductase system. In addition, the group provided a further application of thiosubtilisin as a peptide ligase. The modified protein described by Koshland Jr and Bender lacked the endopeptidase activity but retained the ability to bind activated esters to its chemically inserted cysteine residue. This functionality was explored in amide bond formation by acylating the active site thiol with a peptide containing a C-terminal p-chlorophenyl ester [22]. The resulting thioester is prone to undergo aminolysis by the free N-terminal amino group of another peptide, thus leading to the formation of an amide bond between the two segments. This approach was successfully applied to the synthesis of peptides, including [Leu5]-enkephalin and a segment of ribonuclease T1.
Along with these selective approaches, early reports on more stochastic modifications, targeting more abundant nucleophilic residues (e.g., carboxylates and amino groups) also resulted in novel biocatalysts with interesting properties. For example, targeting carboxylates, Shen & Ryser applied carbodiimide chemistry to conjugate proteins to poly-L-lysine [23]. Conjugated albumin and horseradish peroxidase displayed increased internalization by fibroblast, which represented a safe method to promote enzyme uptake by living cells. Chen & Sigman developed a semisynthetic nuclease by modifying primary amines of the tryptophan repressor (Trp) from Escherichia coli [24]. The protein amino groups were first labeled with 2-iminothiolane hydrochloride. The resulting sulfhydryl groups were then alkylated with iodoacetamide containing a 1,10-phenantroline moiety. Coordination of copper ions resulted in Trp-Phenantroline-Cu complexes which retained DNA binding properties and promoted oxidative cleavage in a sequence-specific fashion. Inada and co-workers have reported the PEGylation of amino groups of different enzymes (lipase, catalase, chymotrypsin, and a peroxidase) with choro-triazine activated-polyethylene glycol (PEG) [25]. All conjugated enzymes were soluble and remained active in benzene. Particularly for lipase and catalase, modified enzymes were active in all tested organic solvents and even displayed activity enhancement in 1,1,1-tricholorethane.
A rich past in the development of novel biocatalysts through chemical modification strategies (Table 1) highlights a diverse set of application possibilities. It is important to note that even with a limited chemical toolbox, the early mentioned examples still provide a solid base for future developments. In the next sessions we discuss the challenges inherent to native protein modification as well as the ever-growing panel of selective labeling reagents and their functional applications.

Table 1.
Historical enzyme modification examples.
3. The Challenges of Biocompatibility and Selectivity
Protein modification strategies that target canonical amino acids rely ultimately on the intrinsic reactivity and nucleophilicity of naturally occurring residues, most notably cysteine and lysine [14,26]. Targeting natural amino acids offers immediate accessibility to modifications, relative to techniques such as genetic codon expansion or enzymatic labeling. In addition, protein modification in a genetically intact system provides better models to investigate functionality and offers the possibility to be performed in vivo [27]. A major drawback, however, is selectivity, since many of protein functional groups are nucleophiles. Also, target abundance plays its role in defining the precision of a modification [28,29]. Thus, protein modification approaches must address two main challenges: bio-compatible reactions and selectivity (Figure 1).
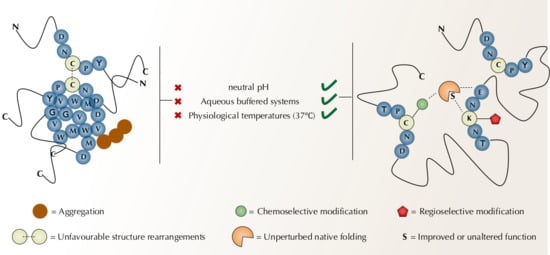
Figure 1.
A schematic representation of conditions required for protein modification strategies targeting canonical residues. Conditions to determine specific modifications and stable products are highlighted.
To retain enzymatic function, chemical modifications of proteins require a tight control over the reaction conditions. Modifications must proceed in aqueous buffered systems and organic solvents should be used with discretion to avoid interfering in protein folding. For the same reasons, pH should be kept near neutrality (6.0 to 8.0) and temperatures below 37 °C. Stoichiometry and concentration are also an issue, as proteins tend to aggregate and thus, should be handled at the µM range (or less) [29,30,31].
Proteins contain multiple copies of reactive amino acids and modification methods must discriminate among similar reacting groups. In this context two features are essential. Chemoselectivity, which is the selectivity of an electrophile to react with a specific nucleophilic functional group (e.g., cysteine thiol, lysine ε-amino group and N-terminus α-amino group) and site-selectivity (or regioselectivity), which requires the electrophile to further distinguish among different copies of the same functional group [30]. The complex nature of a protein provides therein a great challenge but also opportunities. Protein folding promotes interactions among residues that are not evident in the primary structure. The result is a landscape of microenvironments that can shape the reactivity of functional groups creating reactivity hotspots [32]
In a native protein modification perspective, target residues should be solvent exposed and located, preferentially, in the protein surface to avoid disturbances in the hydrophobic core and overall conformation. Chemoselectivity can be achieved by careful selection of targets and reagents. Harnessing the unique properties of functional groups (nucleophilicity, hardness/softness, acidity/basicity, oxidation state), as well as their location, is detrimental to avoid heterogenous and undesired products [33,34]. Site-selectivity, on the other hand, is by no means a trivial task. Because of the complex nature of proteins, microenvironment tuning of pKa (charge-charge interactions, hydrogen bonds and desolvation) profoundly affect the reactivity of certain residues [35]. Moreover, the differences in reactivity among different copies of the same residue can be so subtle that the challenge escalates exponentially with the frequency of the amino acid [30]. Nonetheless, chemical probes can and have already been designed to explore these reactivity differences and selectively modify specific residues. In the next session, we discuss established and new approaches to achieve chemo and site-selectivity in protein modification.
4. Chemical Aspects of Selective Protein Modification
4.1. Cysteines
In proteins, cysteines are multifunctional residues, which display catalytic, regulatory, structural, and metal-binding properties [36]. This wide array of functions is provided by the unique features of the sulfur atom in the thiol group (R-SH) [37]. Properties as valence shell radius, electronegativity, electron polarizability and multiple oxidation states combine to make cysteine thiol into a redox-active soft nucleophile with immense chemical versatility [38,39,40]. Cysteine is also a relatively rare type of residue found in proteins, with a maximum occurrence of 2.3% in mammalian proteomes (Homo sapiens, Bos taurus and Mus musculus), a number that is reduced to 1.27% in yeast (Saccharomyces cerevisiae) and 1.16% in bacteria (Escherichia coli) [41,42]. Interestingly, despite low abundance in protein surfaces, solvent exposed cysteines are among the most reactive ones (together with buried active site cysteines) [43,44]. Altogether, chemical reactivity and relative scarcity render cysteines a very suited target for selective modification of proteins (Figure 2).
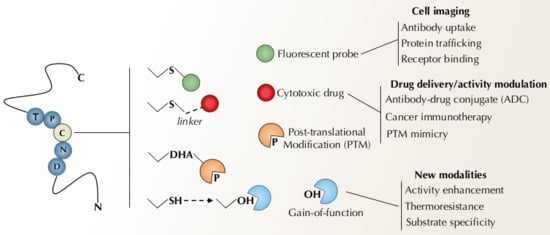
Figure 2.
A schematic representation of protein modification strategies targeting cysteine residues and their application in biomedicine and biotechnology.
In general, under physiological pH cysteine thiols are readily deprotonated to the thiolate anion (R-S-), one of the most reactive functional groups in proteins. Harboring three lone pairs of electrons that are highly polarizable, cysteine thiolates are potent soft nucleophiles that react, preferentially, with electrophilic species of the same nature [39,45,46]. These features provide useful ways to selective modify cysteines over other nucleophilic residues, by adjusting reaction pH to favor the deprotonated thiolate and selecting soft electrophilic probes to avoid reactions with harder nucleophilic side chains (e.g., Lys, Arg, His).
The search for electrophilic cysteine targeting reactants has been fueled by the need for selective cysteine labeling in chemical proteomics, production of protein bioconjugates and targeted covalent inhibitors [47,48,49,50,51,52,53,54]. Haloacetamides (e.g., iodoacetamide and chloroacetamide) have long been used as cysteine tagging-reagents in proteomic research [50]. Maleimides (e.g., N-ethylmaleimide) display higher chemoselectivity towards cysteines and are still the chemistry of choice for the preparation fluorescent-labeled biomolecules, PEGylated proteins, and antibody-drug conjugates (ADCs) (including several FDA approved compounds for cancer treatment) [28,55,56]. Other Michael acceptors as α,β-unsaturated carbonyls (e.g., acrylamides), targeting non-catalytic cysteines, are the most prevalent electrophilic warheads in the development of covalent drugs [49,57]. Sulfur containing electrophiles (e.g., sulfones and sulfonate esters) have also been widely used as cysteine selective probes, whilst activated thiols (e.g., thiosulfates, thiosulfonates and TNB-thiols) provide good chemoselectivity and offer a handle for further modifications through thiol-disulfide exchange reactions [30,50,54]. In recent years a panel of structurally diverse electrophiles has emerged for cysteine selective labeling and functionalization, including: bromo-oxetanes [58]; quaternized viny/alkynil pyridines [59]; dinitroimidazoles [60]; isoxazoliniums [61]; cyclopropenyl ketones [62]; 3-bromo-5-methylene pyrrolones [63]; phosphonamidates [64,65]; vinylphosphonothiolates [66].
4.2. Lysines
Lysine residues in proteins display structural, regulatory, and catalytic functions. Those are mainly related to the electrostatic and nucleophilic properties of its sidechain. Bearing an aliphatic primary amine (ε-amine) that is protonated at physiological pH (7.4), lysine is one of the positively charged (basic) amino acids together with arginine and histidine. The protonated lysine sidechain has a net charge of +1 and is usually present at the protein surface, exposed to the solvent [67]. This positive charge favors protein solubility and provides the basis for functionally important intra-intermolecular interactions such as cation-π, hydrogen bonds and salt bridges [68,69]. Also a target of a wide range of post-translational modifications (e.g., methylation, acylation, and ubiquitination), lysine residues play a crucial role in protein stability, protein-protein interactions, chromatin structure and enzymatic activity [60,70,71]. Lysines occur at a frequency of 5.19% when considering proteomes from all domains of life [72]. Thus, compared to cysteines, lysine residues are much more available for protein modification reactions.
Lysine reactivity as a nucleophile largely depends on its protonation state. The protonated ε-amino group (-NH3+) lacks the lone pair of electrons that makes its neutral counterpart (-NH2) a potent nucleophile [73]. Amino group pKa plays an important role when developing lysine selective reactions. Solvent exposed lysine residues have an overall pKa of ~10, whilst N-termini α-amino groups have a pK of ~8 [74,75,76]. Optimal pH for lysine modification ranges between 8.5 and 9.5. Being more nucleophilic than α-amino groups, the ε-amino group from lysines will be more reactive towards electrophiles in these conditions [34]. Also important is the fact that primary amines are harder nucleophiles than cysteinyl thiols and will react preferentially with harder electrophiles [46,77]. Thus, controlling reaction pH and carefully selecting the electrophilic reagents is crucial to achieve chemoselectivity.
Due to the high nucleophilicity and abundance in protein surfaces lysine residues are a popular target for protein modification [28]. Main applications include the production of bioconjugates such as fluorescent labels (e.g., fluorescein isothiocyanate—FITC), PEGylation, glycoprotein vaccines and antibody-drug conjugates [78,79,80]. Among lysine chemoselective probes, activated esters (mainly N-hydroxysuccinimide—NHS –esters) are the most widely used reagents. Other reagents include isocyanates and isothiocyanates, sulfonyl chlorides, reductive amination of aldehydes, anhydrides, squaric acids and 2-amino-2-methoxyethyl [13,28,34,81]. In contrast to cysteine, lysine ubiquity makes it difficult to develop site-selective labeling strategies. However, a few successful attempts are paving the way for further development. Chen and co-workers were able to label single lysine residues in RNase A and Lysozyme using substoichiometric amounts of a biotin containing NHS-ester reagent [82]. A so-called linchpin directed modification strategy was described by Adusumalli and co-workers using a bifunctional aldehyde reagent [83]. Capable of tagging the most reactive lysine residues in a reversible manner the reagent led to the modification of less reactive adjacent lysines in a site-selective fashion. The method was successfully applied to seven different protein scaffolds. In a different approach, Matos and coworkers described a computer assisted strategy to design a site-selective lysine labeling reagent [84]. Using quantum mechanical calculations to model aza-Michael addition reactions, a sulfonyl acrylate (methyl-2-sulfonylmethyl acrylate) was identified to react with a ΔG‡ of 17.5 kcal mol−1. Hydrogen bonding to the reaction transition state was proven to be the key feature to enhance lysine nucleophilicity, lower the energy barrier and exclude cysteine from the reaction. The reagent was used to label single lysine residues in several proteins (human serum albumin, lyzozyme, C2A domain from synaptotagmin-I and annexin V) without structural or functional impairing. Computational prediction of protein pKa revealed that modified lysine residues were slightly more acidic than the others.
4.3. N-Terminus
Compared to the ubiquity of other reactive nucleophiles, N-terminal α-amine is appealing to site-selective chemical modification for being present in every protein and for consisting of a single reactive handle. Under the influence of the backbone amide, the α-amino group has a reduced pKa (6.0 to 8.0) compared to the ε-amine from lysines (~10.5), being highly reactive at near neutral conditions [85]. This difference in pKa can be exploited to achieve site-selective N-terminal labeling using amine modifying approaches, such as NHS-esters and reductive amination with aldehydes, under appropriate pH control [27,86,87]. Other reagents applied to site-selective N-terminus labeling include imidazole-1-sulfonylazide [88], ketenes [89], 2-pyridinecarboxyaldehydes [90] and 2-ethynylbenzaldehyde [91].
4.4. Carboxilates
Carboxylic acids are present in proteins at the side chains of aspartate and glutamate residues, as well as in the C-terminus. Despite characterizing a nucleophilic functional group, carboxylates exhibit poor nucleophilicity and reactivity under physiological conditions. Selective labeling these groups is, therefore, challenging and usually requires activating reagents [30]. An early strategy includes the esterification with diazo compounds, which dates to the 1960s and has been recently revisited for its possible application in therapeutics and as cross-linking reagent [92,93,94]. However, the most widely used carboxylate targeting reagent are carbodiimides. Reaction leads to the formation of activated acylisoureas, which can react with nucleophiles, namely amines, for further conjugation or labeling [34]. A recent report described a 3-phenyl-2H-arizine probe for carboxylate labeling with applications in chemical proteomics [95]. Despite the challenge, two site-selective carboxylate labeling strategies have been reported. Rawale and co-workers employed a linchpin directed modification to label a single glutamate residue in ubiquitin, using a bivalent reagent containing a 2-hydroxybenzaldehyde and an aryl sulfonate ester moiety [96]. Bloom and coworkers described a photoredox catalysis platform for C-terminal modification. Being able to harness differences in oxidation potential among carboxylate residues, the selective decarboxylative labeling and conjugation at the C-terminus was achieved in model peptides and insulin [97].
5. Selective Protein Modification and Novel Semisynthetic Biocatalysts
5.1. Modification at Cysteines
Thiol labeling chemistry has been primarily used in the development of protein-based therapeutics as protein-drug conjugates (e.g., ADCs) and protein-polymer conjugates (e.g., PEGylated proteins) [54,80]. However, recent examples of enzymes modified at cysteine residues provide good evidence towards the utility of this canonical residue as a reactive handle for enzyme functionalization.
López-Gallego and colleagues have characterized the chemical modification and activity enhancement of a lipase (BTL2) from Geobacillus thermocatenulatus [98]. Three mutant proteins, containing a single cysteine inside the catalytic cavity, were functionalized via thiol-disulfide exchange reactions. First, cysteines were activated by DTNB, followed by tethering of alkane-thiols (1-dodecanethiol and 1-octanethiol). The I320C mutant containing a C12 alkane displayed a 2.5 increment in activity compared to the wild-type (wt) enzyme. Tethering of alkanes to the enzyme active site is supposed to imitate the interfacial activation effect, in which detergents and hydrophobic interfaces enhance enzyme activity by promoting an open conformation of the protein lid module. Insensitivity to Triton-X activation, a reduced Km, increased susceptibility to inhibitors and intrinsic fluorescence changes supports the hypothesis of a permanent conformational change promoted by the modification. Besides higher activity, the semisynthetic enzyme also displayed different substrate specificity (short-chain esters over long-chains). More importantly, the I320-C12 variant of the enzyme displayed higher enantioselectivity (92% enantiomeric excess compared to 79% in wt) in the resolution of a chiral secondary alcohol (synthesis of 3-oxyran-2-yl butyrate). Also using a lipase (from Burkholderia cepacia) as protein scaffold, Ema & Inoue described a semisynthetic biocatalyst modified at a mutated cysteine residue by 2-Iodo-N-phenylacetamide [99]. It allowed a specific modification of a surface exposed cysteine (I287C) over the disulfide forming C190 and C270. Modified enzymes displayed enhanced enantioselectivity in the kinetic resolution of various secondary alcohols with vinyl acetate.
Darby and coworkers developed an inventive approach to enzyme chemical modification, using as model a glycoside hydrolase (BtG84, a O-GlcNAcase) from Bacteroides thetaiotaomicron [100]. In a previous work, the group described a small molecule activator of the enzyme through a fragment-based lead discovery (FBLD) strategy [101]. The 4-etoxyquinazoline compound stabilized the enzyme in an active closed conformation, which promoted an increase in the catalytic efficiency (kcat/Km). Further development of the compound included an acrylamide functional group at position 2 of the quinazoline ring for protein modification. By producing a mutant enzyme containing a single exposed cysteine residue, carefully placed at a flexible loop near the active site (Y550C), the authors were able to covalently attach the activator molecule through a Michael addition reaction to the cysteine thiol. Compared to the wt protein, the functionalized enzyme displayed a 10-fold increase in the catalytic efficiency, including a 5-fold reduction in Km for the synthetic substrate used. Further modifications of the tethered activator molecules resulted in an even more active enzyme (18 and 11-fold increase in kcat/Km, compared to wt, for the strongest activators). A thorough investigation on the mechanism of activation concluded that tethered molecules increase the enzyme pH optima, in addition to altering substrate binding affinity. This example provides a proof of principle in which small fragment-based approaches can be used for enzyme functionalization.
Cysteine residues have also been explored as a reactive handle for the selective anchoring of metal-coordinating ligands or complexes. As a result, different proteins scaffolds can be converted into semi-synthetic hybrid catalysts, known as artificial metalloenzymes [102]. Steroid Carrier Protein 2L mutant (A100C) was converted into a metalloenzyme through tethering of iron-binding nitrogen cofactor containing a maleimide linker [103]. The modified protein was able to catalyze the oxidation of β-O-4 linkage in benzylic alcohols from model substrates of lignin processing. In a similar way, the β-barrel protein nitrobindin was mutated to generate variants of different cavity sizes and single cysteine mutant (Q96C). A Grubbs-Hoveyda type catalyst was anchored to the cysteine thiol through a maleimide linker in the N-heterocycle carbene ligand [104]. The resulting hybrid catalyst was able to perform olefin metathesis reactions in water in the absence of organic solvents.
Another versatile and general strategy for protein modification at cysteine residues consists of its chemical conversion to dehydroalanine (DHA), as described by the Benjamin Davis research group [105,106,107,108]. In terms of protein modification, dehydroalanine (an α-β-unsaturated amino acid) provides a unique electrophilic handle. It can undergo Michael addition reactions with nucleophiles or carbon-based free radical additions in a site-selective manner, to generate functionalized proteins bearing non-canonical sidechains [26,109]. DHA can be obtained from natural or mutated cysteine residues through the chemoselective bisalkylation-elimination reaction with brominated electrophiles (e.g., 2,5-dibromohexanediamide and metyl-2,5-dibromopentanoate) in native conditions [107,110]. Functionalization of proteins through DHA has mainly been applied to the in vitro recreation of post-translational modifications (PTMs) and insertion of non-canonical sidechains [111,112,113,114,115]. Nonetheless its application for the modification of native enzymes has been accomplished and highlights the utility of a DHA-based strategy for mechanistic studies and enzyme engineering.
A well characterized example was described by Alan Berry research group using the enzyme N-acetylneuraminic acid lyase (NAL) from Staphylococcus aureus. The enzyme catalyzes the aldol reaction between N-acetyl-D-mannosamine (ManNAc) and pyruvate to form N-acetyl neuraminic acid (a sialic acid). While being highly specific for its ketone donor (pyruvate) NAL accepts different aldehydes substrates, displaying a strong preference for longer molecules. The first attempt to chemically modify NAL included de substitution of the catalytic Lys165 by a cysteine (the wt protein contains no cysteine residues). The conversion of cysteine to DHA followed by the conjugate addition of 2-aminoethanol resulted in a γ-thialysine containing protein [116]. Crystallographic studies revealed that the modified enzyme exhibits the same structure as the wt counterpart. However, despite binding pyruvate, it displayed reduced activity (30% of wt) and a shift in the pH optima (7.4 to 6.8). The pH profiling and analysis of theoretical titration curves indicated that imbalances in the ionization states of residues within the modified active site prevents a fully active catalyst. In the second attempt, a single cysteine was inserted in various locations inside the enzyme active site, followed by the conversion to DHA and conjugate addition of 13 different thiol containing molecules. For each modified enzyme activity was tested for the condensation of pyruvate with several aldehydes [117]. Most notably, modification of the F190C mutant with three hydroxylated thiols (1-mercapto-2,3-propanediol, 1-mercapto-2-propanol and 1-mercapto-4-butanol) resulted in modified enzymes capable of accepting erythrose as substrate. The C4 aldehyde is poorly metabolized by the wt enzyme, thus the modifications represent a radical change in substrate specificity. The kinetic characterization of the F190Dpc variant (2,3-dihydroxypropyl cysteine) revealed a 10-fold increase in kcat/Km, compared to the wt, for the condensation of erythrose. Comparing to ManNAc, it represented a 30-fold switch in substrate specificity. This is largely attributed to kcat (15-fold increase compared to wt) and not to Km. Structural characterization and molecular modeling revealed that the Dpc sidechain alters the conformation and interaction network in the active site, which promotes a better stabilization of the transition state. Altogether, both reports provide a good highlight on how chemical modification through DHA can be applied to engineer enzyme function.
5.2. Modification at Lysines
Enzyme chemical modification through lysine residues has been limited to the production of heterogenous conjugates due to multiple site labeling. However, it is important to highlight the fact that lysine is a readily accessible and versatile target in native proteins. Its modification by chemical methods is particularly suited when site-selectivity is not detrimental and offers a valuable tool for enzyme functionalization [14,29]. For instance, the chemoselective reaction of ε-amino groups with acid anhydrides (e.g., acetic, citraconic, maleic, phthalic, pyromelltic and succinic). The acetylation of lysines by anhydrides neutralizes its positive charge and introduce extra-hydrophilic groups to the protein surface [14,73]. The effects on enzyme activity vary among different protein scaffolds, however an improvement in overall thermal stability and resistance to detergents and solvents is well documented. This approach has been employed to enzymes with industrial and biotechnological application, namely amylases [118,119,120,121]; bromelain [122]; α-chimotrypsin [123]; horseradish peroxidase [124]; and lypases [123,125,126].
Grafting enzymes to polymers through lysine residues also represent a popular functionalization strategy to create semisynthetic biocatalysts with enhanced properties. Enzyme-polymer conjugates may display enhanced reactivity due to polymer mediated alterations of microenvironment. Important local effects include pH alterations and increases in substrate concentration [127]. The former was demonstrated by Benson and co-workers using Cytochrome C as a protein scaffold that was grafted to poly (acrylic acid) polymers (PAA) [128]. Using carbodiimide chemistry, polymers with different molecular weights were cross-linked to the protein primary amines and effects on the peroxidase activity were evaluated. Bearing a weakly acidic COOH groups, PAA modulation of local pH and destabilization of a wasteful reaction intermediate accounted for the 34-fold increase on kcat of the most active conjugate. In a similar fashion Kovaliov and co-workers reported an increase in lipolytic activity for Candida antarctica lipase-B (CALB) and Thermomyces lanuginosus lipase (TLL) modified with a hydrophobic N-(iso-butoxymethyl) acrylamide (NIBMA) polymer [129]. Monomers were conjugated to the protein lysine residues through a trithiocarbonate chain transfer reagent containing an NHS moiety. Polymerization was performed by a photoinduced electron transfer reversible addition-fragmentation method. Conjugated CALB and TLL displayed a respective 6-fold and 3-fold increase in enzymatic activity. This was related to a possible increase in the local concentration of substrate (p-nitrophenyl palmitate) mediated by the hydrophobic polymer shell.
Another interesting application of protein modification strategies is the production of biocatalysts by ionic liquids. Ionic liquids (ILs) are cationic and anionic low melting salts that are liquid at temperatures below 100 °C [130]. ILs have hybrid physicochemical properties and stand somewhere in between molten inorganic salts and organic solvents [131]. Simply grafting carbonyldiimidazole activated ionic liquids onto lysine residues from lipases (Porcine Pancreatic Lipases—PPL and CALB) has been shown to enhance enzyme properties. Modified proteins exhibited improved enzymatic activity (hydrolysis of p-nitrophenyl palmitate), enantioselectivity (hydrolysis of racemic 1-phenethyl acetate), thermal stability and resistance to organic solvents (methanol and dimethyl sulfoxide—DMSO) [132,133].
Their dual nature makes ILs particularly appropriate in some biocatalysis applications as they can provide both a protein stabilizing environment and, unlike aqueous solutions, are capable of dissolving hydrophobic molecules [134,135,136]. However, finding biocompatible ILs can be a challenge, as they can both enhance or disturb enzymatic functioning, due to the extensive molecular interactions these solvents establish with proteins [137,138]. Grafting IL-analogous cations onto proteins also offers a possibility to make enzymes more stable in ILs. Using carbonyldiimidazole as a linking agent, Bekhouche and coworkers described the grafting of hydroxylated alkyl imidazolium and cholinium salts to lysine residues of formate dehydrogenase (FHD) from Candida boidinii [139]. Modified enzymes displayed reduced activity compared to wt in aqueous solution. In the presence of 30% [MMIm][MeSO4] (1-methyl-3methyl imidazolium/dimethyl sulfate), however, wt enzymes were deactivated whilst modified variants retained their activity. More importantly, in the IL-containing media, FDH bearing hydroxypropylmethyl imidazolium grafts displayed a 30% increase in catalytic efficiency compared to the wt enzyme in aqueous conditions. Alternatively, Nordwald and co-workers reported chemical modification strategy to engineer the surface charge of a commercial Trichoderma reesei cellulase cocktail (TRC) [140]. Primary amines were modified by succinic anhydride changing the negative-to-positive charge ratio. Succinylation of the enzyme preparation resulted in a near 2-fold increment in cellulose conversion in the presence of 15% [BMIm][Cl] (1-butyl-3-methylimidazolium chloride), a condition that inactivated the wt cocktail. Fluorescence quenching assays suggest that stability and activity improvement correlated to reduced disrupting interactions with the Cl− anion.
5.3. Modification at the N-Terminus
Despite being less common than lysine modification, chemically targeting the N-terminal α-amino group offers instant access to site-selectivity. Examples of enzyme functionalization through their N-terminus are discussed below.
Perez-Rizquez and co-workers modified three glycosidases from Lactobacillus plantarum with hyaluronic acid (HA) containing a N-hydroxysuccinimide reactive group (installed by pre-activation using carbodiimide) [141]. Having performed the reaction at pH 7.0, the authors claim the HA molecules were selectively attached to the N-terminal residues of the enzymes. Despite reduced activity, modified enzymes displayed altered substrate specificity for the hydrolysis of p-nitrophenylglicosides and p-nitrophenyl-6-phosphate glycosides (glucose and galactose) tested. Most notably, the attachment of HA to the variant Lp_3525 switched the specificity towards phosphorylated glucose (compared to the more promiscuous wt enzyme), while retaining 80% of the original enzyme activity.
Schoffelen and coworkers described a copper-free diazotransfer reaction on amines using imidazole-1-sulfonylazide in aqueous media [88]. Performing the reaction at pH 8.5 allowed the selective modification of the solvent exposed primary amine with the lowest pKa. For Candida antarctica lipase B (CALB) and elastin-like polypeptide selective labeling of the N-terminal primary amine was achieved. For CALB, labeling resulted in a functional enzyme retaining 97% of its activity compared to control. Moreover, the azide moiety of the labeling reagent allowed for further modification in a copper-free click reaction, with polyethyleneglycol containing a reactive aza-dibenzo-cyclooctine group.
Using an alkyne-functionalized ketene reagent, Chan and co-workers were able to selectively label the N-terminus of enzymes as lysozyme, RNase A and the therapeutic arginase from Bacillus caldovelox (BCArg) [89]. Lysozyme activity was tested and showed that labeling did not interfere with enzyme functioning. The ketene reagent was proved to be superior to NHS-esters in N-terminal selective labeling, while also offering an alkyne reactive handle for further modification reactions through copper-catalyzed cycloaddition. Similarly, the same proteins were modified by Deng and co-workers using 2-ethynylbenzaldehydes [91]. Under neutral or slightly acidic pH (6.5 to 7.4) the reaction is selective towards the N-terminal primary amine, with the formation of an isoquinolinium heterocycle. A modified variant of BCArg was shown to retain most of its enzymatic and cytotoxic activities compared to the wt enzyme.
5.4. Modification at Carboxilates
As for lysines, carboxylates offer abundant and solvent exposed sites for protein modification approaches that do not require site-selectivity. The two examples discussed below highlight the applicability of carbodiimide activation on carboxylate labeling and its potential to enhance enzyme function.
Using a glycosidase from Aspergillus niger, Brogan and co-workers reported the development of an enzyme-surfactant conjugate nanoconstruct [142]. Activation of carboxylates with a carbodiimide, followed by cationization with a diamine reagent (N,N′-bis(2-aminoethyl)-1,3-propanediamine) allowed for electrostatic conjugation with an anionic surfactant (glycolic acid ethoxylate lauril ether). Conjugates were stable in aqueous solution and several ionic liquids (ILs). Moreover, modified proteins displayed an increased half denaturation temperature (up to 79 °C) in tested ILs. Glycosidase activity of both enzymes were similar in aqueous solution. However, in near anhydrous IL (1-ethyl-3-methylimidazolium ethyl sulfate), the modified variant was 10-fold more active than the wt enzyme. Modification also allowed the enzyme to remain highly active at 115 °C with a 10-fold activity increment compared to the same variant at 50 °C.
Krishnamurthy and coworkers described a modified variant of a fibrinolytic enzyme from Serratia marcescens [143]. Carboxylate modifications proceed through a simple labeling with different amounts of 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) at pH 4.1. Treatment with 2.5 mM EDAC resulted in a modified enzyme with a 19-fold increase in specific activity. A pointed 500-fold reduction in Km suggest improved affinity for the substrate (fibrin) and resulted in a near 220-fold increase in catalytic efficiency.
6. Perspectives
The examples discussed above highlight the potential of enzyme chemical modifications as versatile tools for the development of biocatalysts displaying enhanced and novel functionalities (Table 2). Despite significant advances in the development of biocompatible reactions, selectivity remains a major issue to be addressed in the coming years. This last session will provide a brief outlook of what are possible perspectives for each target residues discussed so far.

Table 2.
Recent examples of novel biocatalysts created by chemical modification of protein scaffolds.
For reactions targeting cysteines, chemoselectivity is being addressed with an ever-growing panel of cysteine selective labeling reactions that benefits from advances in chemical proteomics, bioconjugates and other biomaterials. Site-selectivity, however, has mainly been achieved through site directed mutagenesis, by including new cysteine residues or removing existing ones to avoid cross-reactivity. To our understanding a major challenge in this context is identifying the most reactive cysteine residues and developing methods for selectively modification of multiple cysteine-containing proteins. In the last decade, proteomic techniques as isoTOP-ABPP (isotopic tandem orthogonal proteolysis-activity-based protein profiling) have been applied to identify reactive cysteines in human proteomes [47,53,144,145]. The resulting databases fueled the development of structure and sequence-based computational methods for determining cysteine pKa, a major indicator of its reactivity [146,147]. Other physics-based computational methods also have provided new tools to investigate the pKa of cysteine thiols in proteins [148]. Therefore, further developments in the ability to predict cysteine reactivity will be detrimental to the field of enzyme modification. Altogether it could provide valuable information on where to properly place or remove cysteine residues and how to target a single residue when multiple copies are available. Also, it can be applied to the search for new protein scaffolds in less known proteomes.
Regarding lysines, available chemistry offers plenty of options for chemoselective probes, applicable when site-selectivity is not required. However, similar to cysteines, the major challenge regarding lysine modification stands in the grounds of site-selectivity. A challenge that is amplified by the residue ubiquity. As mentioned before, new strategies have been recently described for lysine site-selective targeting. These developments could arguably be extended to the scope of enzyme functionalization. In this context, the research group from Vishal Ray has provided inventive and valuable tools to site-specific protein modification [149]. Beside their previously referenced reviews on bioconjugation strategies [27,30,32] and their linchpin directed modification [83], the group has developed different aldehyde-based two-step methods for site-selective modification of lysines. These included approaches as aldehyde auto-oxidation [150], a copper assisted amine, aldehyde, alkyne coupling [151] and a metal-free phospho-Mannich reaction [152]. All three methods were successfully applied to multiple protein scaffolds, including three enzymes (RNase A, lysozyme and chymotrypsinogen A). Most importantly, RNase A and Lysozyme remained fully active after labeling and thus, represent a future prospect for the application of these approaches to engineering other enzymes. Likewise, the also previously mentioned report by Matos and co-workers on the computer assisted design a sulfonyl acrylate reagent provides exciting developments to the topic [84]. Moreover, these results corroborate the combination of chemical and computational tools for envisioning novel ways to harness microenvironment tuned reactivity of functional groups. If extended to enzyme modification, those concepts represent valuable tools for de development of rationally designed biocatalyst with a diverse panel of applications.
The N-terminal primary amine, when accessible, is inherently a target for site-selective reactions. One of the issues concerning it, however, is developing chemoselective probes that can discriminate between the α-amino group and other nucleophiles, namely the other primary amines. As reviewed by De Rosa et al. [85], several chemical strategies are already available for bioconjugation at the N-terminus. Thus, the remaining challenge is to expand the application of theses selective probes to the modifications of enzymes. In this direction, previously discussed examples provide good evidence that the N-terminus is a well-suited target for the design of novel biocatalysts and holds potential for future enzyme functionalization strategies.
Finally, among the residues explored in this article, carboxylates are the ones to offer a greater stage to innovation. Somehow, they resemble lysines, as their high frequency and presence in the protein surface make them an easily accessible target. On the other hand, the low reactivity of carboxylic acids represents both a challenge and an opportunity for the development of novel chemoselective probes. Previously mentioned application of carbodiimide chemistry demonstrate the versatility of carboxylates in enzyme functionalization. Therefore, expanding beyond this class of reagents could bring a new set of options for the enzyme modification toolbox. The possibility of site-selective reactions has been demonstrated for the C-terminal residue in peptides and proteins [97,153]. Being able to harness the reactivity of the C-terminus could provide an important handle for site-selective functionalization.
7. Conclusions
In this review we discussed chemical aspects and the challenges regarding selective enzyme modification strategies and their applications. Examples of novel semi-synthetic biocatalysts are manyfold and strongly validate the pursuit for improvement and new developments. Within this framework, selective labeling, and protein bioconjugation have provided an ever-growing panel of chemo and site-selective reagents. Therefore, along with the development of chemical probes, the next steps would be further extending these new chemical tools to enzyme functionalization approaches.
Author Contributions
Conceptualization, J.B.B., T.B.P. and H.T.; methodology, T.B.P.; writing—original draft preparation, T.B.P.; writing—review and editing, H.T. and J.B.B.; project administration, J.B.B.; funding acquisition, J.B.B. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledged financial support by the Wilhelm-Roux program, Martin-Luther University of Halle-Wittenberg (grant FKZ 31/09 to J.B.B.), CAPES (fellowship to T.B.P.), CNPq (grants to H.T.) the Open access publishing program from the German Research Foundation (DFG).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no financial interests or personal relationships that could have appeared to influence the work reported in this review paper.
References
- Truppo, M.D. Biocatalysis in the Pharmaceutical Industry: The Need for Speed. ACS Med. Chem. Lett. 2017, 8, 476–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clouthier, C.M.; Pelletier, J.N. Expanding the Organic Toolbox: A Guide to Integrating Biocatalysis in Synthesis. Chem. Soc. Rev. 2012, 41, 1585–1605. [Google Scholar] [CrossRef]
- Bell, E.L.; Finnigan, W.; France, S.P.; Green, A.P.; Hayes, M.A.; Hepworth, L.J.; Lovelock, S.L.; Niikura, H.; Osuna, S.; Romero, E.; et al. Biocatalysis. Nat. Rev. Methods Primers 2021, 1, 46. [Google Scholar] [CrossRef]
- Pyser, J.B.; Chakrabarty, S.; Romero, E.O.; Narayan, A.R.H. State-of-the-Art Biocatalysis. ACS Cent. Sci. 2021, 7, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-M.; Han, S.-S.; Kim, H.-S. Industrial Applications of Enzyme Biocatalysis: Current Status and Future Aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef]
- Sandoval, B.A.; Hyster, T.K. Emerging Strategies for Expanding the Toolbox of Enzymes in Biocatalysis. Curr. Opin. Chem. Biol. 2020, 55, 45–51. [Google Scholar] [CrossRef]
- Woodley, J.M. New Frontiers in Biocatalysis for Sustainable Synthesis. Curr. Opin. Green Sustain. Chem. 2020, 21, 22–26. [Google Scholar] [CrossRef]
- Davis, B.G. Chemical Modification of Biocatalysts. Curr. Opin. Biotechnol. 2003, 14, 379–386. [Google Scholar] [CrossRef]
- DeSantis, G.; Jones, J.B. Chemical Modification of Enzymes for Enhanced Functionality. Curr. Opin. Biotechnol. 1999, 10, 324–330. [Google Scholar] [CrossRef]
- Díaz-Rodríguez, A.; Davis, B.G. Chemical Modification in the Creation of Novel Biocatalysts. Curr. Opin. Chem. Biol. 2011, 15, 211–219. [Google Scholar] [CrossRef]
- Kaushik, M.; Sinha, P.; Jaiswal, P.; Mahendru, S.; Roy, K.; Kukreti, S. Protein Engineering and de Novo Designing of a Biocatalyst. J. Mol. Recognit. 2016, 29, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Hamachi, I. Recent Progress in Chemical Modification of Proteins. Anal. Sci. 2019, 35, 5–27. [Google Scholar] [CrossRef] [Green Version]
- Pagar, A.D.; Patil, M.D.; Flood, D.T.; Yoo, T.H.; Dawson, P.E.; Yun, H. Recent Advances in Biocatalysis with Chemical Modification and Expanded Amino Acid Alphabet. Chem. Rev. 2021, 121, 6173–6245. [Google Scholar] [CrossRef]
- Neet, K.E.; Koshland, D.E. The Conversion of Serine at the Active Site of Subtilisin to Cysteine: A “Chemical Mutation”. Proc. Natl. Acad. Sci. USA 1966, 56, 1606–1611. [Google Scholar] [CrossRef] [Green Version]
- Polgar, L.; Bender, M.L. A New Enzyme Containing a Synthetically Formed Active Site. Thiol-Subtilisin. J. Am. Chem. Soc. 1966, 88, 3153–3154. [Google Scholar] [CrossRef]
- Yokosawa, H.; Ojima, S.; Ishii, S. Thioltrypsin: Chemical Transformation of the Active-Site Serine Residue of Streptomyces griseus Trypsin to a Cysteine Residue. J. Biochem. 1977, 82, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.I.; Lowe, G. Chemical Mutations of Papain. The Preparation of Ser 25- and Gly 25-Papain. J. Chem. Soc. Chem. Commun. 1977, 24, 923–924. [Google Scholar] [CrossRef]
- Kaiser, E.T. Catalytic Activity of Enzymes Altered at Their Active Sites. Angew. Chem. Int. Ed. Engl. 1988, 27, 913–922. [Google Scholar] [CrossRef]
- Kaiser, E.T.; Lawrence, D.S. Chemical Mutation of Enzyme Active Sites. Science 1984, 226, 505–511. [Google Scholar] [CrossRef]
- Kokubo, T.; Sassa, S.; Kaiser, E.T. Flavohemoglobin: A Semisynthetic Hydroxylase Acting in the Absence of Reductase. J. Am. Chem. Soc. 1987, 109, 606–607. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Sasaki, T.; Kaiser, E.T. Peptide Segment Synthesis Catalyzed by the Semisynthetic Enzyme Thiolsubtilisin. J. Am. Chem. Soc. 1987, 109, 3808–3810. [Google Scholar] [CrossRef]
- Shen, W.C.; Ryser, H.J. Conjugation of Poly-L-Lysine to Albumin and Horseradish Peroxidase: A Novel Method of Enhancing the Cellular Uptake of Proteins. Proc. Natl. Acad. Sci. USA 1978, 75, 1872–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-H.; Sigman, D.S. Chemical Conversion of a DNA-Binding Protein into a Site-Specific Nuclease. Science 1987, 237, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Inada, Y.; Takahashi, K.; Yoshimoto, T.; Ajima, A.; Matsushima, A.; Saito, Y. Application of Polyethylene Glycol-Modified Enzymes in Biotechnological Processes: Organic Solvent-Soluble Enzymes. Trends Biotechnol. 1986, 4, 190–194. [Google Scholar] [CrossRef]
- Dadová, J.; Galan, S.R.; Davis, B.G. Synthesis of Modified Proteins via Functionalization of Dehydroalanine. Curr. Opin. Chem. Biol. 2018, 46, 71–81. [Google Scholar] [CrossRef]
- Rawale, D.G.; Thakur, K.; Adusumalli, S.R.; Rai, V. Chemical Methods for Selective Labeling of Proteins. Eur. J. Org. Chem. 2019, 2019, 6749–6763. [Google Scholar] [CrossRef]
- Shadish, J.A.; DeForest, C.A. Site-Selective Protein Modification: From Functionalized Proteins to Functional Biomaterials. Matter 2020, 2, 50–77. [Google Scholar] [CrossRef]
- Spicer, C.D.; Davis, B.G. Selective Chemical Protein Modification. Nat. Commun. 2014, 5, 4740. [Google Scholar] [CrossRef] [Green Version]
- Reddy, N.C.; Kumar, M.; Molla, R.; Rai, V. Chemical Methods for Modification of Proteins. Org. Biomol. Chem. 2020, 18, 4669–4691. [Google Scholar] [CrossRef]
- Tamura, T.; Hamachi, I. Chemistry for Covalent Modification of Endogenous/Native Proteins: From Test Tubes to Complex Biological Systems. J. Am. Chem. Soc. 2019, 141, 2782–2799. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Reddy, N.C.; Rai, V. Chemical Technologies for Precise Protein Bioconjugation Interfacing Biology and Medicine. Chem. Commun. 2021, 57, 7083–7095. [Google Scholar] [CrossRef]
- Shannon, D.A.; Weerapana, E. Covalent Protein Modification: The Current Landscape of Residue-Specific Electrophiles. Curr. Opin. Chem. Biol. 2015, 24, 18–26. [Google Scholar] [CrossRef]
- Koniev, O.; Wagner, A. Developments and Recent Advancements in the Field of Endogenous Amino Acid Selective Bond Forming Reactions for Bioconjugation. Chem. Soc. Rev. 2015, 44, 5495–5551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, T.K.; Turner, G.J. Structural Basis of Perturbed PKa Values of Catalytic Groups in Enzyme Active Sites. IUBMB Life 2002, 53, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Pace, N.J.; Weerapana, E. Diverse Functional Roles of Reactive Cysteines. ACS Chem. Biol. 2013, 8, 283–296. [Google Scholar] [CrossRef]
- Bischoff, R.; Schlüter, H. Amino Acids: Chemistry, Functionality and Selected Non-Enzymatic Post-Translational Modifications. J. Proteom. 2012, 75, 2275–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bak, D.W.; Bechtel, T.J.; Falco, J.A.; Weerapana, E. Cysteine Reactivity across the Subcellular Universe. Curr Opin. Chem. Biol. 2019, 48, 96–105. [Google Scholar] [CrossRef]
- Go, Y.M.; Chandler, J.D.; Jones, D.P. The Cysteine Proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef] [PubMed]
- Miseta, A.; Csutora, P. Relationship Between the Occurrence of Cysteine in Proteins and the Complexity of Organisms. Mol. Biol. Evol. 2000, 17, 1232–1239. [Google Scholar] [CrossRef] [Green Version]
- Wiedemann, C.; Kumar, A.; Lang, A.; Ohlenschläger, O. Cysteines and Disulfide Bonds as Structure-Forming Units: Insights From Different Domains of Life and the Potential for Characterization by NMR. Front. Chem. 2020, 8, 280. [Google Scholar] [CrossRef]
- Marino, S.M.; Gladyshev, V.N. Cysteine Function Governs Its Conservation and Degeneration and Restricts Its Utilization on Protein Surfaces. J. Mol. Biol. 2010, 404, 902–916. [Google Scholar] [CrossRef] [Green Version]
- Wible, R.S.; Sutter, T.R. Soft Cysteine Signaling Network: The Functional Significance of Cysteine in Protein Function and the Soft Acids/Bases Thiol Chemistry That Facilitates Cysteine Modification. Chem. Res. Toxicol. 2017, 30, 729–762. [Google Scholar] [CrossRef]
- Bulaj, G.; Kortemme, T.; Goldenberg, D.P. Ionization-Reactivity Relationships for Cysteine Thiols in Polypeptides. Biochemistry 1998, 37, 8965–8972. [Google Scholar] [CrossRef] [PubMed]
- LoPachin, R.M.; Gavin, T. Reactions of Electrophiles with Nucleophilic Thiolate Sites: Relevance to Pathophysiological Mechanisms and Remediation. Free Radic. Res. 2016, 50, 195–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backus, K.M.; Correia, B.E.; Lum, K.M.; Forli, S.; Horning, B.D.; González-Páez, G.E.; Chatterjee, S.; Lanning, B.R.; Teijaro, J.R.; Olson, A.J.; et al. Proteome-Wide Covalent Ligand Discovery in Native Biological Systems. Nature 2016, 534, 570–574. [Google Scholar] [CrossRef] [Green Version]
- Boutureira, O.; Bernardes, G.J.L. Advances in Chemical Protein Modification. Chem. Rev. 2015, 115, 2174–2195. [Google Scholar] [CrossRef] [PubMed]
- Baillie, T.A. Targeted Covalent Inhibitors for Drug Design. Angew. Chem. Int. Ed. 2016, 55, 13408–13421. [Google Scholar] [CrossRef]
- Hoch, D.G.; Abegg, D.; Adibekian, A. Cysteine-Reactive Probes and Their Use in Chemical Proteomics. Chem. Commun. 2018, 54, 4501–4512. [Google Scholar] [CrossRef]
- Hoyt, E.A.; Cal, P.M.S.D.; Oliveira, B.L.; Bernardes, G.J.L. Contemporary Approaches to Site-Selective Protein Modification. Nat. Rev. Chem. 2019, 3, 147–171. [Google Scholar] [CrossRef] [Green Version]
- Litwin, K.; Crowley, V.M.; Suciu, R.M.; Boger, D.L.; Cravatt, B.F. Chemical Proteomic Identification of Functional Cysteines with Atypical Electrophile Reactivities. Tetrahedron Lett. 2021, 67, 152861. [Google Scholar] [CrossRef] [PubMed]
- Maurais, A.J.; Weerapana, E. Reactive-Cysteine Profiling for Drug Discovery. Curr. Opin. Chem. Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef]
- Ochtrop, P.; Hackenberger, C.P.R. Recent Advances of Thiol-Selective Bioconjugation Reactions. Curr. Opin. Chem. Biol. 2020, 58, 28–36. [Google Scholar] [CrossRef]
- Lahnsteiner, M.; Kastner, A.; Mayr, J.; Roller, A.; Keppler, B.K.; Kowol, C.R. Improving the Stability of Maleimide–Thiol Conjugation for Drug Targeting. Chem. Eur. J. 2020, 26, 15867–15870. [Google Scholar] [CrossRef] [PubMed]
- Renault, K.; Fredy, J.W.; Renard, P.Y.; Sabot, C. Covalent Modification of Biomolecules through Maleimide-Based Labeling Strategies. Bioconjugate Chem. 2018, 29, 2497–2513. [Google Scholar] [CrossRef]
- Jackson, P.A.; Widen, J.C.; Harki, D.A.; Brummond, K.M. Covalent Modifiers: A Chemical Perspective on the Reactivity of α,β-Unsaturated Carbonyls with Thiols via Hetero-Michael Addition Reactions. J. Med. Chem. 2017, 60, 839–885. [Google Scholar] [CrossRef] [PubMed]
- Boutureira, O.; Martínez-Sáez, N.; Brindle, K.M.; Neves, A.A.; Corzana, F.; Bernardes, G.J.L. Site-Selective Modification of Proteins with Oxetanes. Chem. Eur. J. 2017, 23, 6483–6489. [Google Scholar] [CrossRef] [Green Version]
- Matos, M.J.; Navo, C.D.; Hakala, T.; Ferhati, X.; Guerreiro, A.; Hartmann, D.; Bernardim, B.; Saar, K.L.; Compañón, I.; Corzana, F.; et al. Quaternization of Vinyl/Alkynyl Pyridine Enables Ultrafast Cysteine-Selective Protein Modification and Charge Modulation. Angew. Chem. Int. Ed. 2019, 58, 6640–6644. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Tao, Y.; Sheng, W.; Lu, J.; Wang, H. Dinitroimidazoles as Bifunctional Bioconjugation Reagents for Protein Functionalization and Peptide Macrocyclization. Nat. Commun. 2019, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.R.; Chung, S.F.; Leung, A.S.L.; Yip, W.M.; Yang, B.; Choi, M.C.; Cui, J.F.; Kung, K.K.Y.; Zhang, Z.; Lo, K.W.; et al. Chemoselective and Photocleavable Cysteine Modification of Peptides and Proteins Using Isoxazoliniums. Commun. Chem. 2019, 2, 93. [Google Scholar] [CrossRef]
- Smith, N.J.; Rohlfing, K.; Sawicki, L.A.; Kharkar, P.M.; Boyd, S.J.; Kloxin, A.M.; Fox, J.M. Fast, Irreversible Modification of Cysteines through Strain Releasing Conjugate Additions of Cyclopropenyl Ketones. Org. Biomol. Chem. 2018, 16, 2164–2169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zang, C.; An, G.; Shang, M.; Cui, Z.; Chen, G.; Xi, Z.; Zhou, C. Cysteine-Specific Protein Multi-Functionalization and Disulfide Bridging Using 3-Bromo-5-Methylene Pyrrolones. Nat. Commun. 2020, 11, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasper, M.; Stengl, A.; Ochtrop, P.; Gerlach, M.; Stoschek, T.; Schumacher, D.; Helma, J.; Penkert, M.; Krause, E.; Leonhardt, H.; et al. Ethynylphosphonamidates for the Rapid and Cysteine-Selective Generation of Efficacious Antibody–Drug Conjugates. Angew. Chem. Int. Ed. 2019, 58, 11631–11636. [Google Scholar] [CrossRef] [Green Version]
- Kasper, M.; Glanz, M.; Stengl, A.; Penkert, M.; Klenk, S.; Sauer, T.; Schumacher, D.; Helma, J.; Krause, E.; Cardoso, M.C.; et al. Cysteine-Selective Phosphonamidate Electrophiles for Modular Protein Bioconjugations. Angew. Chem. Int. Ed. 2019, 58, 11625–11630. [Google Scholar] [CrossRef]
- Baumann, A.L.; Schwagerus, S.; Broi, K.; Kemnitz-Hassanin, K.; Stieger, C.E.; Trieloff, N.; Schmieder, P.; Hackenberger, C.P.R. Chemically Induced Vinylphosphonothiolate Electrophiles for Thiol-Thiol Bioconjugations. J. Am. Chem. Soc. 2020, 142, 9544–9552. [Google Scholar] [CrossRef]
- Luo, M. Chemical and Biochemical Perspectives of Protein Lysine Methylation. Chem. Rev. 2018, 118, 6656–6705. [Google Scholar] [CrossRef]
- Gallivan, J.P.; Dougherty, D.A. A Computational Study of Cation-π Interactions vs Salt Bridges in Aqueous Media: Implications for Protein Engineering. J. Am. Chem. Soc. 2000, 122, 870–874. [Google Scholar] [CrossRef]
- Larda, S.T.; Pichugin, D.; Prosser, R.S. Site-Specific Labeling of Protein Lysine Residues and N-Terminal Amino Groups with Indoles and Indole-Derivatives. Bioconjugate Chem. 2015, 26, 2376–2383. [Google Scholar] [CrossRef]
- Al Temimi, A.H.K.; Amatdjais-Groenen, H.I.V.; Reddy, Y.V.; Blaauw, R.H.; Guo, H.; Qian, P.; Mecinović, J. The Nucleophilic Amino Group of Lysine Is Central for Histone Lysine Methyltransferase Catalysis. Commun. Chem. 2019, 2, 112. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.A.; Cole, P.A. The Chemical Biology of Reversible Lysine Post-Translational Modifications. Cell Chem. Biol. 2020, 27, 953–969. [Google Scholar] [CrossRef]
- Kozlowski, L.P. Proteome-pI: Proteome Isolectric Point Database. Nucleic Acid Res. 2016, 45, D1112–D1116. [Google Scholar] [CrossRef]
- Geoghegan, K.F. Modification of Amino Groups. Curr. Protoc. Protein Sci. 2016, 86, 15.2.1–15.2.20. [Google Scholar] [CrossRef]
- Pace, C.N.; Grimsley, G.R.; Scholtz, J.M. Protein Ionizable Groups: PK Values and Their Contribution to Protein Stability and Solubility. J. Biol. Chem. 2009, 284, 13285–13289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pahari, S.; Sun, L.; Alexov, E. PKAD: A Database of Experimentally Measured PKa Values of Ionizable Groups in Proteins. Database 2019, 2019, baz024. [Google Scholar] [CrossRef] [Green Version]
- Platzer, G.; Okon, M.; McIntosh, L.P. PH-Dependent Random Coil 1H, 13C, and 15N Chemical Shifts of the Ionizable Amino Acids: A Guide for Protein PK a Measurements. J. Biomol. NMR 2014, 60, 109–129. [Google Scholar] [CrossRef]
- Pettinger, J.; Jones, K.; Cheeseman, M.D. Lysine-Targeting Covalent Inhibitors. Angew. Chem. Int. Ed. 2017, 56, 15200–15209. [Google Scholar] [CrossRef]
- Adamo, R.; Nilo, A.; Castagner, B.; Boutureira, O.; Berti, F.; Bernardes, G.J.L. Synthetically Defined Glycoprotein Vaccines: Current Status and Future Directions. Chem. Sci. 2013, 4, 2995–3008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banks, P.R.; Paquette, D.M. Comparison of Three Common Amine Reactive Fluorescent Probes Used for Conjugation to Biomolecules by Capillary Zone Electrophoresis. Bioconjugate Chem. 1995, 6, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K.M.; Page, R.C.; Konkolewicz, D. Bioconjugates—From a Specialized Past to a Diverse Future. Polymer 2020, 211, 123062. [Google Scholar] [CrossRef]
- Naowarojna, N.; Cheng, R.; Lopez, J.; Wong, C.; Qiao, L.; Liu, P. Chemical Modifications of Proteins and Their Applications in Metalloenzyme Studies. Synth. Syst. Biothecnol. 2021, 6, 32–49. [Google Scholar] [CrossRef]
- Chen, X.; Muthoosamy, K.; Pfisterer, A.; Neumann, B.; Weil, T. Site-Selective Lysine Modification of Native Proteins and Peptides via Kinetically Controlled Labeling. Bioconjugate Chem. 2012, 23, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Adusumalli, S.R.; Rawale, D.G.; Thakur, K.; Purushottam, L.; Reddy, N.C.; Kalra, N.; Shukla, S.; Rai, V. Chemoselective and Site-Selective Lysine-Directed Lysine Modification Enables Single-Site Labeling of Native Proteins. Angew. Chem. Int. Ed. 2020, 59, 10332–10336. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.J.; Oliveira, B.L.; Martínez-Sáez, N.; Guerreiro, A.; Cal, P.M.S.D.; Bertoldo, J.; Maneiro, M.; Perkins, E.; Howard, J.; Deery, M.J.; et al. Chemo- and Regioselective Lysine Modification on Native Proteins. J. Am. Chem. Soc. 2018, 140, 4004–4017. [Google Scholar] [CrossRef]
- De Rosa, L.; Di Stasi, R.; Romanelli, A.; D’Andrea, L.D. Exploiting Protein N-Terminus for Site-Specific Bioconjugation. Molecules 2021, 26, 3521. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Disotuar, M.M.; Xiong, X.; Wang, Y.; Chou, D.H.C. Selective N-Terminal Functionalization of Native Peptides and Proteins. Chem. Sci. 2017, 8, 2717–2722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, C.B.; Francis, M.B. Targeting the N Terminus for Site-Selective Protein Modification. Nat. Chem. Biol. 2017, 13, 697–705. [Google Scholar] [CrossRef]
- Schoffelen, S.; van Eldijk, M.B.; Rooijakkers, B.; Raijmakers, R.; Heck, A.J.R.; van Hest, J.C.M. Metal-Free and PH-Controlled Introduction of Azides in Proteins. Chem. Sci. 2011, 2, 701–705. [Google Scholar] [CrossRef]
- Chan, A.O.Y.; Ho, C.M.; Chong, H.C.; Leung, Y.C.; Huang, J.S.; Wong, M.K.; Che, C.M. Modification of N-Terminal α-Amino Groups of Peptides and Proteins Using Ketenes. J. Am. Chem. Soc. 2012, 134, 2589–2598. [Google Scholar] [CrossRef]
- Macdonald, J.I.; Munch, H.K.; Moore, T.; Francis, M.B. One-Step Site-Specific Modification of Native Proteins with 2-Pyridinecarboxyaldehydes. Nat. Chem. Biol. 2015, 11, 326–331. [Google Scholar] [CrossRef]
- Deng, J.R.; Lai, N.C.H.; Kung, K.K.Y.; Yang, B.; Chung, S.F.; Leung, A.S.L.; Choi, M.C.; Leung, Y.C.; Wong, M.K. N-Terminal Selective Modification of Peptides and Proteins Using 2-Ethynylbenzaldehydes. Commun. Chem. 2020, 3, 67. [Google Scholar] [CrossRef]
- McGrath, N.A.; Andersen, K.A.; Davis, A.K.F.; Lomax, J.E.; Raines, R.T. Diazo Compounds for the Bioreversible Esterification of Proteins. Chem. Sci. 2015, 6, 752–755. [Google Scholar] [CrossRef] [Green Version]
- Mix, K.A.; Raines, R.T. Optimized Diazo Scaffold for Protein Esterification. Org. Lett. 2015, 17, 2358–2361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.H.; Tan, D.; Li, Q.; Li, M.; Gong, Z.; Tang, C.; Liu, Z.; Dong, M.Q.; Lei, X. Carboxylate-Selective Chemical Cross-Linkers for Mass Spectrometric Analysis of Protein Structures. Anal. Chem. 2018, 90, 1195–1201. [Google Scholar] [CrossRef]
- Ma, N.; Hu, J.; Zhang, Z.M.; Liu, W.; Huang, M.; Fan, Y.; Yin, X.; Wang, J.; Ding, K.; Ye, W.; et al. 2 H-Azirine-Based Reagents for Chemoselective Bioconjugation at Carboxyl Residues Inside Live Cells. J. Am. Chem. Soc. 2020, 142, 6051–6059. [Google Scholar] [CrossRef]
- Rawale, D.G.; Thakur, K.; Sreekumar, P.; Sajeev, T.K.; Ramesh, A.; Adusumalli, S.R.; Mishra, R.K.; Rai, V. Linchpins Empower Promiscuous Electrophiles to Enable Site-Selective Modification of Histidine and Aspartic Acid in Proteins. Chem. Sci. 2021, 12, 6732–6736. [Google Scholar] [CrossRef] [PubMed]
- Bloom, S.; Liu, C.; Kölmel, D.K.; Qiao, J.X.; Zhang, Y.; Poss, M.A.; Ewing, W.R.; Macmillan, D.W.C. Decarboxylative Alkylation for Site-Selective Bioconjugation of Native Proteins via Oxidation Potentials. Nat. Chem. 2018, 10, 205–211. [Google Scholar] [CrossRef] [PubMed]
- López-Gallego, F.; Abian, O.; Guisán, J.M. Altering the Interfacial Activation Mechanism of a Lipase by Solid-Phase Selective Chemical Modification. Biochemistry 2012, 51, 7028–7036. [Google Scholar] [CrossRef]
- Ema, T.; Inoue, H. Chemical Modification of Lipase for Rational Enhancement of Enantioselectivity. Chem. Lett. 2015, 44, 1374–1376. [Google Scholar] [CrossRef] [Green Version]
- Darby, J.F.; Atobe, M.; Firth, J.D.; Bond, P.; Davies, G.J.; O’Brien, P.; Hubbard, R.E. Increase of Enzyme Activity through Specific Covalent Modification with Fragments. Chem. Sci. 2017, 8, 7772–7779. [Google Scholar] [CrossRef] [Green Version]
- Darby, J.F.; Landström, J.; Roth, C.; He, Y.; Davies, G.J.; Hubbard, R.E. Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase. Angew. Chem. Int. Ed. 2014, 53, 13419–13423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, W.J.; Yu, J.; Song, W.J. Proteins as Diverse, Efficient, and Evolvable Scaffolds for Artificial Metalloenzymes. Chem. Commun. 2020, 56, 9586–9599. [Google Scholar] [CrossRef]
- Doble, M.V.; Jarvis, A.G.; Ward, A.C.C.; Colburn, J.D.; Götze, J.P.; Bühl, M.; Kamer, P.C.J. Artificial Metalloenzymes as Catalysts for Oxidative Lignin Degradation. ACS Sustain. Chem. Eng. 2018, 6, 15100–15107. [Google Scholar] [CrossRef] [Green Version]
- Sauer, D.F.; Himiyama, T.; Tachikawa, K.; Fukumoto, K.; Onoda, A.; Mizohata, E.; Inoue, T.; Bocola, M.; Schwaneberg, U.; Hayashi, T.; et al. A Highly Active Biohybrid Catalyst for Olefin Metathesis in Water: Impact of a Hydrophobic Cavity in a β-Barrel Protein. ACS Catal. 2015, 5, 7519–7522. [Google Scholar] [CrossRef]
- Bernardes, G.J.L.; Chalker, J.M.; Errey, J.C.; Davis, B.G. Facile Conversion of Cysteine and Alkyl Cysteines to Dehydroalanine on Protein Surfaces: Versatile and Switchable Access to Functionalized Proteins. J. Am. Chem. Soc. 2008, 130, 5052–5053. [Google Scholar] [CrossRef] [PubMed]
- Chalker, J.M.; Bernardes, G.J.L.; Lin, Y.A.; Davis, B.G. Chemical Modification of Proteins at Cysteine: Opportunities in Chemistry and Biology. Chem. Asian J. 2009, 4, 630–640. [Google Scholar] [CrossRef]
- Chalker, J.M.; Bernardes, G.J.L.; Davis, B.G. A “Tag-and-Modify” Approach to Site-Selective Protein Modification. Acc. Chem. Res. 2011, 44, 730–741. [Google Scholar] [CrossRef]
- Chalker, J.M.; Gunnoo, S.B.; Boutureira, O.; Gerstberger, S.C.; Fernández-González, M.; Bernardes, G.J.L.; Griffin, L.; Hailu, H.; Schofield, C.J.; Davis, B.G. Methods for Converting Cysteine to Dehydroalanine on Peptides and Proteins. Chem. Sci. 2011, 2, 1666–1676. [Google Scholar] [CrossRef]
- Jones, L.H. Dehydroamino Acid Chemical Biology: An Example of Functional Group Interconversion on Proteins. RSC Chem. Biol. 2020, 1, 298–304. [Google Scholar] [CrossRef]
- Dadová, J.; Wu, K.J.; Isenegger, P.G.; Errey, J.C.; Bernardes, G.J.L.; Chalker, J.M.; Raich, L.; Rovira, C.; Davis, B.G. Precise Probing of Residue Roles by Post-Translational β,γ-C,N Aza-Michael Mutagenesis in Enzyme Active Sites. ACS Cent. Sci. 2017, 3, 1168–1173. [Google Scholar] [CrossRef]
- Chalker, J.M.; Lercher, L.; Rose, N.R.; Schofield, C.J.; Davis, B.G. Conversion of Cysteine into Dehydroalanine Enables Access to Synthetic Histones Bearing Diverse Post-Translational Modifications. Angew. Chem. Int. Ed. 2012, 51, 1835–1839. [Google Scholar] [CrossRef] [PubMed]
- Chooi, K.P.; Galan, S.R.G.; Raj, R.; McCullagh, J.; Mohammed, S.; Jones, L.H.; Davis, B.G. Synthetic Phosphorylation of P38α Recapitulates Protein Kinase Activity. J. Am. Chem. Soc. 2014, 136, 1698–1701. [Google Scholar] [CrossRef]
- Freedy, A.M.; Matos, M.J.; Boutureira, O.; Corzana, F.; Guerreiro, A.; Akkapeddi, P.; Somovilla, V.J.; Rodrigues, T.; Nicholls, K.; Xie, B.; et al. Chemoselective Installation of Amine Bonds on Proteins Through Aza-Michael Ligation. J. Am. Chem. Soc. 2017, 139, 18365–18375. [Google Scholar] [CrossRef] [Green Version]
- Wright, T.H.; Vallée, M.R.J.; Davis, B.G. From Chemical Mutagenesis to Post-Expression Mutagenesis: A 50 Year Odyssey. Angew. Chem. Int. Ed. 2016, 55, 5896–5903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.; Ha, S.; Ahn, J.; Kim, R.; Kim, S.; Lee, Y.; Kim, J.; Söll, D.; Lee, H.-Y.; Park, H.-S. A Chemical Biology Route to Site-Specific Authentic Protein Modifications. Science 2016, 354, 623–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timms, N.; Windle, C.L.; Polyakova, A.; Ault, J.R.; Trinh, C.H.; Pearson, A.R.; Nelson, A.; Berry, A. Structural Insights into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with γ-Thialysine by Using a Chemical Mutagenesis Strategy. ChemBioChem 2013, 14, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Windle, C.L.; Simmons, K.J.; Ault, J.R.; Trinh, C.H.; Nelson, A.; Pearson, A.R.; Berry, A. Extending Enzyme Molecular Recognition with an Expanded Amino Acid Alphabet. Proc. Natl. Acad. Sci. USA 2017, 114, 2610–2615. [Google Scholar] [CrossRef] [Green Version]
- Shaw, B.F.; Schneider, G.F.; Bilgiçer, B.; Kaufman, G.K.; Neveu, J.M.; Lane, W.S.; Whitelegge, J.P.; Whitesides, G.M. Lysine Acetylation Can Generate Highly Charged Enzymes with Increased Resistance toward Irreversible Inactivation. Protein Sci. 2008, 17, 1446–1455. [Google Scholar] [CrossRef] [Green Version]
- Ismaya, W.T.; Hasan, K.; Kardi, I.; Zainuri, A.; Rahmawaty, R.I.; Permanahadi, S.; el Viera, B.V.; Harinanto, G.; Gaffar, S.; Natalia, D.; et al. Chemical Modification of Saccharomycopsis Fibuligera R64 α-Amylase to Improve Its Stability against Thermal, Chelator, and Proteolytic Inactivation. Appl. Biochem. Biotechnol. 2013, 170, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Nwagu, T.N.; Okolo, B.; Aoyagi, H.; Yoshida, S. Chemical Modification with Phthalic Anhydride and Chitosan: Viable Options for the Stabilization of Raw Starch Digesting Amylase from Aspergillus Carbonarius. Int. J. Biol. Macromol. 2017, 99, 641–647. [Google Scholar] [CrossRef]
- Nwagu, T.N.; Aoyagi, H.; Okolo, B.; Moneke, A.; Yoshida, S. Citraconylation and Maleylation on the Catalytic and Thermodynamic Properties of Raw Starch Saccharifying Amylase from Aspergillus Carbonarius. Heliyon 2020, 6, e04351. [Google Scholar] [CrossRef]
- Xue, Y.; Wu, C.Y.; Branford-White, C.J.; Ning, X.; Nie, H.L.; Zhu, L.M. Chemical Modification of Stem Bromelain with Anhydride Groups to Enhance Its Stability and Catalytic Activity. J. Mol. Catal. B Enzym. 2010, 63, 188–193. [Google Scholar] [CrossRef]
- Matsumoto, M.; Nakagawa, T.; Uchida, Y.; Seki, K.; Ohba, M.; Kondo, K. Effect of Modification of Citraconic Anhydrides on Catalytic Activity and Thermostability of Enzymes. J. Chem. Technol. Biotechnol. 2016, 91, 59–64. [Google Scholar] [CrossRef]
- Hassani, L.; Nourozi, R. Modification of Lysine Residues of Horseradish Peroxidase and Its Effect on Stability and Structure of the Enzyme. Appl. Biochem. Biotechnol. 2014, 172, 3558–3569. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, M.B.; Yee, L.H.; Poljak, A.; Cavicchioli, R.; Kjelleberg, S.J.; Siddiqui, K.S. Enhancement of Lipase Stability and Productivity through Chemical Modification and Its Application to Latex-Based Polymer Emulsions. Process Biochem. 2017, 57, 131–140. [Google Scholar] [CrossRef]
- Solanki, K.; Gupta, M.N. A Chemically Modified Lipase Preparation for Catalyzing the Transesterification Reaction in Even Highly Polar Organic Solvents. Bioorg. Med. Chem. Lett. 2011, 21, 2934–2936. [Google Scholar] [CrossRef]
- Gauthier, M.A.; Klok, H.A. Polymer-Protein Conjugates: An Enzymatic Activity Perspective. Polym. Chem. 2010, 1, 1352–1373. [Google Scholar] [CrossRef]
- Benson, K.R.; Gorecki, J.; Nikiforov, A.; Tsui, W.; Kasi, R.M.; Kumar, C.V. Cytochrome: C-Poly(Acrylic Acid) Conjugates with Improved Peroxidase Turnover Number. Org. Biomol. Chem. 2019, 17, 4043–4048. [Google Scholar] [CrossRef]
- Kovaliov, M.; Allegrezza, M.L.; Richter, B.; Konkolewicz, D.; Averick, S. Synthesis of Lipase Polymer Hybrids with Retained or Enhanced Activity Using the Grafting-from Strategy. Polymer 2018, 137, 338–345. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 177, 7132–7189. [Google Scholar] [CrossRef]
- Shi, R.; Wang, Y. Dual Ionic and Organic Nature of Ionic Liquids. Sci. Rep. 2016, 6, 19644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, R.; Hu, Y.; Liu, L.; Jiang, L.; Zou, B.; Huang, H. Enhancing Catalytic Performance of Porcine Pancreatic Lipase by Covalent Modification Using Functional Ionic Liquids. ACS Catal. 2013, 3, 1976–1983. [Google Scholar] [CrossRef]
- Xu, C.; Suo, H.; Xue, Y.; Qin, J.; Chen, H.; Hu, Y. Experimental and Theoretical Evidence of Enhanced Catalytic Performance of Lipase B from Candida Antarctica Acquired by the Chemical Modification with Amino Acid Ionic Liquids. Mol. Catal. 2021, 501, 111355. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Kamiya, N.; Goto, M. Activation and Stabilization of Enzymes in Ionic Liquids. Org. Biomol. Chem. 2010, 8, 2887–2899. [Google Scholar] [CrossRef]
- Pramanik, S.; Dhoke, G.V.; Jaeger, K.E.; Schwaneberg, U.; Davari, M.D. How to Engineer Ionic Liquids Resistant Enzymes: Insights from Combined Molecular Dynamics and Directed Evolution Study. ACS Sustain. Chem. Eng. 2019, 7, 11293–11302. [Google Scholar] [CrossRef]
- Weingärtner, H.; Cabrele, C.; Herrmann, C. How Ionic Liquids Can Help to Stabilize Native Proteins. Phys. Chem. Chem. Phys. 2012, 14, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Schindl, A.; Hagen, M.L.; Muzammal, S.; Gunasekera, H.A.D.; Croft, A.K. Proteins in Ionic Liquids: Reactions, Applications, and Futures. Front. Chem. 2019, 7, 347. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Liang, S.; Zong, M.H.; Lou, W.Y. Ionic Liquids for Regulating Biocatalytic Process: Achievements and Perspectives. Biotechnol. Adv. 2021, 51, 107702. [Google Scholar] [CrossRef]
- Bekhouche, M.; Blum, L.J.; Doumèche, B. Ionic Liquid-Inspired Cations Covalently Bound to Formate Dehydrogenase Improve its Stability and Activity in Ionic Liquids. ChemCatChem 2011, 3, 875–882. [Google Scholar] [CrossRef]
- Nordwald, E.M.; Brunecky, R.; Himmel, M.E.; Beckham, G.T.; Kaar, J.L. Charge Engineering of Cellulases Improves Ionic Liquid Tolerance and Reduces Lignin Inhibition. Biotechnol. Bioeng. 2014, 111, 1541–1549. [Google Scholar] [CrossRef]
- Perez-Rizquez, C.; Lopez-Tejedor, D.; Plaza-Vinuesa, L.; de las Rivas, B.; Muñoz, R.; Cumella, J.; Palomo, J.M. Chemical Modification of Novel Glycosidases from Lactobacillus Plantarum Using Hyaluronic Acid: Effects on High Specificity against 6-Phosphate Glucopyranoside. Coatings 2019, 9, 311. [Google Scholar] [CrossRef] [Green Version]
- Brogan, A.P.S.; Bui-Le, L.; Hallett, J.P. Non-Aqueous Homogenous Biocatalytic Conversion of Polysaccharides in Ionic Liquids Using Chemically Modified Glucosidase. Nat. Chem. 2018, 10, 859–865. [Google Scholar] [CrossRef]
- Krishnamurthy, A.; Mundra, S.; Belur, P.D. Improving the Catalytic Efficiency of Fibrinolytic Enzyme from Serratia Marcescens Subsp. Sakuensis by Chemical Modification. Process Biochem. 2018, 72, 79–85. [Google Scholar] [CrossRef]
- Wang, C.; Weerapana, E.; Blewett, M.M.; Cravatt, B.F. A Chemoproteomic Platform to Quantitatively Map Targets of Lipid-Derived Electrophiles. Nat. Methods 2014, 11, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Weerapana, E.; Wang, C.; Simon, G.M.; Richter, F.; Khare, S.; Dillon, M.B.D.; Bachovchin, D.A.; Mowen, K.; Baker, D.; Cravatt, B.F. Quantitative Reactivity Profiling Predicts Functional Cysteines in Proteomes. Nature 2010, 468, 790–797. [Google Scholar] [CrossRef] [Green Version]
- Soylu, I.; Marino, S.M. Cy-Preds: An Algorithm and a Web Service for the Analysis and Prediction of Cysteine Reactivity. Proteins 2016, 84, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Li, C.; Liu, Y.; Yang, F.; Wang, C. Sequence-Based Prediction of Cysteine Reactivity Using Machine Learning. Biochemistry 2018, 57, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.C.; Liu, R.; Shen, J. Predicting Reactive Cysteines with Implicit-Solvent-Based Continuous Constant PH Molecular Dynamics in Amber. J. Chem. Theory Comput. 2020, 16, 3689–3698. [Google Scholar] [CrossRef]
- Adakkattil, R.; Thakur, K.; Rai, V. Reactivity and Selectivity Principles in Native Protein Bioconjugation. Chem. Rec. 2021, 21, 1941–1956. [Google Scholar] [CrossRef]
- Purushottam, L.; Adusumalli, S.R.; Chilamari, M.; Rai, V. Chemoselective and Site-Selective Peptide and Native Protein Modification Enabled by Aldehyde Auto-Oxidation. Chem. Commun. 2017, 53, 959–962. [Google Scholar] [CrossRef] [Green Version]
- Chilamari, M.; Purushottam, L.; Rai, V. Site-Selective Labeling of Native Proteins by a Multicomponent Approach. Chem. Eur. J. 2017, 23, 3819–3823. [Google Scholar] [CrossRef] [PubMed]
- Chilamari, M.; Kalra, N.; Shukla, S.; Rai, V. Single-Site Labeling of Lysine in Proteins through a Metal-Free Multicomponent Approach. Chem. Commun. 2018, 54, 7302–7305. [Google Scholar] [CrossRef] [PubMed]
- Garreau, M.; Le Vaillant, F.; Waser, J. C-Terminal Bioconjugation of Peptides through Photoredox Catalyzed Decarboxylative Alkynylation. Angew. Chem. Int. Ed. 2019, 58, 8182–8186. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).